Abstract
Accurate and timely diagnosis of bacterial infection is crucial for effective and targeted treatment, yet routine microbiological identification is inefficient and often delayed to an extent that makes it clinically unhelpful. The immune system is capable of a rapid, sensitive and specific detection of a broad spectrum of microbes, which has been optimized over millions of years of evolution. A patient's early immune response is therefore likely to provide far better insight into the true nature and severity of microbial infections than conventional tests. To assess the diagnostic potential of pathogen-specific immune responses, we characterized the local responses of 52 adult patients during episodes of acute peritoneal dialysis (PD)–associated peritonitis by multicolor flow cytometry and multiplex ELISA, and defined the immunologic signatures in relation to standard microbiological culture results and to clinical outcomes. We provide evidence that unique local “immune fingerprints” characteristic of individual organisms are evident in PD patients on the day of presentation with acute peritonitis and discriminate between culture-negative, Gram-positive, and Gram-negative episodes of infection. Those humoral and cellular parameters with the most promise for defining disease-specific immune fingerprints include the local levels of IL-1β, IL-10, IL-22, TNF-α, and CXCL10, as well as the frequency of local γδ T cells and the relative proportion of neutrophils and monocytes/macrophages among total peritoneal cells. Our data provide proof of concept for the feasibility of using immune fingerprints to inform the design of point-of-care tests that will allow rapid and accurate infection identification and facilitate targeted antibiotic prescription and improved patient management.
Bacterial infections remain a leading cause of morbidity and mortality worldwide and pose a critical challenge for public health in the 21st century, not the least of which is due to the unprecedented spread of antibiotic resistance.1–5 Effective management of infected patients is hampered by the performance of standard diagnostics and inappropriate or delayed choice of treatments. There remains a lack of appreciation of how the body senses and fights different bacterial pathogens. Diagnosis of suspected infections depends largely on the positive identification of the likely pathogen in biologic fluids, a concept that was introduced by Robert Koch more than a century ago. However, microbiological culture methods are inherently slow and inefficient. In many cases, no organism can be identified despite clinical signs of infection.6,7 Even in the case of positive results, neither microbiological cultures nor state-of-the-art molecular methods such as PCR or mass spectrometry–based techniques discriminate between pathogens causing disease, asymptomatic carriage, and sample contaminants.
In the absence of accurate and rapid point-of-care methods that direct therapy, especially in cases in which organism virulence is a determinant of outcome, treatment of most infections remains largely empirical.5,8–12 Ineffective and delayed therapy can lead to chronic or recurrent infections that are typically more serious and more difficult to treat. As a result, there is a low threshold for prescribing broad-spectrum antibiotics, although many patients receiving such treatments actually may not have an infection, or could safely be treated with better targeted drugs.13 Of note, inappropriate therapy and overuse of antibiotics are the main causes driving multidrug resistance, which is one of the major global threats for human health as identified by the World Health Organization.
It is increasingly clear that the nature of the inflammatory response to infection is a major determinant of outcome.14 This is especially true in vulnerable individuals such as patients with acute peritoneal dialysis (PD)–associated peritonitis, in which the effect of host responses has detrimental consequences due to inflammation-related tissue damage.15,16 Key to developing better and stratified approaches to treating infections is a detailed understanding of the intricate host-pathogen relationships in disease, and the physiologic and pathophysiologic events driving early inflammatory responses and pathogen clearance. The most meticulous cross-sectional and longitudinal investigations in this respect are being made in PD patients.17–20 The peritoneal catheter in PD serves as a unique window to inflammatory scenarios that can be observed continuously in vivo. It affords noninvasive access to all relevant cellular components and humoral mediators involved in local inflammation, as well as direct insight into how treatment and type of infection modulate disease-specific pathways in a clinically relevant manner.
The immune system has evolved to survey the body constantly for potentially hazardous structures.21 Different pathogens express different molecular patterns and hence interact uniquely with different components of the immune system. The type of infection is therefore likely to evoke distinct immunologic signatures, or “immune fingerprints,” that can be assessed quantitatively and qualitatively.22,23 However, to our knowledge, no experimental attempt has been made to systematically characterize cellular and humoral responses to defined pathogens in a human infection scenario and to translate the notion of pathogen-specific immune responses into the clinic to assess its diagnostic value. In this study, we performed a detailed immunologic and microbiological analysis of samples obtained from PD patients presenting with acute peritonitis, and provide evidence that each bacterial infection leaves a characteristic local immune fingerprint. These proof-of-concept findings may have applicability for rapid, accurate, and differentiating point-of-care diagnosis of patients with suspected infections.
