Abstract
Rev-Erbα and Rev-Erbβ are nuclear receptors that regulate the expression of genes involved in the control of circadian rhythm1,2, metabolism3,4, and inflammatory responses5. Rev-Erbs function as transcriptional repressors by recruiting NCoR/HDAC3 co-repressor complexes to Rev-Erb response elements in enhancers and promoters of target genes6-8, but the molecular basis for cell-specific programs of repression is not known. Here, we present evidence that in macrophages, Rev-Erbs regulate target gene expression by inhibiting the functions of distal enhancers that are selected by macrophage lineage-determining factors, thereby establishing a macrophage-specific program of repression. Remarkably, the repressive functions of Rev-Erbs are associated with their ability to inhibit the transcription of enhancer-derived RNAs (eRNAs). Furthermore, targeted degradation of eRNAs at two enhancers subject to negative regulation by Rev-Erbs resulted in reduced expression of nearby mRNAs, implying a direct role of these eRNAs in enhancer function. By precisely defining eRNA start sites using a method that quantifies nascent 5′ ends (5′-GRO-Seq), we show that transfer of full enhancer activity to a target promoter requires both the sequences mediating transcription factor binding and the specific sequences encoding the eRNA transcript. These studies provide evidence for direct roles of eRNAs in contributing to enhancer functions and suggest that Rev-Erbs act to suppress gene expression at a distance by repressing eRNA transcription.
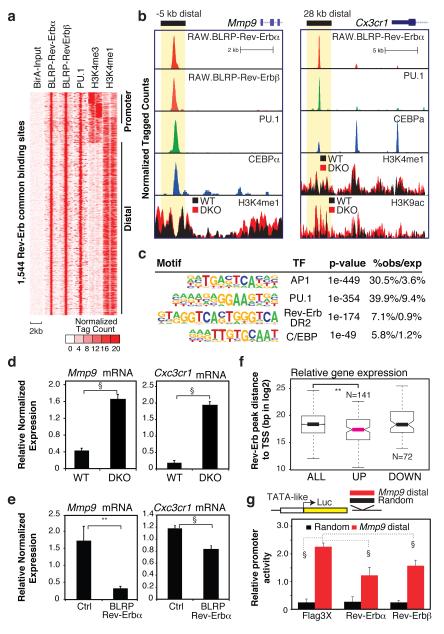
To study mechanisms underlying Rev-Erb regulation of macrophage gene expression, we first determined genome-wide binding profiles in RAW264.7 macrophages engineered to contain biotin-tagged Rev-Erbα and Rev-Erbβ. Chromatin immunoprecipitation linked to deep sequencing (ChIP-Seq) indicated enrichment for both Rev-Erbα and Rev-Erbβ at the promoter of the circadian target gene BmalI (Supplementary Fig. 1a), in accordance with previous studies1,7. We focused on a core set of highest confidence peaks (n = 1,544) occupied by both proteins for detailed analysis (Fig. 1a, Supplementary Fig. 1b). The majority (~90%) of Rev-Erb peaks were in intra- and intergenic regions at least 1 kilobase (kb) away from annotated transcription start sites (Supplementary Fig. 1c), exemplified by binding sites vicinal to the Mmp9 and Cx3cr1 genes (Fig. 1b). In addition, ~70% of Rev-Erb bound sites were in regions demarcated by high H3K4me1 and low H3K4me3, a combination characteristic of enhancer elements9 (Fig. 1a). De novo motif discovery of Rev-Erb-bound loci returned significant enrichment for binding sites for Rev-Erb, PU.1, AP1 and C/EBP (Fig. 1c). PU.1, AP-1 and C/EBP transcription factors are required for macrophage differentiation10 and have recently been shown to select the majority of the enhancer-like elements in macrophages11. Co-localization of Rev-Erbs with PU.1 and C/EBP in macrophages was confirmed by comparison with direct binding data for these factors11 (Fig. 1a, b). Consistent with these findings, a majority of the Rev-Erb bound sites defined here localize to enhancer-like elements specific for macrophages (Supplementary Fig. 2a,b).
Figure 1. Rev-Erb binding and function at macrophage-specific enhancers.
a, Cluster plot of ChIP-Seq signals for the indicated transcription factors and histone marks flanking 2 kb from the center of 1,544 Rev-Erb binding sites. b, Genomic loci of Mmp9 and Cx3cr1 with ChIP-Seq signals for the indicated transcription factors and histone marks. The locations of the −5 kb Mmp9 and 28 kb Cx3cr1 enhancers are indicated at top. c, Top-enriched transcription factor motifs identified by de novo motif discovery at Rev-Erb bound loci. d, Q-PCR analysis of Mmp9 and Cx3cr1 mRNA in Rev-Erb DKO macrophages (N = 8, and N WT DKO = 7) and e, in RAW264.7 macrophages engineered to stably express BLRP-Rev-Erbα (Nctrl = 17, Nalpha = 18 independent lines). Data represent mean + s.e.m. P-value **, P < 0.01, §, P < 0.005 versus control by two tail Student’s t-test.. f, Box and whisker plot of distances of nearest Rev-Erb binding sites to genes exhibiting significant up or down regulation in Rev-Erb DKO macrophages in comparison to all genes. The edges of the box represent the first and third quartile, and the whiskers indicated 1.5X of the interquartile range. ** p<0.005. g, Assessment of enhancer activity of the −5 kb Mmp9 Rev-Erb binding region. Luciferase reporter was cotransfected with an empty (Flag3X), Rev-Erbα or Rev-Erbβ expression construct. §, P < 0.005, ANOVA by Tukey HSD test.
We next performed Global Run-On sequencing (GRO-Seq)12 in Rev-Erbα/Rev-Erbβ-deficient and wild type bone marrow derived macrophages (BMDMs) from Tie2-Cre; Rev-Erbαflox/flox; Rev-Erbβflox/flox (DKO) animals and Cre-negative littermates (WT). Tie2-Cre expression in hematopoietic stem cells13 resulted in excision efficiencies in DKO macrophages of 85% for Rev-Erbα and 92% for Rev-Erbβ (Supplementary Fig. 3). GRO-Seq analysis indicated that 142 mRNAs were significantly up-regulated in DKO macrophages (p-value < 0.005), while 71 genes were down-regulated (p-value < 0.005) (Supplemental Table 1). Quantitative reverse transcriptase-dependent PCR confirmed up-regulation of Mmp9 and Cx3cr1 mRNAs in Rev-Erb DKO macrophages (Fig. 1d). Conversely, constitutive expression of either Rev-Erbα or Rev-Erbβ in RAW264.7 macrophages resulted in repression of Mmp9 and Cx3cr1 expression (Fig 1e, Supplementary Fig. 4a,b). Analysis of multiple independent clones indicated that the extent of Mmp9 and Cx3cr1 repression correlated with Rev-Erb expression levels (Supplementary Fig 4c-f). Genes that were up-regulated in DKO macrophages were significantly closer to Rev-Erb binding sites than down-regulated genes (Fig. 1f), consistent with primary roles of Rev-Erbs as transcriptional repressors. However, only 3 of the 142 up-regulated genes had Rev-Erb peaks within 2kb of annotated transcription start sites, suggesting that Rev-Erbs primarily act to repress gene expression at distant enhancer-like elements.
