SUMMARY
Insulin homeostasis in pancreatic β-cell is now recognized as a critical element in the progression of obesity and type II diabetes (T2D). Proteins that interact with insulin to direct its sequential synthesis, folding, trafficking and packaging into reserve granules to manage release in response to elevated glucose remains largely unknown. Using a conformation based approach combined with mass spectrometry we have now generated the insulin biosynthetic interaction network (insulin BIN), a proteomic roadmap in the β-cell that describes the sequential interacting partners of insulin along the secretory axis. The insulin BIN revealed an abundant C2 domain-containing transmembrane protein 24 (TMEM24) that manages glucose-stimulated insulin secretion from reserve pool of granules, a critical event impaired in patients with T2D. Identification of TMEM24 in the context of a comprehensive set of sequential insulin binding partners provides a molecular description of the insulin secretory pathway in β-cells.
INTRODUCTION
Given the impact of a high fat, high sugar diet leading to obesity, metabolic syndrome and type II diabetes (T2D) (Ashcroft and Rorsman, 2012), it is crucial to understand the protein interactions that govern the biogenesis and release of insulin into plasma. Orci and colleagues initially described the trafficking itinerary of insulin through the secretory pathway of the pancreatic β-cell using electron microscopy (Orci et al., 1986; Orci et al., 1985). Mature insulin is synthesized in the ER, transported through the secretory pathway and then stored in two functionally distinct pools of granules: the reserve pool (RP) and the readily releasable pool (RRP) from which insulin is released in a biphasic fashion upon stimulation with glucose (Seino et al., 2011). We now know that folding in the ER, ER-to-Golgi transport and secretion of proteins to the extracellular milieu in response to stimulus is aided and monitored by the proteostasis network that make continual adjustments to the protein fold (Balch et al., 2008; Hutt and Balch, 2013; Powers and Balch, 2013). In human disease, early components of the folding machinery play a critical role in β-cell biology (Back and Kaufman, 2012). Surprisingly, little is known about the proteins that interact with insulin directly or indirectly to promote conformational maturation required to maintain the biphasic insulin release that is defective in T2D.
To explore the proteins promoting insulin synthesis and folding, granule maturation and secretion, previous proteomics studies focused on whole human pancreatic islets under basal (Waanders et al., 2009) and elevated glucose levels (Schrimpe-Rutledge et al., 2012). Using mass spectrometry of purified intact islets (which contain multiple cell types), these studies identified a large number of metabolic and signaling pathway components. Studies using insulin granules purified from a rat insulinoma cell line, INS1-E, by subcellular fractionation with or without immuno-affinity purification of the granule surface-associated protein VAMP-2 (Brunner et al., 2007; Hickey et al., 2009), identified a smaller subset of proteins enriched in granules, as well as lysosomal and mitochondrial proteins likely due to co-sedimentation of these organelles during fractionation (Suckale and Solimena, 2010). Secretory granule membranes purified from mouse insulinoma MIN6 cells showed that p23, a p24 family protein involved in vesicular transport, was localized to the insulin secretory granule membrane (Hosaka et al., 2007). While these data provide a glimpse of the proteomic composition of islet cell types and the insulin secretory granule, they do not identify the proteins that interact directly or indirectly with insulin to manage its synthesis, folding, trafficking and packaging for storage in granules - the central axis dictating β-cell function.
Herein, we report an insulin biosynthetic interaction network (insulin BIN) that describes in molecular terms the secretory pathway of the islet β-cell. Using a conformation based approach coupled to mass spectrometry, we characterized protein interactions for the immature insulin separately from interactions found with the mature insulin. To address the importance of highly abundant proteins recovered in the insulin BIN in β-cell biology, we characterized one prominent insulin BIN protein, transmembrane protein 24 (TMEM24) (also referred to as C2 domain containing protein 2 like (C2CD2L)). Molecular analysis of TMEM24 identify it as a pancreatic islet enriched protein that plays a key role in regulating glucose sensitive insulin release from the reserve pool of granules. Our results now provide a database that can now be used to dissect the biochemical and molecular mechanisms responsible for β-cell secretory pathway function in insulin biogenesis and release.