Results
Local Inflammatory Markers in Acute Peritonitis
Episodes of acute peritonitis in PD patients were characterized by a range of cellular and soluble biomarkers associated with inflammatory responses (Supplemental Table 1). This was true for culture-negative episodes of peritonitis as well as cases of confirmed infection by Gram-negative or Gram-positive bacteria (Figures 1 and 2). Although peritoneal leukocytes in stable patients were relatively low in numbers (typically <107 cells per bag) and were composed mainly of monocytes/macrophages and T cells, acute peritonitis was dominated by a massive recruitment of neutrophils, at times reaching >95% of all peritoneal cells and >1011 cells in total. Detailed analyses of the peritoneal leukocyte subpopulations revealed a preferential increase in Vδ2+ γδ T cells within the local T cell population in acute peritonitis, whereas the percentages of CD4+ and CD8+ T cells remained virtually unchanged (Supplemental Table 1). Soluble mediators increased in acute peritonitis included IL-1β, IL-6, soluble IL-6 receptor (sIL-6R), IL-10, IL-22, CXCL8 (IL-8), CXCL10, TNF-α, TGF-β, and matrix metalloproteinase-3 (MMP-3) (Supplemental Table 1).
Figure 1.
Cellular immune biomarkers in PD patients. (A) Total number and relative frequency (%) of peritoneal neutrophils (CD15+), monocytes/macrophages (CD14+), and T cells (CD3+) in stable PD patients and on day 1 of acute peritonitis. (B) Total number and proportion of helper T cells (CD4+), cytotoxic T cells (CD8+), and γδ T cells (Vδ2+) within the peritoneal CD3+ T cell population in stable PD patients and on day 1 of acute peritonitis. Patients presenting with a cloudy bag are grouped according to the microbiological culture results into patients with culture-negative peritonitis (n/c) or confirmed Gram-positive or Gram-negative infection. Data points represent individual patients, and horizontal lines represent mean values per group.
Figure 2.
Peritoneal levels (in nanograms per milliliter) of IL-1β, IL-2, IL-6, sIL-6R, IL-10, IL-12p70, IL-17A, IL-22, CXCL8, CXCL10, IFN-γ, TNF-α, GM-CSF, TGF-β1, and MMP-3 in stable PD patients and on day 1 of acute peritonitis. Patients presenting with a cloudy bag are grouped according to the microbiological culture results into patients with culture-negative peritonitis (n/c) or confirmed Gram-positive or Gram-negative infection. Data points represent individual patients, and horizontal lines represent mean values per group.
IL-1β and IL-10 Predict Culture-Negative Peritonitis
We next stratified patients into distinct subgroups according to the microbiological culture results provided by the routine diagnostic laboratory. These analyses revealed that the immune responses were markedly different between patients with culture-negative peritonitis and those with confirmed culture-positive infection. This latter group had significantly higher numbers of all leukocyte subpopulations analyzed. However, in culture-positive episodes of infection, a larger proportion of the infiltrating immune cells were neutrophils, whereas a smaller proportion were monocytes/macrophages, suggesting that the ratio of neutrophils to monocytes/macrophages might be of relevance for diagnostic purposes (Supplemental Table 2). Culture-positive patients also had higher peritoneal levels of IL-1β, IL-2, IL-6, IL-10, IL-22, and TNF-α (as well as sIL-6R and MMP-3 with borderline significance) (Supplemental Table 2), indicating a more severe inflammatory course than in patients during culture-negative peritonitis.
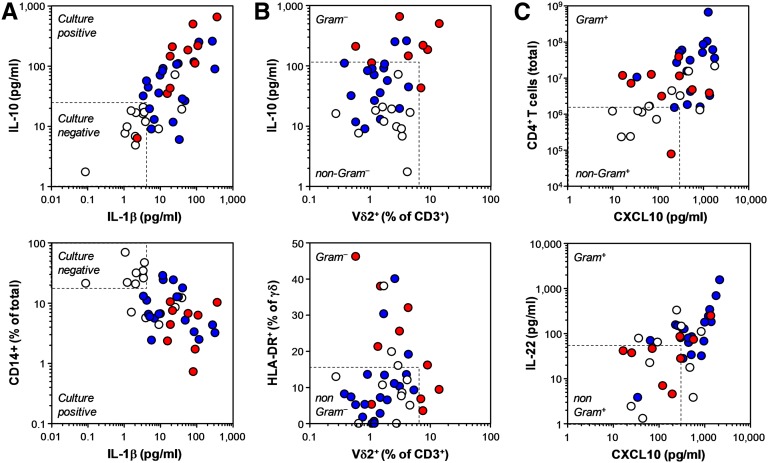
Given the distinct immune responses observed in the different patient groups, we tested whether specific immune fingerprints on day 1 could predict the microbiological culture results. Area under the receiver operating characteristic curve (AUROC) calculations identified a number of parameters with discriminatory power, such as the proportion of neutrophils among peritoneal cells as well as the levels of IL-1β, IL-10, TNF-α, and IL-6 (Supplemental Table 3). Levels of IL-1β and IL-10 gave the best Youden index (= sensitivity + specificity – 1) and highest overall correctness as individual parameters, for prediction of either culture-positive or culture-negative peritonitis (Supplemental Table 4). A simple model for the discriminatory potential between culture-negative and culture-positive peritonitis was developed by combining biomarkers according to their cut-off values (Figure 3A). When combined together, IL-1β (<4.1 pg/ml) and IL-10 (<23.3 pg/ml) resulted in 79% sensitivity and 97% specificity for the correct prediction of culture-negative peritonitis (Supplemental Table 4). Univariate analyses indicated that the proportion of neutrophils and monocytes/macrophages among total peritoneal cells as well as the levels of IL-1β and IL-10 had prognostic value. Further multivariate analyses of these four parameters alone and in combination confirmed that the combination of IL-1β and IL-10 had independent prognostic significance for the prediction of a culture-negative peritonitis episode on the day of presentation (Supplemental Table 5).