We next tested genomic regions containing Rev-Erb binding sites for enhancer activity. A 983bp region surrounding the Rev-Erb-bound site at −5kb from the Mmp9 transcription start site (TSS) was cloned downstream of a luciferase reporter driven by a TATA-like promoter (Fig 1g). This region increased reporter gene activity in RAW264.7 macrophages and was sensitive to Rev-Erb repression (Fig 1g). In contrast, this element is inactive in a hepatoma cell line that lacks expression of PU.1 (Supplementary Fig. 5). RAR-related orphan nuclear receptors (RORs) also bind to Rev-Erb response elements and constitutively activate gene expression14. Consistent with this, we found that constitutive expression of RORα increased activity of the Mmp9 enhancer element (Supplementary Fig. 6). Co-expression of wild type Rev-Erbβ, but not Rev-Erbβ with a mutation disrupting sequence-specific DNA binding, antagonized RORα activation (Supplementary Fig. 6). Six of six other Rev-Erb-bound distal regions chosen for analysis were activated by RORα, four of which were antagonized by Rev-Erb co-transfection.
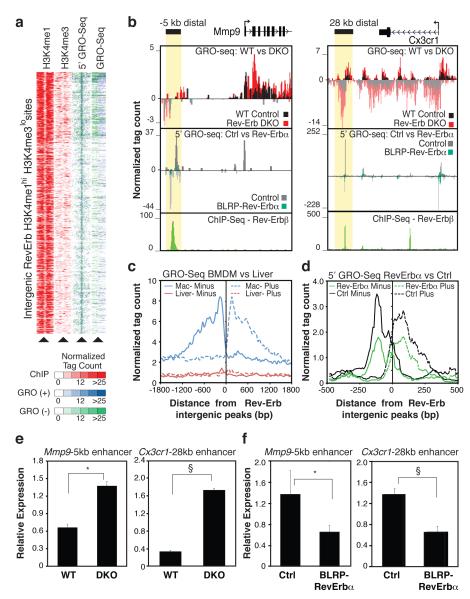
Examination of GRO-Seq data at intergenic Rev-Erb binding sites exhibiting the enhancer histone signature H3K4me1hi/H3K4me3lo, indicated the presence of bi-directional transcripts (Fig. 2a-c), consistent with recent studies indicating that RNAs are transcribed from distal enhancer elements on a genome-wide scale15-17. To determine whether transcripts were being initiated at enhancers, we modified the GRO-Seq protocol to detect nascent RNA with a 5′ 7-methylguanylated cap (5′-GRO-Seq). This methodology precisely localized start sites of well-characterized mRNAs (Supplementary Fig. 7) and identified eRNA initiation at 76% of the RevErb binding sites at enhancer-like regions of the genome (Fig. 2a). The majority (56%) of these sites direct bi-directional transcription, exemplified by the Mmp9 −5kb and Cx3cr1 28kb enhancers (Fig. 2b). No significant GRO-Seq signal was detected in macrophages at locations of intergenic Rev-Erbα peaks in liver8 (Fig. 2c), consistent with cell type specific eRNA expression.
Figure 2. Rev-Erb negatively regulates enhancer transcription.
a, Cluster plot of tag counts for H3K4me1 and H3K4me3 ChIP-Seq, 5’GRO-Seq and GRO-Seq at 544 intergenic Rev-Erb bound, H3K4me1hi H3K4me3lo regions. b. Genomic loci for Mmp9 and Cx3cr1 showing indicated tag counts for GRO-Seq, 5’GRO-Seq and Rev-Erbβ. c, Distribution of averaged macrophage GRO-Seq eRNA signal in macrophages flanking Rev-Erb intergenic sites defined in macrophages (n = 544, blue) and Rev-Erb intergenic sites defined in liver (n = 521, red8). d, Distribution of average 5’GRO-Seq signal from Rev-Erbα overexpressing (green) and control RAW264.7 macrophages (black) flanking the top 100 Rev-Erb-sites. e, Q-PCR analysis of the −5kb Mmp9 and 28kb Cx3cr1 eRNAs in Rev-Erb DKO (top, N WT = 6, and N DKO = 5) and f, Rev-Erbα overexpressing RAW264.7 macrophages (bottom, Nctrl = 13, Nalpha = 14 independent lines). Data represent mean + s.e.m. *, P < 0.01, § P < 0.005, versus control by two tail Student’s t-test.
To determine whether Rev-Erbs regulate eRNA expression, transcription of nascent RNA at Rev-Erb bound enhancers was examined in both loss and gain of function models. Rev-Erbβ binding was strongly associated with reduced 5’GRO-Seq signal at the most confident Rev-Erbβ binding sites (Supplementary Fig. 8a). Analysis of averaged 5’GRO-Seq signal at the top 100 Rev-Erb intergenic enhancers showed a marked decrease of eRNA initiation in macrophages overexpressing Rev-Erbα compared to control macrophages (Fig 2d). Conversely, these same intergenic enhancers exhibited an overall increase of GRO-Seq RNA signal in Rev-Erb DKO macrophages (Supplementary Fig. 9a). In either loss or gain of function experiment, the eRNA signal at the top 100 PU.1-bound enhancers showed no significant changes (e.g., Supplementary Fig. 9b), indicating that changes in eRNA are specific to Rev-Erb-bound elements. Effects of gain or loss of Rev-Erb function on eRNA expression at the Mmp9 −5kb and Cx3cr1 28kb enhancers were confirmed by RT-PCR (Fig 2e,f). Overall, levels of de-repressed eRNAs in Rev-Erb DKO were inversely correlated to levels of eRNA repression upon constitutive expression of Rev-Erbα (Supplementary Fig 8b).