RESULTS
Defining the Insulin Biosynthetic Interaction Network (insulin BIN)
During granule maturation, the conformational precursor of insulin (proinsulin) generated by co-translational insertion into the endoplasmic reticulum (ER) is transported to the Golgi where it is proteolytically cleaved to yield the mature insulin. (Furuta et al., 1998; Zhu et al., 2002) Insulin is then stored in two pools of secretory granules leading to biphasic, glucose stimulated insulin secretion (GSIS) (Figure 1A). The vast majority of granules belong to the slow releasing reserve pool (RP) responsible for the sustained second phase of GSIS while a small fraction (5%) exist in the readily releasable pool (RRP) mediating the first phase of GSIS (Ohara-Imaizumi et al., 2002; Wang and Thurmond, 2009).
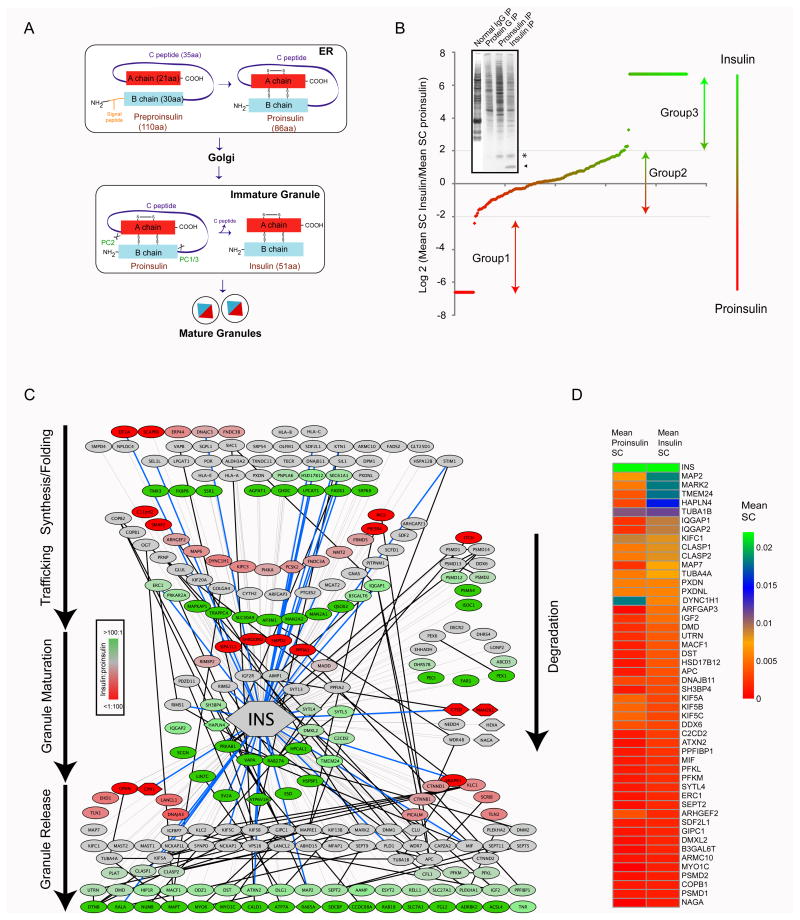
Figure 1. Insulin Biosynthetic Interaction Network (insulin BIN).
(A) Schematic for insulin maturation pathway in pancreatic β-cell. (B) Human orthologs of secretory proteins are identified, insulin: proinsulin binding ratios are calculated and plotted on a logarithmic (base2) scale. In the inset, silver stain of eluate from MIN6 cells immunoprecipitated with Control IgG (lane 1) or Protein G beads alone (lane 2) or with antibodies to proinsulin (18D2, lane 3) or insulin (K36ac10, lane 4) is shown. (C) Proteins that belong to the secretory compartment of insulin BIN are denoted as nodes (ovals; hexagons were used for proteins previously identified in proteomic studies of insulin secretory granules from INS1-E cells (Brunner et al., 2007; Hickey et al., 2009)). Proteins are arranged according to their location in the secretory pathway. Color of the node represent more binding to insulin (green), proinsulin (red) or to both (grey) based on log2 values of insulin to proinsulin binding ratios and statistical confidence (proteins with p ≥ 0.5 for differential binding to insulin/proinsulin based on a t-test are always grey). Grey lines represent the edges in the network that show direct or indirect interaction with insulin identified by insulin BIN, black straight lines represent interactions between nodes in the interactome based on PINA and blue lines connect insulin to proteins that have previous described function in insulin folding, trafficking or secretion or an association with diabetic conditions. (D) Heatmap of fifty proteins with highest fractional spectral coverage in insulin and proinsulin immunoprecipitate is shown. See also Figure S1 and S2.