Figure 3.
Examples of immunological biomarkers in patients presenting with a cloudy bag on day 1, depending on the microbiological culture results. (A) Discrimination between culture-negative and culture-positive peritonitis. (B) Discrimination between Gram-negative and non-Gram–negative infections. (C) Discrimination between Gram-positive and non-Gram–positive infections. Data points represent individual episodes (white, culture-negative peritonitis; blue, confirmed Gram-positive infection; red, confirmed Gram-negative infection). Dashed lines indicate calculated cut-off values for positive or negative discrimination.
Vδ2+ T Cells Predict Gram-Negative Infections
We next tested whether pathogen-specific immune fingerprints existed that could predict infections by certain groups of bacteria. PD effluent from patients with microbiologically confirmed Gram-negative infections contained larger numbers of infiltrating neutrophils and consequently lower proportions of monocytes/macrophages and T cells than effluent from patients with either culture-negative or Gram-positive peritonitis (Supplemental Tables 6 and 7). Within the T cell population, Vδ2+ T cells were significantly increased and expressed higher levels of the activation marker HLA-DR in Gram-negative infections. In addition, peritoneal monocytes/macrophages expressed lower levels of CD86 in Gram-negative infections compared with the rest of the patients. Inflammatory markers that significantly increased in Gram-negative infections included IL-1β, IL-10, and TNF-α (Figure 3B and Supplemental Table 7). AUROC calculations identified the combination of Vδ2+ T cell frequencies and peritoneal levels of IL-10 as an excellent discriminator that predicted Gram-negative infections (Supplemental Table 8). This combination had the highest overall correctness, with 100% sensitivity and 93% specificity for the prediction of Gram-negative infections (Supplemental Table 9). The prognostic value of the proportion of monocytes/macrophages among all cells (<10.7%) and their expression of CD86 (<67.9%), the frequency of Vδ2+ T cells within the T cell population (≥6.3%) and their expression of HLA-DR (≥16.1%), and peritoneal IL-10 levels (≥110.1 pg/ml) were all confirmed by univariate analyses. Multivariate analyses identified the Vδ2+ T cell frequency as an independent prognostic biomarker for the prediction of Gram-negative peritonitis in all patients presenting with a cloudy bag (Supplemental Table 10).
CD4+ T Cells and CXCL10 Predict Gram-Positive Infections
We next attempted to identify predictors of Gram-positive infections in an analogous way. Among all patients presenting with acute peritonitis, patients with confirmed Gram-positive infections displayed larger numbers of infiltrating CD4+ and CD8+ T cells than the rest of the patients (i.e., individuals with culture-negative or Gram-negative peritonitis) (Supplemental Table 11). In addition, the proportion of Vδ2+ T cells within the T cell population was significantly lower in Gram-positive infections. Those inflammatory markers that significantly increased in Gram-positive infections included IL-22 and CXCL10 (Figure 3C and Supplemental Table 12). AUROC calculations identified the combination of CXCL10 (≥301.2 pg/ml) and IL-22 levels (≥54.3 pg/ml) and Vδ2+ T cell frequencies (<2.7%) as an excellent discriminator that predicted Gram-positive infections (Supplemental Table 12). This combination also had the highest overall correctness, with 89% sensitivity and 67% specificity for the prediction of Gram-positive peritonitis. In addition, CXCL10 levels (≥301.2 pg/ml) combined with total CD4+ T cell counts (≥15.2×106) had a sensitivity of only 53% yet a specificity of 95% for the prediction of Gram-positive peritonitis (Supplemental Table 13). CD4+ T cell counts as well as Vδ2+ T cell frequencies and CXCL10 levels had prognostic value as confirmed by univariate analyses. Multivariate analysis of these parameters alone and in combination identified the combination of CD4+ T cell counts and CXCL10 levels as independent predictors of Gram-positive peritonitis in patients presenting with a cloudy bag (Supplemental Table 14).