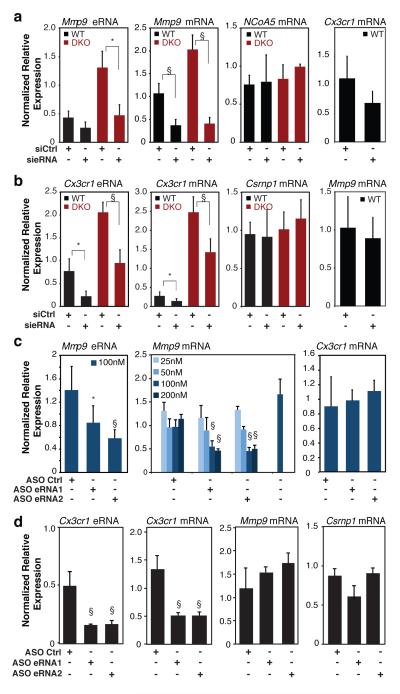
ChIP-Seq experiments demonstrated that gain or loss of Rev-Erb function also resulted in reciprocal loss or gain of H3K9 acetylation (H3K9ac) at Rev-Erb-occupied enhancers, respectively, (Fig 1b, Supplementary Fig. 10a,c, e-g), consistent with Rev-Erb-mediated recruitment of NCoR/HDAC3 complexes7. In contrast, H3K9ac was not changed at the global set of PU.1-enhancers (Fig. S10b, d). Notably, constitutive expression of Rev-Erbα had no significant effect on H3K4me1 or PU.1 binding at Rev-Erb bound enhancer elements (Supplementary Fig. 11a, b), despite the profound changes in eRNA initiation (Fig. 2b,d). Collectively, these results raised the possibility that Rev-Erbs repressed gene expression at a distance by regulating enhancer-directed transcription. Consistent with this possibility, changes in eRNA expression at Rev-Erb-bound sites due to gain or loss of Rev-Erb function were associated with changes in expression of the nearest mRNA (Supplementary Fig. 12a, b) and were better predictors than Rev-Erb binding itself (Supplementary Fig. 12c, d). In addition, although levels of eRNAs are low at steady state15, 5’GRO-Seq data suggest that the extent of engaged RNA Pol II at enhancers is often comparable to that at promoters, as exemplified by Cx3cr1 and Mmp9 (Fig. 2b). Three experimental approaches were used to investigate whether the synthesis of enhancer-directed RNA transcripts contributed to enhancer activity. First, we designed siRNAs that specifically reduced expression of eRNAs associated with the Mmp9 or Cx3cr1 enhancers in primary WT macrophages. Reduced eRNA expression was associated with a corresponding reduction of Mmp9 and Cx3cr1 mRNAs, but not mRNAs from nearest expressing genes such as NCoA5 and Csrnp1, respectively (Fig. 3a-b). Furthermore, these siRNAs reversed the de-repression phenotype associated with increased eRNA expression in Rev-Erb DKO macrophages. Importantly, the siRNA directed against the plus strand Mmp9 eRNA had no effect on expression of the minus strand eRNA or binding of PU.1 to the Mmp9 −5kb enhancer (Supplementary Fig. 13a, b), thereby excluding potential silencing effects of the siRNA on the transcriptional activity of the −5kb enhancer itself.
Figure 3. Reduction of eRNA expression results in reduced expression of nearby mRNAs.
a, Q-PCR analysis of Mmp9 eRNA, and Mmp9, NCoA5 and Cx3cr1 mRNAs for wildtype and Rev-Erb DKO thioglycollate-elicited macrophages transfected with Ctrl or Mmp9 eRNA siRNA (N WT = 4, and N DKO = 4). b, Q-PCR analysis of Cx3cr1 eRNA, and Cx3cr1, Csrnp1 and Mmp9 mRNAs for wildtype and Rev-Erb DKO bone marrow-derived macrophages transfected with siRNA targeting Cx3cr1 eRNA (N WT = 6, and N DKO = 5). c, Q-PCR analysis of Mmp9 eRNA and Mmp9 and Cx3cr1 mRNAs in thioglycollate-elicited macrophages transfected with the indicated antisense oligonucleotides (ASO, n = 3-7 per condition). d, Q-PCR analysis of Cx3cr1 eRNA and Cx3cr1, Mmp9 and Csrnpl mRNAs in BMDMs transfected with the indicated antisense oligonucleotides (ASO, n = 3-7 per condition). Data in a-d represent mean + s.d., with expression normalized to 36B4 in all cases. For a-b, statistical significance was determined by two tails Student’s t-test; for c-d, one-way ANOVA with Tukey HSD test. P value, * P < 0.05, § P < 0.005 versus control.
As a second approach, we used antisense oligonucleotides (ASO) to knock down Mmp9 −5kb and Cx3cr1 28 kb enhancer eRNAs. ASOs mediate nuclear RNA degradation via an RNase H pathway18. This provides an independent method for eRNA targeting, as siRNA-directed silencing may alter enhancer function through ways other than RNA degradation19,20. We systematically screened ASOs targeting the Mmp9 −5kb and Cx3cr1 28kb eRNAs (Supplementary Fig. 14) and selected subsets of the most effective ASOs for detailed analysis. ASOs exhibiting the ability to reduce Mmp9 −5kb plus strand eRNA expression resulted in dose-dependent reduction of the corresponding Mmp9 mRNA, but did not affect the Cx3cr1 mRNA (Fig. 3c). ASOs exhibiting the ability to knock down the minus strand Cx3cr1 28kb eRNA reduced Cx3cr1, but not Mmp9 or Csrnp1 expression (Fig. 3d).
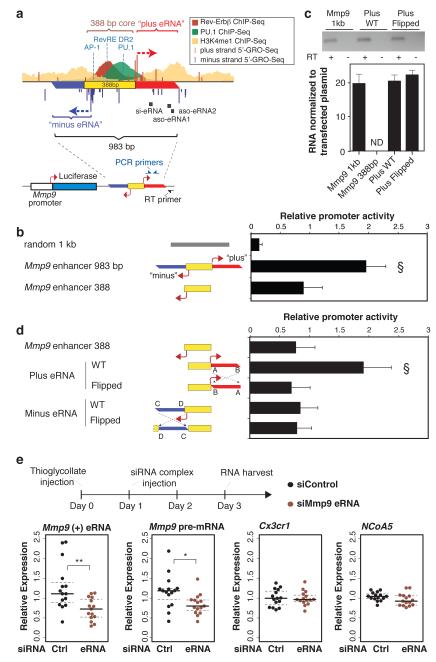
As a third approach, we examined the functional significance of the Mmp9 and Cx3cr1 eRNAs using an enhancer assay guided by 5’GRO-Seq definition of eRNA start sites. The 983 bp sequence upstream of Mmp9 that confers Rev-Erb-regulated enhancer activity in RAW264.7 cells encompasses a 388 bp central region containing the binding sites for PU.1, C/EBPs, AP-1 and Rev-Erbs, as well as start sites of plus and minus-strand eRNAs (Fig. 4a). Notably, the 388 bp core was significantly less active than the 983 bp sequence, which encodes the eRNAs (Fig. 4b). Expression of the plus-strand eRNA from the 983 bp enhancer was confirmed by RT-PCR using a reporter-specific primer for first strand cDNA synthesis (Fig. 4c). Addition of DNA encoding the plus-strand eRNA, but not the minus-strand eRNA, restored transcriptional activity to the 388 bp core (Fig 4d), consistent with the finding that siRNAs and ASOs directed against the plus-strand eRNA resulted in reduction of Mmp9 mRNA expression. A similar activity of the plus strand eRNA was observed when the 983 bp or core enhancer elements were inserted in the reverse orientation (Supplementary Fig. 15). Similarly, the central 210 bp of the Cx3cr1 28 kb enhancer containing PU.1 and Rev-Erb binding sites was less active than a 967 bp fragment encoding plus and minus strand eRNAs. Adding back the minus strand eRNA, but not the plus strand, restored the activity of the enhancer core (Supplementary Fig. 16), consistent with the results of the siRNA and ASO experiments.