To define the sequential interacting partners that bind to insulin during its maturation from the ER to storage granules, we immunopurified proinsulin alone and proinsulin/insulin containing protein complexes from mouse insulinoma MIN6 cells, a validated model of insulin maturation and secretion. We used two monoclonal antibodies that differentiate between the maturation states of insulin during its transit from the ER to post-Golgi granules. The first conformation specific antibody binds to the C-peptide region present only in mouse proinsulin and not in mature insulin (Figure S1A) while the second antibody binds to a common epitope present in both proinsulin and mature insulin (Figure 1B, inset). Proteins that differentially bound to proinsulin and insulin were identified by comparing the two protein complexes using mass spectrometry. High confidence proteins were then organized by IPI accession number and subcellular compartment as reported by the Gene Ontology (GO) (Ashburner et al., 2000), followed by manual curation based on known protein-protein interactions and published literature. Final lists of proteins associated with the secretory pathway are listed in supplementary Table S1 and S2.
Using normalized spectral counts assigned to statistically significant protein-protein interactions, insulin: proinsulin binding ratios were computed and plotted on a logarithmic (base 2) scale to highlight preferential binding partners to either proinsulin, insulin or both (Figure 1B and Table S3). The insulin BIN contains proteins from multiple subcellular compartments whose functions include protein synthesis, folding, trafficking and secretion. We used the comprehensive list of identified secretory pathway proteins (Table S3) in order to validate known features and to elucidate additional players involved in insulin maturation and secretion. Including insulin, we found 230 insulin BIN proteins to be annotated by GO and/or previously published literature as localized to secretory compartments and reflect interactions that could occur within the lumen of secretory compartments, or through transmembrane proteins that link cargo proteins in the lumen to cytosolic structural components facilitating insulin synthesis, folding, trafficking and secretion. We further compared our dataset with two recent human islet proteomic (Schrimpe-Rutledge et al., 2012; Waanders et al., 2009) and gene expression analyses (Mahdi et al., 2012; Marselli et al., 2010). Of the 230 proteins we had an overlap of 83% and 100% with the islet proteome and gene expression arrays, respectively, validating the relevance of our dataset in human islets (Table S3 and Figure S1B) (see Discussion). Based on the insulin: proinsulin binding ratios (Figure 1B), proteins in insulin BIN could be divided into three broad groups. Group 1 had log2 ratios smaller than −2.0 indicating at least four-fold (2−2.0) stronger interaction with proinsulin over insulin. This ratio would suggest roles for these proteins in ER to early Golgi function. Group 2 had ratios between 2.0 and −2.0 suggesting their potential to interact with both proinsulin and insulin, and therefore likely to include intermediate steps in maturation through later Golgi steps. Group 3 proteins had log2 ratios greater than 2.0, indicating at least a four-fold preferential binding to insulin over proinsulin and therefore potentially components that are found and function in late secretory pathway/granule maturation. Differential interaction of select proteins in each group with proinsulin and/or insulin was further validated by Western blotting of independent experimental replicates (Figure S1C).