Vδ2+ T Cells Predict Clinical Outcome
Gram-negative infections are typically associated with worse outcomes, including higher rates of technique failure and mortality.24–26 Our earlier findings indicated that elevated peritoneal frequencies of γδ T cells within the total T cell population on the day of presentation were associated with subsequent technique failure.20 Here, AUROC calculations identified Vδ2+ T cell frequencies ≥4.3% as excellent discriminator that predicted short-term technique failure within the first 1–3 months (Figure 4), with an overall correctness of 94% at day 30 (4 of 48 patients with technique failure) and 77% at day 90 (9 of 48 patients) (Supplemental Tables 15–18). No other immune parameters tested, including the numbers and frequencies of neutrophils, monocytes, or CD4+ and CD8+ T cells, reached statistical significance. Taken together, these data indicate that disease-specific local immune fingerprints predict not only the type of pathogen but also the associated risk of downstream complications (Table 1).
Figure 4.
Kaplan–Meier plot showing cumulative technique survival of patients with acute peritonitis, depending on the frequency of Vδ2+ T cells among all peritoneal T cells on day 1. The cut-off value is determined by receiver operating characteristic analysis, and the statistical difference between the two curves is analyzed by the log-rank test.
Table 1.
Biomarkers of potential diagnostic and prognostic value in patients presenting with a cloudy bag on day 1, depending on the microbiological culture results and clinical outcome
| Biomarker | Culture-Negative Peritonitis | Culture-Positive Infection | Gram-Positive Infection | Gram-Negative Infection | Technique Failure |
|---|---|---|---|---|---|
| Monocytes (% of total) | ≥19.1 | <19.1 | <10.7 | ||
| CD4+ T cells (×106 cells) | ≥15.2 | ||||
| Vδ2+ (% of T cells) | <2.7 | ≥6.3 | ≥4.3 | ||
| HLA-DR+ (% of γδ T cells) | ≥16.1 | ||||
| CD86+ (% of monocytes) | <67.9 | ||||
| IL-1β (pg/ml) | <4.1 | ≥4.1 | ≥14.0 | ||
| IL-10 (pg/ml) | <23.3 | ≥23.3 | ≥110.1 | ||
| IL-22 (pg/ml) | ≥54.3 | ||||
| CXCL10 (pg/ml) | ≥301.2 | ||||
| TNF-α (pg/ml) | <19.5 | ≥19.5 | ≥32.8 |
Discussion
In a radical departure from current practice, our novel approach to diagnosing infections exploits, for the first time, the concept that microorganisms display distinct sets of pathogen-associated patterns and interact with the immune system in a unique and specific manner. Conventionally, patient management is determined on the basis of symptoms, initial clinical findings, and basic laboratory markers. Yet the two most widely advocated inflammatory biomarkers, C-reactive protein and procalcitonin, remain of limited usefulness in the clinic because they often do not reach satisfying discriminatory power to distinguish infectious from noninfectious inflammation,27,28 and have questionable relevance in patients with PD-related peritonitis.29 Our present findings demonstrate that different types of pathogens leave characteristic immune fingerprints early during infection that may be of diagnostic and prognostic relevance, with far-reaching implications for our view of acute responses in infectious scenarios in which early and accurate biomarkers are urgently needed.5,30
Culture-negative peritonitis in PD patients is associated with a fairly benign clinical course with low rates of technique failure.6,31 Prediction of culture-negative episodes would thus have important implications because it might allow the identification of low-risk patients and patients who might not require antimicrobial therapy, thereby helping to reduce unnecessary exposure to antibiotics.32,33 Our results demonstrate that designated episodes of culture-negative peritonitis can be distinguished from overt infection and have significantly different immune fingerprints that are characterized by lower numbers (and proportions) of infiltrating neutrophils and less severe inflammation, as indicated by lower peritoneal cytokine levels compared with culture-positive infections. In this respect, the cytokines IL-1β, IL-10, and TNF-α and the ratio of neutrophils to monocytes/macrophages are particularly promising candidates for a future biomarker-based diagnosis of culture-negative peritonitis.