Figure 4. eRNA contribution to enhancer activity and consequences of knockdown in vivo.
a, Experimental design for testing eRNA coding sequences. 983 bp of the Mmp9 enhancer was cloned downstream of the luciferase reporter gene driven by the Mmp9 promoter. The yellow box represents the 388 bp core mediating transcription factor binding as indicated by the ChIP-Seq tracks. Positions of transcription factor motifs are indicated in blue. Directional arrows represent eRNA transcription start sites defined by 5’GRO-Seq that give rise to ‘Plus eRNA’ and ‘minus eRNA’ as indicated. The locations of RT and PCR primers for detecting plasmid-directed eRNAs are indicated. b, Luciferase activity of the indicated reporter constructs in RAW264.7 macrophages. Bars represent mean normalized values from 8 independent experiments + s.d, (§ P < 0.005 versus all other indicated conditions). c, RT-PCR of Mmp9 plus eRNA normalized to the copy number of the indicated transfected plasmid DNA. Gel electrophoresis of PCR with or without RT is indicated on top. ND = not detected. d, Enhancer reporter assays performed as in b with the indicated luciferase reporters. Dashed lines represent inversions of the plus or minus DNA sequence relative to the core enhancer element (yellow) (bars represent mean normalized values from 5 independent experiments + s.d., § P < 0.005 versus all other indicated conditions). Statistical significance was determined by one-way ANOVA followed by Tukey HSD test. e, Sterile peritonitis was initiated by intraperitoneal injection of thioglycollate on day 0. Mice were injected with lipofectamine-siRNA complexes on day 1, and peritoneal exudate cells were recovered for analysis of Mmp9 eRNA, Mmp9 primary RNA, NCoA5, and Cx3cr1 mRNA on day 3 (n = 15 per condition). Values were normalized to the average of 36B4 and cyclopilin A mRNA. *, P < 0.05 versus siControl as determined by two tailed Student’s t-test.
We next reversed the orientation of the eRNA-coding sequences relative to the enhancer cores, thereby retaining any putative transcription factor binding sites but completely changing the sequence of any potential eRNA product. In the “flipped” Mmp9 plus eRNA construct, Mmp9 enhancer activity was reduced to a level comparable to the 388 bp-core despite production of an ‘antisense’ eRNA (Fig 4c and S15). Corresponding results were obtained for the Cx3cr1 enhancer (Supplementary Fig. 16). As even broadly expressed genes are often under the control of cell-specific enhancers, these findings raised the question of whether enhancers might be considered as targets for cell-specific manipulation of gene expression in vivo. To explore this possibility, we induced sterile peritonitis in mice and investigated the ability of siRNAs directed against the Mmp9 −5kb plus strand eRNA to alter Mmp9 mRNA expression. Using lipofectamine-siRNA delivery21, the eRNA-specific siRNA, but not a control siRNA, reduced expression of the −5kb plus strand eRNA and the Mmp9 primary transcript, as was observed in in vitro (Fig. 4e).
In concert, we provide evidence that Rev-Erbs function to repress macrophage gene expression by repressing transcription from enhancers that are selected by macrophage lineage-determining factors. The recent finding of widespread enhancer transcription raises the question of whether eRNAs represent ‘noise’ due to spurious transcription from regions of open chromatin, reflect an important role of enhancer transcription itself, or directly contribute to enhancer function. Our findings suggest that at least some eRNAs make a quantitative contribution to enhancer function, in agreement with findings for noncoding RNAs expressed in the vicinity of the p53 and SNAI1 genes22,23. These results do not exclude transcription-independent functions of the enhancer core or roles of enhancer transcription unrelated to the eRNA product. A major goal for the future will be to establish functional relevance of eRNAs in vivo. As the expression of many widely expressed genes appears to be controlled by cell-restricted enhancers, the expression of such genes might be altered in a cell-restricted manner by targeting corresponding functional eRNAs. Recent advances in development of chemically modified antisense oligonucleotides that can effectively reduce RNA expression in vivo24 could potentially enable this effort, suggesting the possibility of an ‘enhancer therapy’ approach to the treatment of disease.
Full Methods
Reagents and expression plasmids
Rabbit anti-PU.1 (SC-352) was from Santa Cruz Biotechnology. Rabbit anti-H3K4me1 (8895) was purchased from abcam. Rabbit anti-H3K9ac (07-352) was purchased from Millipore. Expression constructs for Rev-Erbα (amino acid 1-614), Rev-Erbβ (amino acid 1-576), and RORα (amino acid 1-460) were cloned into p3XFlag-CMV7.1 (Sigma) at NotI and BamHI sites. The following primers were used. Rev-Erbα: 5′-AGCTTGCGGCCGCTATGACGACCCTGGACTCC-3′, 5′-ATTACGGATCCTCACTGGGCGTCCACCCG-3′; Rev-Erbβ: 5′-AGCTTGCGGCCGCTATGGAGCTGAACGCAGGA-3′, 5′-ATTACGGATCCTTAAGGATGAACTTTAA-3′; RORα: 5′-AGCTTGCGGCCGCTATGAAAGCTCAAATTGAA-3′, 5′ ACCCGGGATCCTTACCCATCGATTTGCATG-3′. Mutation of the DNA binding domain of Rev-Erbβ was generated using QuickChange II site directed mutagenesis (Stratagene). The cysteines in the zinc finger domain (Rev-Erbβ, amino acid 133 and 136) were mutated to alanines using the following oligos, with mutated sequence underlined: Rev-Erbβ sense 5′-AGTGGCATGGTTCTACTGGCTAAAGTCGCTGGGGATGTGGCATCAGG-3′ antisense, 5′-CCTGATGCCACATCCCCAGCGACTTTAGCCAGTAGAACCATGCCACT-3′.
Rev-Erb DKO mice and genotyping
Mice with floxed alleles for Rev-Erbα and Rev-Erbβ were prepared as described 25. Rev-Erbαflox/flox; Rev-Erbβflox/flox mice were crossed with Tie2-Cre; Rev-Erbαflox/flox; Rev-Erbβflox/flox to obtain hematopoietic specific knockout of Rev-Erbs deletion. Littermates without Tie2-Cre transgene were used as control groups. DNA were harvested from tail and macrophages, and standard PCR protocol was used for genotyping. The Tie2-Cre transgene was detected using primers 5’GCATTACCGGTCGATGCAACGAGTGATGAG-3′ and 5′-GAGTGAACGAACCTGGTCGAAATCAGTGCG-3′, yielding a 408 bp PCR product. Genotyping PCR primers 5′-TCTCCGTTGGCATGTCTAGAGATGG-3′ and 5′-GAAGAGTGTGTGTTTGCCCAAGAGG-3′ distinguish wild type (191 bp) and floxed (332 bp) alleles in Rev-Erbα locus. Genotyping PCR primers 5′-GGTTAGGTTTGTGAGTGTCCACAGC-3′ and 5′-AAGTGCTCCAACAAGGTAGTGCA-3′ distinguish wild type (236 bp) and floxed (377 bp) alleles in Rev-Erbβ locus.