We are aware that in order to establish robust pathogen-specific immune fingerprints, accurate identification of the causative agents is critical. Thus, for optimum stratification in future studies and to circumvent the limitations of conventional culture techniques, state-of-the-art molecular methods such as 16S and 23S rDNA amplification, mass spectrometry, and/or whole-sample sequencing and metagenomics might be utilized.7–11,34,35 Such approaches, however, add a further level of complication in the analysis of patient results when the data produced are inconsistent or conflicting. Consequently, there is currently not sufficient evidence for recommending the routine use of molecular techniques for the diagnosis of peritonitis.36 Our own findings suggest that there is a clear microbiological and immunologic difference between patients with culture-negative and culture-positive peritonitis, and argue against a serious bias due to potentially suboptimal culture techniques. It is conceivable that a proportion of the culture-negative episodes in this study might have been triggered by infectious agents (e.g., subclinical bacterial or viral infection) or conversely by chemical irritation. Because the improved outcome in that patient group does not necessarily reflect antibiotic responsiveness, it is not possible at this stage to speculate on the potential causes. This will require a considerably larger cohort to determine whether responses to treatment in the culture-negative group can be further stratified and highlights the need for a more accurate diagnostic approach to therapy including immune fingerprinting.
Although it is well established that patients with Gram-negative and Gram-positive infections present with different clinical signs and outcomes,24–26 there is no rapid diagnostic test that would guide treatment and allow predictive risk modeling at the time of presentation. Here, immune fingerprints in Gram-positive infections were markedly different from those in Gram-negative infections and were indicative of a relatively large underlying T cell component with higher numbers of CD4+ and CD8+ T cells and elevated levels of CXCL10, IFN-γ, and IL-22. The chemokine CXCL10 appeared to be a particularly good predictor of Gram-positive infections in this study, which might have stemmed from stimulation of monocytes/macrophages or mesothelial cells by certain microbial compounds such as staphylococcal α-toxin and Toll-like receptor 2 ligands.37–39
Gram-negative infections were dominated by large numbers of infiltrating neutrophils and elevated levels of cytokines such as IL-1β, IL-10, and TNF-α. A proportion of these immune fingerprints were likely due to pronounced responses to bacterial LPS, which is present in the outer cell wall of Gram-negative organisms but absent from Gram-positive organisms. LPS is a powerful inducer of proinflammatory cytokines and chemokines by monocytes/macrophages and neutrophils, including IL-1β, TNF-α, and CXCL8. At the same time, LPS stimulation also leads to secretion or large amounts of IL-6 and IL-10 by monocytes/macrophages (but not human neutrophils).40
Another powerful microbial factor present in the majority of Gram-negative bacteria (including the pathogens identified in this study) is the isoprenoid precursor (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate (HMB-PP), the natural activator of human Vγ9/Vδ2 T cells. In contrast, many Gram-positive bacteria do not produce HMB-PP and consequently do not activate Vγ9/Vδ2 T cells.20 Despite the very low proportion of T cells in acute Gram-negative infections, Vγ9/Vδ2 T cells were selectively enriched among peritoneal T cells in those patients, thereby singling out Vγ9/Vδ2 T cells as highly sensitive and selective responder cells of diagnostic and prognostic value in acute peritonitis. This observation also has direct therapeutic implications because fosmidomycin, a natural antibiotic that specifically targets the HMB-PP pathway in many pathogens including multidrug-resistant Gram-negative bacteria,41 possesses anti-inflammatory properties by inhibiting Vγ9/Vδ2 T cell–driven immune responses to HMB-PP–producing bacteria.20
Taken together, this study provides proof of concept for measuring disease-specific immune responses as discriminatory markers of early infection. These responses are likely to be triggered by unique combinations of pathogen-associated molecular patterns interacting with distinct aspects of the human immune system. At the same time, these results lay the foundation for targeted therapies that modulate disease-specific pathways to fight infection and reduce inflammation-related tissue damage on the day of presentation. Our research highlights the importance of detailed immune monitoring comprising both humoral and cellular parameters to establish accurate immune fingerprints. Although cellular analyses typically require trained laboratory staff and central equipment such as flow cytometers, recent advances in microfluidics have made the development of accurate and affordable single-use chip devices a realistic option.42–44 This promising technological progress is likely to allow routine monitoring of complex immune fingerprints in infected patients in the near future, suggesting that rapid differential biomarker-based diagnosis and guided management of noninfectious and infectious syndromes at the point of care are feasible.
Concise Methods
Patients
The study cohort comprised 52 adult PD patients admitted on day 1 of acute peritonitis between September 2008 and January 2012; 15 age- and sex-matched stable patients with no infection in the previous 3 months were included as controls. Diagnosis of acute peritonitis was based on the presence of abdominal pain and cloudy peritoneal effluent with >100 white blood cells/mm3. Infections were grouped into culture-negative, Gram-positive, and Gram-negative episodes, according to the result of the microbiological analysis of the effluent by the routine Microbiology Laboratory, Public Health Wales; cases of fungal infection (n=1) and mixed Gram-negative/Gram-positive culture results (n=1) were excluded from this analysis. End points of outcome analyses were 30th-day and 90th-day technique failure (catheter removal, transfer to hemodialysis, and/or patient death); two patients who stopped their treatment because of noninfection-related reasons were removed from the outcome analysis. This study was approved by the South East Wales Local Ethics Committee (04WSE04/27), and conducted according to the principles expressed in the Declaration of Helsinki. All patients provided written informed consent.
Flow Cytometry
Peritoneal cells were acquired on an eight-color FACSCanto II (BD Biosciences) and analyzed with FlowJo (Tree Star), using mAbs against CD3 (UCHT1), CD4 (SK3), CD8 (RPA-T8), CD15 (HI98), CD69 (FN50), CD86 (2331-FUN1), HLA-DR (L234), and TCR-Vδ2 (B6.1) from BD Biosciences; CD14 (61D3) from eBioscience; and TCR-Vγ9 (Immu360) from Beckman Coulter, together with appropriate isotype controls. Leukocyte populations were gated based on their appearance in side scatter and forward scatter area/height, and exclusion of live/dead staining (fixable Aqua; Invitrogen).
ELISA
Cell-free peritoneal effluents were analyzed for IL-1β, IL-2, IL-6, sIL-6R, IL-10, IL-12p70, CXCL8, IFN-γ, TNF-α, GM-CSF, and MMP-3 on a SECTOR Imager 6000 (Meso Scale Discovery). IL-17A, IL-22, and CXCL10 (R&D Systems) as well as TGF-β1 (eBioscience) were measured on a Dynex MRX II reader, using conventional ELISA kits.
Statistical Analyses
Statistical analyses were performed using SPSS 16.0 and GraphPad Prism 4.0 software. All variables were tested for normal distributions using the Kolmogorov–Smirnov test. Differences between patient groups were analyzed using t tests for normally distributed data or Mann–Whitney U tests for nonparametric data. Predictive biomarkers were assessed using univariate analyses; statistically significant (P<0.05) variables from the univariate analysis were included in a multivariate analysis. Multiple logistic regression analyses were conducted based on forward and/or backward elimination of data, as indicated in the tables. AUROC analyses were used to assess discrimination using nonparametric approaches, and to calculate cut-off values as well as sensitivity and specificity. Cumulative survival curves were generated using the Kaplan-Meier approach and compared using log-rank tests. Differences were considered statistically significant as indicated in the figures and tables (*P<0.05; **P<0.01; ***P<0.001).
Disclosures
None.
Acknowledgments
We are grateful to all patients for participating in this study. We also thank the staff on the B5 ward for their cooperation, and James Chess, Chantal Colmont, Eryl Cox, Donald Fraser, David Johnson, Bernhard Moser, Ian Weeks, and John Williams for their input.
This research was supported by the National Institute for Social Care and Health Research (NISCHR) Clinical Research Centre (UKCRN ID #11838, PERIT-PD), the NISCHR Academic Health Science Collaboration, Welsh Assembly Government (grants RFS/H07-03-18 and RFS/HA09-009), and Baxter Healthcare (Renal Discoveries extramural grant #08AP002).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Cloudy Peritoneal Dialysate: In Search of a Clear Cause?,” on pages 1929–1931.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013040332/-/DCSupplemental.