Generation of Biotin-Tagged Rev-Erbα and Rev-Erbβ
Details on generating biotin-tagged protein stably expressed in RAW264.7 macrophages was previously published 11. Briefly, Rev-Erbs were fused in frame at the N terminus with amino acid peptide MAGGLNDIFEAQKIEWHEDTGGGGSGGGGSGENLYFQS containing a biotin ligase recognition peptide (BLRP) and a TEV protease specific cleavage sequence (underlined). BLRP-empty, -Rev-Erbα, or -Rev-Erbβ expression plasmid were transfected into RAW264.7 macrophages engineered to stably express BirA. G418 (275 ng/mL) and puromycin (2.5 μg/mL) were used for stable selection. Multiple stable cell lines were isolated and screened for BLRP Rev-Erb expression and biotinylation by western blot using anti Avi-tag antibody specifically recognizing the BLRP tag (Genscript) or HRP-streptavidin (Jackson Immunoresearch), respectively.
The following primers were used for cloning full length Rev-Erbα and Rev-Erbβ at the NotI/PmelI sites in BLRP-expression construct. BLRP-Rev-Erbα, AGCTTGCGGCCGCTATGACGACCCTGGACTCC, AGCTTTGTTTAAACTCACTGGGCGTCCACCCG; BLRP-Rev-Erbβ, AGCTTGCGGCCGCTATGGAGCTGAACGCAGGA, AGCTTTGTTTAAACTTAAGGATGAACTTTAA.
ChIP-Seq
Detailed protocols for antibody based-ChIP experiments were published 11. ChIP for biotinylated Rev-Erbs was performed as described 11 with the following modifications. Sonicated chromatin was incubated with BSA-blocked streptavidin T1 Dynabeads (Invitrogen) overnight at 4 °C with rotation. The captured biotin-streptavidin complex were washed, and samples were equilibrated in TEV buffer (50 mM Tris/HCl pH 8.0, 100 mM NaCl, 0.1% NP-40 (CA630), 0.5 mM EDTA) for 5 min at RT, followed by AcTEV protease (Invitrogen) digestion (5-10 U) for 1 hr at RT in 40 μL TEV buffer. The streptavidin-conjugated beads were eluted again with TEV buffer with 10 min incubation at RT. Eluted samples were reverse cross-linked, RNAseA and Proteinase K treated following standard ChIP protocol11. ChIP fragments were ligated to Genomic adaptor (Illumina) or NEXTflex DNA barcodes adaptors (BioO Scientific). Adaptor ligated DNA fragments were size selected (150-250 bp), PCR amplified, and sequenced on Illumina Genome Analyzer or HiSeq system according to the manufacturer’s instructions.
HOMER for ChIP-Seq analysis and de novo motif discovery
ChIPseq peak identification, quality control, and motif analysis were performed using HOMER (http://biowhat.ucsd.edu/homer) as described 11,16. Peaks from separate experiments were considered co-bound if their peak centers were located within 200 bp of each other. For de novo motif analysis, transcription factor motif finding was performed on + 100 bp relative to the peak center defined from ChIP-Seq. Peak sequences were compared to random genomic fragments of the same size and normalized G+C content to identify motifs enriched in ChIP-Seq targeted sequence. To generate histogram for average distribution of tag densities, position corrected, normalized tags in 40 bp windows were tabulated within the indicated distance + from specific sites in the genome (i.e. Rev-Erb binding sites). Clustering plots for normalized tag densities at each genomic region were generated using HOMER and then clustered using Cluster (http://bonsai.hgc.jp/~mdehoon/software/cluster/) and visualized using Java TreeView 26.
Global Run-On Sequencing
Global run-on and library preparation for sequencing was described previously 16. Four 10-cm plates of confluent BMDM from WT control Rev-Erb DKO were used as starting material. Two biological samples were used per group. Approximately 10 million nuclei per sample were extracted and used for run-on and BrU incorporation.
BrU labeled nascent transcripts were immunoprecipiated with anti-BrU agarose beads (Santa Cruz Biotech), washed, eluted and precipitated in ethanol. BrU precipitated RNA were subjected for first strand complementary DNA synthesis. First, polyA tailed was added using Poly(A)-polymerase (NEB). Reverse transcription was then performed using Scriptscript III Reverse Transcriptase (Invitrogen) with oNTI223 primer (5′-pGATCGTCGGACTGTAGAACTCT;CAAGCAGA AGACGGCATACGATTTTTTTTTTTTTTTTTTTTVN-3′) where the p indicates 5′ phosphorylation, ‘;’ indicates the abasic dSpacer furan and VN indicates degenerate nucleotides. Subsequently, excess oNTI223 primers were removed by Exonuclease I (Fermentas). First-strand cDNA products were fragment with basic hydrolysis and size-selected (105-400 nt) in a 10% polyacrylamide TBE-urea gel (Invitrogen).
cDNA was subsequently circularlized using CircLigase (Epicentre), and relinearized at the basic dSpacer furan with ApeI (NEB). The ssDNA template was amplified to generate DNA for sequencing by Phusion Hig-Fidelity DNA Polymerase (Thermo Science) with primers oNTI200 (5′-CAAGCAGAAGACGGCATA-3′) and oNTI201 (5′-AATG ATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGACG-3′). PCR product was purified and size selected (140-225 bp) by gel electrophoresis on a non-denaturing 8% polyacrylamide TBE gel (Invitrogen). Purified DNA was then sequence on Illumina Genome Analyzer II according to the manuactuere’s instructions with small RNA sequencing primer 5′-CGACAGG TTCAGAGTTCTACAGTCCGACGATC-3′.
5′-GRO-Seq
For each RAW264.7 cell line, 20×106 nuclei were prepared from approximately 30×106 cells. Nuclear run-ons were performed in parallel on 100 μl aliquots containing 5×106 nuclei as for conventional GRO-Seq. Reactions were stopped and RNA was extracted with 450 μl TRIzol LS reagent (Invitrogen) each according to the manufacturer’s instructions. Following DNase treatment, the RNA was hydrolyzed in 20 μl total volume with 2 μl RNA fragmentation buffer (Ambion) for 10 minutes, and divalent cations were removed by gel filtration. Fragmented RNA was then 3′-dephosphorylated with polynucleotide kinase (Enzymatics), for 2 h at 37°C. The reaction was stopped with EDTA and PNK was inactivated and RNA denatured by heating the reaction to 75°C for 5 minutes, then cooled on ice for 2 minutes. BrdU-containing RNA fragments were precipitated using anti-BrdU agarose beads. The resulting RNA was dephosphorylated with calf intestinal phosphatase (NEB) and 5′-de-capped with tobacco acid pyrophosphatase (Epicentre). The reaction was stopped and RNA was extracted with Trizol LS, and libraries were prepared by ligating Illumina TruSeq-compatible adapters to the RNA 3′ and 5′ ends with truncated mutant RNA ligase 2 (K227Q) and RNA ligase 1 (NEB), respectively, followed by reverse transcription, cDNA isolation, and PCR amplification for 12 cycles. Final libraries were size-selected on PAGE/TBE gels to 60-110 bp insert size. A detailed protocol is available on request.