References
- 1.Binder S, Levitt AM, Sacks JJ, Hughes JM: Emerging infectious diseases: Public health issues for the 21st century. Science 284: 1311–1313, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Livermore DM: Has the era of untreatable infections arrived? J Antimicrob Chemother 64[Suppl 1]: i29–i36, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Arias CA, Murray BE: Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N Engl J Med 360: 439–443, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Fauci AS, Morens DM: The perpetual challenge of infectious diseases. N Engl J Med 366: 454–461, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Davies SC: 2011 Annual Report of the Chief Medical Officer: Infections and the Rise of Antimicrobial Resistance, Vol. 2, London, Department of Health, 2013. Available at: https://www.gov.uk/government/publications/chief-medical-officer-annual-report-volume-2 Accessed August 14, 2013 [DOI] [PubMed]
- 6.Fahim M, Hawley CM, McDonald SP, Brown FG, Rosman JB, Wiggins KJ, Bannister KM, Johnson DW: Culture-negative peritonitis in peritoneal dialysis patients in Australia: Predictors, treatment, and outcomes in 435 cases. Am J Kidney Dis 55: 690–697, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Shafazand S, Weinacker AB: Blood cultures in the critical care unit: Improving utilization and yield. Chest 122: 1727–1736, 2002 [DOI] [PubMed] [Google Scholar]
- 8.Ince J, McNally A: Development of rapid, automated diagnostics for infectious disease: Advances and challenges. Expert Rev Med Devices 6: 641–651, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Claus RA, Otto GP, Deigner HP, Bauer M: Approaching clinical reality: markers for monitoring systemic inflammation and sepsis. Curr Mol Med 10: 227–235, 2010 [DOI] [PubMed] [Google Scholar]
- 10.Clerc O, Greub G: Routine use of point-of-care tests: Usefulness and application in clinical microbiology. Clin Microbiol Infect 16: 1054–1061, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Leggieri N, Rida A, François P, Schrenzel J: Molecular diagnosis of bloodstream infections: Planning to (physically) reach the bedside. Curr Opin Infect Dis 23: 311–319, 2010 [DOI] [PubMed] [Google Scholar]
- 12.Chan T, Gu F: Early diagnosis of sepsis using serum biomarkers. Expert Rev Mol Diagn 11: 487–496, 2011 [DOI] [PubMed] [Google Scholar]
- 13.Yu VL: Guidelines for hospital-acquired pneumonia and health-care-associated pneumonia: A vulnerability, a pitfall, and a fatal flaw. Lancet Infect Dis 11: 248–252, 2011 [DOI] [PubMed] [Google Scholar]
- 14.Llewelyn MJ, Cohen J: Tracking the microbes in sepsis: Advancements in treatment bring challenges for microbial epidemiology. Clin Infect Dis 44: 1343–1348, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Brown MC, Simpson K, Kerssens JJ, Mactier RA, Scottish Renal Registry : Peritoneal dialysis-associated peritonitis rates and outcomes in a national cohort are not improving in the post-millennium (2000-2007). Perit Dial Int 31: 639–650, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Li PK, Chow KM: Infectious complications in dialysis—epidemiology and outcomes. Nat Rev Nephrol 8: 77–88, 2012 [DOI] [PubMed] [Google Scholar]
- 17.Hurst SM, Wilkinson TS, McLoughlin RM, Jones S, Horiuchi S, Yamamoto N, Rose-John S, Fuller GM, Topley N, Jones SA: Il-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity 14: 705–714, 2001 [DOI] [PubMed] [Google Scholar]
- 18.Roberts GW, Baird D, Gallagher K, Jones RE, Pepper CJ, Williams JD, Topley N: Functional effector memory T cells enrich the peritoneal cavity of patients treated with peritoneal dialysis. J Am Soc Nephrol 20: 1895–1900, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberl M, Roberts GW, Meuter S, Williams JD, Topley N, Moser B: A rapid crosstalk of human gammadelta T cells and monocytes drives the acute inflammation in bacterial infections. PLoS Pathog 5: e1000308, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey MS, Lin CY, Roberts GW, Heuston S, Brown AC, Chess JA, Toleman MA, Gahan CG, Hill C, Parish T, Williams JD, Davies SJ, Johnson DW, Topley N, Moser B, Eberl M: Human neutrophil clearance of bacterial pathogens triggers anti-microbial γδ T cell responses in early infection. PLoS Pathog 7: e1002040, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blander JM, Sander LE: Beyond pattern recognition: Five immune checkpoints for scaling the microbial threat. Nat Rev Immunol 12: 215–225, 2012 [DOI] [PubMed] [Google Scholar]
- 22.Feezor RJ, Oberholzer C, Baker HV, Novick D, Rubinstein M, Moldawer LL, Pribble J, Souza S, Dinarello CA, Ertel W, Oberholzer A: Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun 71: 5803–5813, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D: Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 109: 2066–2077, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe R, Oda S, Sadahiro T, Nakamura M, Hirayama Y, Tateishi Y, Shinozaki K, Hirasawa H: Gram-negative bacteremia induces greater magnitude of inflammatory response than Gram-positive bacteremia. Crit Care 14: R27, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan M, Thodis E, Ikonomopoulos D, Vidgen E, Chu M, Bargman JM, Vas SI, Oreopoulos DG: Predictors of outcome following bacterial peritonitis in peritoneal dialysis. Perit Dial Int 22: 573–581, 2002 [PubMed] [Google Scholar]