Genome-wide gene expression analysis with GRO-Seq
GRO-Seq analysis of genome-wide gene expression was performed by HOMER followed by edgeR 27. Briefly, HOMER was used to generate a gene expression matrix by identifying uniquely mapped RNA tags to gene bodies based on RefSeq annotation for the mouse genome (mm9). Statistical analysis for differential expression was performed using edgeR 27 on raw sequencing reads from WT and Rev-Erb DKO macrophages with two biological replicates per group. Genes with p < 0.005 were considered as differentially expressed.
Enhancer-associated RNA analysis
To examine regulation of eRNA expression, putative enhancers sites were first defined based on ChIP-Seq enrichment of H3K4me1 and H3K4me3 flanking + 1000 bp from the center of the transcription factor of interest. Putative enhancers were defined by the following criteria: 1) Regions are at least 2 kb away from annotated transcription start sites. 2) Regions have at least 16 tags from H3K4me1 ChIP-Seq normalized to 10 million tags. And 3) normalized ChIP-Seq tag count for H3K4me1 is greater than H3K4me3. HOMER was used to quantitate eRNA expression by tabulating normalized GRO-Seq tags within + 800 bp from the center of Rev-Erb or PU.1-bound intergenic enhancers. For 5’GRO Seq experiment, tag counts within + 250 bp of enhancers were tabulated. Histograms of RNA distribution at indicated enhancers were generated by tabulating average normalized RNA tag counts in resolution of 40 bp within 2 kb + from centers of specified genomic sites (e.g. Rev-Erb enhancers). Only enhancers with > 4 tags from GRO-Seq or 5’GRO-Seq within the specified window were included in expression analysis.
Analysis of correlation between eRNA to nearby protein coding gene was performed by examining differential expression of eRNA to that of the nearest expressing protein-coding genes. Briefly, Rev-Erb bound enhancers were assigned to the nearest expressing annotated genes defined by having at least 20 sequencing tags normalized to the length of the gene body. Differential expression of eRNA was determined by GRO-Seq from Rev-Erb DKO versus WT control, or by 5’GRO-Seq from Rev-Erbα overexpressing macrophages to control. Differential expression of annotated protein coding genes was determined by GRO-Seq in Rev-Erb DKO experiments. The data set was categorized as UP, NO CHANGE, and DOWN based on the differential expression of the eRNA. For Rev-Erb DKO GRO-Seq, eRNA with > 1.5-fold changes in GRO-Seq signal was considered differentially expressed; for Rev-Erbα overexpression experiment, 2.0-fold in 5’GRO-Seq signal. Spearmen rank correlation was used to test whether changes in eRNA and the corresponding protein coding gene covary.
Construction of enhancer reporters
For construction of Rev-Erb enhancer reporter plasmids, 900-1100 bp of sequence centered on Rev-Erb-bound sites flanked by demarcation of H3K4me1 were PCR amplified and cloned into the pGL4-TATA-TK at the BamHI/SalI sites downstream of the luciferase reporter gene as previously described 11. The following primers were used to PCR amplify enhancer sequences from mouse genomic DNA. Arhgap25 33 kb enhancer: 5′-GATCGTCGACTTTCCATGGGTCCAGAGATG-3′; 5′-GATCGGATCCAGCAGGCTGGGATATGAGTG-3′. Cx3cr1 9.8kb enhancer: 5′-GATCGGATCCTACACCTGCACAAGCACACA-3′; 5′-GATCGTCGACAACTGGGCGGAAATTGTAAA-3′. Cx3cr1 28 kb enhancer: 5′-GATCGGATCCGACCCTGGGTTGTCAGTAGG; 5′-GATCGTCGACACTTATGGGGGAGGATCTGG-3′. Eif2c4 -20 kb enhancer: 5′-GATCGTCGACCCCTCAAAGCTAACCATCCA-3′; 5′-GATCGGATCCAAAGTCATGCGAGACCTGAAA-3′. Mmp9 −5kb enhancer: 5′-GATCGGATCCGTGGCTCAGCATCAGGAAAT-3′; 5′-GATCGTCGACACTTGGCAGGCAGAGTGAGT-3′. P4Ha2 55 kb enhancer: 5′-GATCGGATCCGCCACAGCTCTGCTTTATGG-3′; 5′-GATCGTCGACGCTCACTGGCCTTGCTAACT-3′. Slc7a8 27 kb enhancer: 5′-GATCGTCGACCTGCATCCCGACTCATACCT-3′; 5′-GATCGGATCCTTCCAGCAAGCACTCTTTCA-3′. As negative control, a genomic region devoid of Rev-Erb binding and other enhancer-like features was used (chr5:30,278,928-30,279,847: 5′-gatcGGATCCAGTTCCATGTCCAGCGAATC-3′; 5′-gatcGTCGACGGAGCAAGGAGGGAGAGAG-3′).
For experiments testing functional significance of eRNA coding sequences for Mmp9 and Cx3cr1, various alterations of the enhancers were cloned into BamHI/SalI site downstream of the luciferase gene in pGL4 reporter driven by Mmp9 and Cx3cr1 promoter, respectively. Inversion of the eRNA coding sequence relative to the core element of Mmp9 (388 bp) and Cx3cr1 (210 bp) was achieved by Flip-PCR 28.
The following primers were used for constructing each enhancer variants. Mmp9 −5kb, 388 bp core enhancer: 5′-GATCGGATCCGGAAGCCGTTCCTTATCTCC-3′, 5′-GATCGTCGACTACACCCTGCTCACCAACAC-3′. Plus eRNA WT: 5′-GATCGGATCCGACTCAGGACTCCAGGTCTAG-3′, 5′-GATCGTCGACACTTGGCAGGCAGAGTGAGT-3′. Minus eRNA WT: 5′-GATCGGATCCGTGGCTCAGCATCAGGAAAT-3′, 5′-GATCGTCGACTACACCCTGCTCACCAACAC-3′. Plus eRNA flipped: flanking primer 1, 5′-GATCGTCGACTGTGTGGGGGTGGCAATGGA-3′, internal primer 1, 5′-GTGTTGGTGAGCAGGGTGTAACTTGGCAGGCAGAGTGAGT-3′; flanking primer 2, 5′-GATCGGATCCGACTCAGGACTCCAGGTCTAG-3′, internal primer 2 5′-ACTCACTCTGCCTGCCAAGTTACACCCTGCTCACCAACAC-3′. Minus eRNA flipped: flanking primer 3, 5′-GATCGGATCCACTCACTGGCAGATTACACAGC-3′, internal primer 3 5′-CTAGACCTGGAGTCCTGAGTCGTGGCTCAGCATCAGGAAAT-3′; flanking primer 4, 5′-GATCGTCGACTACACCCTGCTCACCAACAC-3′, internal primer 4 5′-ATTTCCTGATGCTGAGCCACGACTCAGGACTCCAGGTCTAG-3′. Cx3cr1 28 kb, 210 bp core enhancer: 5′-TTCAGGGATCCGCTGAGAGTTGCAGCATTGC-3′, 5′-TATTTGTCGACCTTGCTTGTTTCTTAAGCTCC-3′; Plus eRNA WT: 5′-TTCAGGGATCCGCTGAGAGTTGCAGCATTGC-3′, 5′-AGCATGTCGACACTTATGGGGGAGGATCTGG-3′; Minus eRNA WT: 5′-ATCGTGGATCCGACCCTGGGTTGTCAGTAGG-3′, 5′-TATTTGTCGACCTTGCTTGTTTCTTAAGCTCC-3′; Plus eRNA flipped: flanking primer 1, 5′-TTCAGGGATCCGCTGAGAGTTGCAGCATTGC-3′, internal primer 1, 5′-ATCCTCCCCCATAAGTCTTGCTGTTTCTTAAGCTCC-3′; flanking primer 2, 5′-TAAGAAACAGCAAGACTTATGGGGGAGGATCTGG-3′. Internal primer 2, 5′-ATTATGTCGACGCTTAAAAATAAACCTC-3′. Minus eRNA flipped: flanking primer 3, 5′-TATTTGTCGACCTTGCTTGTTTCTTAAGCTCC-3′, internal primer 3, 5′-CTGACAACCCAGGGTCGCTGAGAGTTGCAGCA-3′, flanking primer 4, 5′-TGCTGCAACTCTCAGCGACCCTGGGTTGTCAG, internal primer 4, 5′-AAGATGGATCCAACCAGCTGAAGATCAGCAG-3′. The promoter of Mmp9 and Cx3cr1 were each cloned into XhoI/BglII sites upstream of the luciferase gene in, respectively, pGL4-11 and pGL4-10 based vector with neomycin resistance gene. The following primers was used for promoter PCR amplification: Mmp9: 5′-GATCCTCGAGTGCCAAAGCTTTCCTGAGTG-3′, 5′-GATCAGATCTGGTGAGGACCGCAGCTTCT-3′. Cx3cr1: 5′-ATCGGCTAGCTCTAGCCTCCCTGGGCACAT-3′, 5′-TGCAAAGCTTCTCAAGTTACGAGCGTGCAA-3′.
Enhancer reporter experiments
Enhancer reporters were transfected into RAW264.7 macrophages using SuperFect (Qiagen) as described previously 11, using 300 ng of enhancer reporter and 200 ng of beta-actin promoter driven beta-galactosidase. Plasmids were complexed with 5 μL of SuperFect per 400 μL of medium in 24-well culture plate (Corning) seeded with 1 x 105 cells 24 hr prior to transfection. Luciferase activity was measured 24 hr post transfection using a Veritas microplate luminometer (Turner Biosystems) and normalized to beta-galactosidase activity (Applied Biosystem) for transfection efficiency. Each experiment was performed at least three independent times, with each reaction done in triplicates. Data represented mean + s.d., and statistical significance was determined by one-way ANOVA followed by Tukey HSD test.
RNA isolation and RT-PCR
Total RNA from macrophages was harvested using RNEasy Kit (Qiagen) followed by DNAse treatment using DNase (Qiagen) or TurboDNAse (Ambion) according to the manufacturer’s instruction. Total RNA (0.5-2 μg) was used for complementary DNA synthesis using Superscript III Reverse Transcriptase (Invitrogen) with random hexamers. No template controls were prepared by excluding reverse transcriptase from first strand cDNA synthesis. Quantitative transcript analysis was performed on an Applied Biosystems 7300 Real-time PCR system or Step One Plus using SYBR GreenER qPCR mastermix (Invitrogen). Values are normalized for 36B4 mRNA content. A modified ΔΔCT method, which incorporates PCR efficiencies, was used to determine relative expression of RNA.
For detection of RNA expression from enhancer reporter, strand-specific primers were designed to anneal at plasmid-specific sequence for first strand cDNA synthesis. These primers anneal to 22 and 87 base pair downstream of the SalI cloning site – pGL4_RT_en3 (CGAGTTGCATGATAAAGAAGA) and pGL4_RT_en3a (AGGAGCTGACTGGGTTGAAG). Total RNA harvested from RAW264.7 macrophages stably transfected with enhancer reporter (500 μg/mL G418) were isolated as described above. DNA was harvested, and primers annealing to the luciferase gene were use for Q-PCR to control for transfection efficiency.
Primers for quantitative PCR are as follows: Mmp9 mRNA (CATTCGCGTGGATAAGGAGT, GAAACTCACACGCCAGAAGA); Mmp9 plus eRNA 1 (AAGATGGGGGAAATGGTAGG, ACTTGGCAGGCAGAGTGAGT); Mmp9 plus eRNA 2 (CCCACTGCTTACCCACTGTT, TCGACACAACCTACCATTTCC); Mmp9 minus eRNA (TGGAGTCCCACAAAATCCTC, TAGCTCAACTGTGGGGTGTG); Mmp9 primary RNA transcript (AAGCGGACATTGTCATCCA, CAGGCATAAGAGCGGACAG); Cx3cr1 mRNA (AGTTCCCTTCCCATCTGCTC, AATGTCGCCCAAATAACAGG); Cx3cr1 28kb eRNA 1 (CTGCCTCAGGGAGAAACAAG, CTGCAACTCTCAGCAACCAG); Cx3cr1 28kb eRNA 2 (GCACCTACAATGTAATGACCTCTTTC, GATGCCCTCCGCCATTC); NCoA5 (CCTGGCCCAGAGAGTCAA, ACAGGCCTCTCAGCATCAAA); Csrnp1 (TCCTGTGGCCTCCAGAGTTT, GGCACCGTGGGAAATAGTAG); Rev-Erbα DBD (AGAGATGCTGTGCGTTTTGG, AGGCTGCTCAGTTGGTTGTT); Rev-Erbα C’ (AGGCTTCCGTGACCTTTCTC, TCACTGTCTGGTCCTTCACG); Rev-Erbβ (AGTGGCATGGTTCTACTGTGT, GCTCCTCCGAAAGAAACCCTTA); cyclopilin A (GGGTTCCTCCTTTCACAGAA, GATGCCAGGACCTGTATGCT); 36B4 (AGGGCGACCTGGAAGTCC, CCCACAATGAAGCATTTTGGA); Luciferase (ACGTGCAAAAGAAGCTACCG, ATGGGAAGTCACGAAGGTGT).
siRNA transfection
Non-targeting control and siRNAs directed against Mmp9 and Cx3cr1 eRNA (Dharmacon) were transfected into ThioMacs with DeliverX (Affymetrix) according to manufacturer’s protocol, or into BMDM with Lipofectamine 2000 (Invitrogen) using 100 nM siRNA as described previously 21. RNA was harvested from transfected macrophages 20-24 hr post-transfection.