- 26.Troidle L, Gorban-Brennan N, Kliger A, Finkelstein F: Differing outcomes of gram-positive and gram-negative peritonitis. Am J Kidney Dis 32: 623–628, 1998 [DOI] [PubMed] [Google Scholar]
- 27.Simon L, Saint-Louis P, Amre DK, Lacroix J, Gauvin F: Procalcitonin and C-reactive protein as markers of bacterial infection in critically ill children at onset of systemic inflammatory response syndrome. Pediatr Crit Care Med 9: 407–413, 2008 [DOI] [PubMed] [Google Scholar]
- 28.Tang BM, Eslick GD, Craig JC, McLean AS: Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: Systematic review and meta-analysis. Lancet Infect Dis 7: 210–217, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Lam MF, Leung JC, Lam CW, Tse KC, Lo WK, Lui SL, Chan TM, Tam S, Lai KN: Procalcitonin fails to differentiate inflammatory status or predict long-term outcomes in peritoneal dialysis-associated peritonitis. Perit Dial Int 28: 377–384, 2008 [PubMed] [Google Scholar]
- 30.Kessel AS, Sharland M: The new UK antimicrobial resistance strategy and action plan. BMJ 346: f1601, 2013 [DOI] [PubMed]
- 31.Chen KH, Chang CT, Weng SM, Yu CC, Fang JT, Huang JY, Yang CW, Hung CC: Culture-negative peritonitis: A fifteen-year review. Ren Fail 29: 177–181, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Choi P, Nemati E, Banerjee A, Preston E, Levy J, Brown E: Peritoneal dialysis catheter removal for acute peritonitis: A retrospective analysis of factors associated with catheter removal and prolonged postoperative hospitalization. Am J Kidney Dis 43: 103–111, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Rocklin MA, Teitelbaum I: Noninfectious causes of cloudy peritoneal dialysate. Semin Dial 14: 37–40, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Woo PC, Lau SK, Teng JL, Tse H, Yuen KY: Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 14: 908–934, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Nakamura S, Nakaya T, Iida T: Metagenomic analysis of bacterial infections by means of high-throughput DNA sequencing. Exp Biol Med (Maywood) 236: 968–971, 2011 [DOI] [PubMed] [Google Scholar]
- 36.Li PK, Szeto CC, Piraino B, Bernardini J, Figueiredo AE, Gupta A, Johnson DW, Kuijper EJ, Lye WC, Salzer W, Schaefer F, Struijk DG, International Society for Peritoneal Dialysis : Peritoneal dialysis-related infections recommendations: 2010 update. Perit Dial Int 30: 393–423, 2010 [DOI] [PubMed] [Google Scholar]
- 37.Visser CE, Tekstra J, Brouwer-Steenbergen JJ, Tuk CW, Boorsma DM, Sampat-Sardjoepersad SC, Meijer S, Krediet RT, Beelen RH: Chemokines produced by mesothelial cells: huGRO-alpha, IP-10, MCP-1 and RANTES. Clin Exp Immunol 112: 270–275, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colmont CS, Raby AC, Dioszeghy V, Lebouder E, Foster TL, Jones SA, Labéta MO, Fielding CA, Topley N: Human peritoneal mesothelial cells respond to bacterial ligands through a specific subset of Toll-like receptors. Nephrol Dial Transplant 26: 4079–4090, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasraie S, Niebuhr M, Kopfnagel V, Dittrich-Breiholz O, Kracht M, Werfel T: Macrophages from patients with atopic dermatitis show a reduced CXCL10 expression in response to staphylococcal α-toxin. Allergy 67: 41–49, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Davey MS, Tamassia N, Rossato M, Bazzoni F, Calzetti F, Bruderek K, Sironi M, Zimmer L, Bottazzi B, Mantovani A, Brandau S, Moser B, Eberl M, Cassatella MA: Failure to detect production of IL-10 by activated human neutrophils. Nat Immunol 12: 1017–1018, author reply 1018–1020, 2011 [DOI] [PubMed] [Google Scholar]
- 41.Davey MS, Tyrrell JM, Howe RA, Walsh TR, Moser B, Toleman MA, Eberl M: A promising target for treatment of multidrug-resistant bacterial infections. Antimicrob Agents Chemother 55: 3635–3636, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frankowski M, Bock N, Kummrow A, Schädel-Ebner S, Schmidt M, Tuchscheerer A, Neukammer J: A microflow cytometer exploited for the immunological differentiation of leukocytes. Cytometry A 79: 613–624, 2011 [DOI] [PubMed] [Google Scholar]
- 43.Diaw PA, Daneau G, Coly AA, Ndiaye BP, Wade D, Camara M, Mboup S, Kestens L, Dieye TN: Multisite evaluation of a point-of-care instrument for CD4(+) T-cell enumeration using venous and finger-prick blood: the PIMA CD4. J Acquir Immune Defic Syndr 58: e103–e111, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A, Heath JR: A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med 17: 738–743, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]