The following siRNA oligos were used for in vitro studies, and the underlined siRNA were used for in vivo experiments. siGENOME Non-targeting siRNA pool #2 (Thermo Science, D-001810): UAAGGCUAUGAAGAGAUAC, AUGUAUUGGCCUGUAUUAG, AUGAACGUGAAUUGCUCAA, UGGUUUACAUGUCGACUAA. Custom siRNA for Mmp9 eRNA: GGUCACACAAUGAGCUGAAUU, AGAAAGAACCAGCAGCAAUUU. Custom siRNA for Cx3cr1 eRNA: CCUCUAAGAUGGCCAGUAAUU, UUACUGGCCAUCUUAGAGGUU.
Antisense oligonucleotide design
All ASOs used in this study contained a full phosphorothioate backbone and a 10 base 2′-deoxynucleoside gap flanked by 2’modified nucleosides. These 2′-modified nucleosides were either 2′-O-(2-methoxyethyl) (MOE) or constrained 2′-O-ethyl (cET) modifications. The motif for the ASOs targeting Mmp9 eRNA tested was eek-10-kke where “e” represents MOE, “k” represents cEt and “-10-“ represents the 10 base DNA gap. For the plus and minus Cx3cr1 eRNAs the motif used was kkk-10-kkk. ASOs were synthesized and purified as described previously 29,30. For each target strand, 78 test ASOs were screened for reduction of target RNA. A positive control targeting the coding sequence of MMP9 and a negative control matching no mouse transcripts were included. Two ASOs that resulted in maximum reduction of Mmp9 eRNA: ISIS 566237 (5′-ATTGTGTGACCCCAGC) and ISIS 566241 (5′-CAAGCTTCAGCTCATT) were selected for further studies. In addition, ASOs targeting the Cx3cr1 minus strand eRNA, Isis # 586596 (5′-TATGGCTGCCTCAGGG) and 586600 (5′-TGAGGAGTTTTCCCAT) were used. All oligos were designed to not contain G-strings with 4Gs or 2 sets of 3Gs in a row to prevent non antisense mediated effects 29.
Antisense oligonucleotide (ASO) transfection
For the antisense oligonucleotide experiments, ASOs targeting the Mmp9 plus strand eRNA, Isis #566237 (Mmp9 eRNA 1, GCACCTTTCCCTCGGATGGG ) and 566241 (Mmp9 eRNA 2, ATTGTGTGACCCCAGC), and ASOs targeting the Cx3cr1 minus strand eRNA, Isis # 586596 (Cx3cr1, TATGGCTGCCTCAGGG) and 586600 (Cx3cr1, TGAGGAGTTTTCCCAT) were used. These oligos are an eek-10-kke chemistry where “e” refers to 2′-O-methoxyethyl (2’MOE) modifications and “k” refers to a cET (constrained ethyl) modification. The “10” refers to a gap segment consisting of ten linked deoxynucleosides. A non-targeting ASO, Isis #129700 (TAGTGCGGACCTACCCACGA, a 5-10-5 MOE gapmer), and a Mmp9 mRNA-targeted ASO, Isis # 535522 (GCACCTTTCCCTCGGATGGG, a 5-10-5 MOE gapmer) were used as controls. For the experiments with ASOs directed against the Mmp9 plus strand eRNA, oligos at a concentration of 100 nM were transfected into 1.5-2.5 million ThioMac using Cytofectin (Gene Therapy Systems) in Opti-MEM reduced serum medium (Invitrogen) at a concentration of 3.5ug cytofectin/ml. Experiments with the ASOs targeting the Cx3cr1 minus strand eRNA (40nM) were conducted in confluent WT BMDM also using Cytofectin. Macrophages were incubated with ASO:Cytofectin complex for 4 hrs in 37 °C with 5% CO2, then replaced with RPMI medium containing 10% FBS (Hylcone), 100 U/mL of Penicillin and 100 mg/mL of Streptomycin (Invitrogen). Macrophages were harvested for RNA analysis after 16-24 hr post-transfection.
Supplementary Material
Acknowledgements
We thank Lynn Bautista for assistance with figure preparation. These studies were supported by NIH grants CA17390, U19DK62434, DK091183, DK063491 and CA52599. MGR and RME are Investigators of the Howard Hughes Medical Institute. MTL is supported by UCSD Medical Scientist Training Program T32 GM007198-37, and Genetics Training Program T32 GM008666, National Institute of General Medical Sciences. MUK was supported by a LeDucq Foundation Fellowship. HPL was supported by Finnish Cultural Foundation, Instumentarium Foundation, The Paulo Foundation, Paavo Nurmi Foundation, Finnish Foundation for Cardiovascular Research, The Maud Kuistila Memorial Foundation, and The Fulbright Center.
Footnotes
Author Contributions: M.T.L., S.H., C.B., A.W., T.R.G., M.G.R., R.M.E. and C.K.G. conceived the project and planned experiments, which were performed by M.T.L., H.C., H.P.L., D.G., S.H., Y.T-O., M.U.K., A.S.K., M.K., and C.Y.L., and analyzed by M.T.L., H.P.L., D.G., C.B., and C.K.G. The entire project was supervised by C.K.G., who wrote the manuscript with M.T.L. Competing financial interests: C.Y.L, A.W., and T.R.G. are employees of Isis Pharmaceuticals.
Data availability: Sequencing data can be accessed at GEO using the accession GSE45914
References
- 1.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadiantranscription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 2.Liu AC, et al. Redundant Function of REV-ERBá and â and Non-Essential Role for Bmal1 Cycling in Transcriptional Regulation of Intracellular Circadian Rhythms. PLoS Genetics. 2008;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raspé E, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43(12):2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 4.Le Martelot G, et al. REV-ERBá Participates in Circadian SREBP Signaling and Bile Acid Homeostasis. PLoS Biol. 2009;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbs JE, et al. The nuclear receptor REV-ERBá mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamir I, et al. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16(10):5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19(6):1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 8.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 11.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlaeger TM, Mikkola HK, Gekas C, Helgadottir HB, Orkin SH. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood. 2005;105(10):3871–3874. doi: 10.1182/blood-2004-11-4467. [DOI] [PubMed] [Google Scholar]
- 14.Giguere V, et al. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8(5):538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 15.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011 doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 19.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465(7301):1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu SG, et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nature Genetics. 2012;44(2):157. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang W, et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470(7334):414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013 doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melo CA, et al. eRNAs Are Required for p53-Dependent Enhancer Activity and Gene Transcription. Molecular Cell. 2013;49(3):524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Raal FJ, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375(9719):998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 25.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schanke JT. Sequence inversion by Flip-PCR. Methods Mol Biol. 1997;67:203–208. doi: 10.1385/0-89603-483-6:203. [DOI] [PubMed] [Google Scholar]
- 29.Baker BF, et al. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. The Journal of biological chemistry. 1997;272(18):11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 30.Seth PP, et al. Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symp Ser (Oxf) 2008;(52):553–554. doi: 10.1093/nass/nrn280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.