Arranging all 230 secretory proteins in the insulin BIN according to their likely subcellular localization suggests that proinsulin-binding proteins (red nodes) extend all the way from the ER to the late secretory compartments where processing is completed during granule formation and granules are prepared for regulated release (Figure 1C). The number of proinsulin interacting proteins (Figure 1C, group 1 (red)) found in late compartments agrees with experimental evidence suggesting that proinsulin to insulin conversion, occurs at the acidic clathrin coated compartments derived from trans Golgi Network (Orci et al., 1985). Green nodes reflect proteins likely to function in late steps of maturation and release (group 3; log2 values >2.0). Proteins with log2 values close to zero or p value greater than 0.5 (based on a t-test corrected for multiple testing) are shaded grey to indicate binding to both insulin and proinsulin based on log values or due to lack of statistical confidence in preferential binding. While network analysis reveals the presence of a small subset of proteins currently recognized to play a role in insulin secretion in normal and/or diabetic conditions (nodes connected by blue line to insulin) and previously characterized binding partners for proteins within the insulin BIN (connected by black lines), the majority of interactions are new. Many of these interactors are proteins of unknown function in insulin secretory pathway identified with high spectral counts suggestive of strong interactions (Figures 1C, D and S1C).
Operation of the insulin BIN
In order to explore the functional significance of proinsulin/insulin interaction with proteins recovered in the immunoprecipitates and to validate our approach, select proteins were chosen based on previous knowledge of enrichment in islets or secretion. Sub-networks were created for proteins with known interactions, by choosing a given protein and finding all interactions based on Protein Interaction Network Analysis (PINA) platform (Wu et al., 2009) (Figure S2). Proteins binding only to proinsulin (Group 1) were the smallest group in the insulin BIN. This suggests efficient folding and export of proinsulin from the ER, an interpretation that is in agreement with the pulse-chase and electron microscope autoradiography experiments which show that ~90% of proinsulin is converted to insulin in approximately 1 h (Huang and Arvan, 1995; Orci et al., 1986; Wang et al., 2011). In group 2, we identified a number of proteins likely mediating events leading to insulin maturation following synthesis, folding and export from the ER. Here, for example, we identified COPB2, a subunit of Golgi coatomer coat complex (COPI) involved in retrograde Golgi to ER transport (Cosson and Letourneur, 1994). As expected, the COPB2 subnetwork showed interactions with another component of the complex, COPB1, also found in the insulin BIN (Figure S2A). Also in group 2, we identified one of the two prohormone convertases, PCSK2, which has previously been implicated in the cleavage of proinsulin to insulin (Furuta et al., 1998). The prohormone convertase PCSK1 was not identified in our experiment or in previous proteomic analysis (Suckale and Solimena, 2010), a result potentially reflecting its low abundance, less stable interaction, and/or poor detection by mass spectrometry. However, PCSK1 was present in the sub-network of PCSK2 through its interaction with islet amyloid polypeptide (IAPP) (Westermark et al., 2011) found in our dataset suggesting the discovery of interactome pathways relevant to insulin based β-cell biology (Figure S2C). In group 3, we identified Rab27a, a small cytosolic GTPase associated with the granule membrane. Rab27a showed preferential strong association with insulin over proinsulin (Kasai et al., 2005). A subnetwork consisting of 24 proteins previously reported to interact with Rab27a included five proteins (RIMS1, RIMS2, Sytl4, Sytl5, and Eef1a1) that were identified in the insulin BIN (Figure S2D). Apart from validating the known components of insulin secretory pathway, insulin BIN also revealed proteins previously uncharacterized in insulin maturation. For example, we identified Erp44, a member of the thioredoxin-PDI family (Anelli et al., 2002), with a log2 value of −1.44 indicative of ~2.7 fold more binding to proinsulin. Erp44, localized to ER-Golgi intermediate compartment (ERGIC) and the cis-Golgi, forms mixed disulfide bonds with ER disulfide oxidases Ero1α and Ero1β that could potentially mediate the redox cascade facilitating proinsulin folding. Western blotting of proinsulin or insulin immunoprecipitates with antibodies to the above mentioned proteins largely corroborated their assigned log2 values, thus validating our Insulin BIN dataset (Figure S1C).
In summary, analysis of secretory proteins recovered in the insulin BIN validates our approach to pathway building and helps in the visualization of the potential multi-protein machinery (the insulin BIN) involved in β-cell function. Furthermore, it provides critical insight into a suggested sequential association of previously known and unknown proteins affecting insulin maturation in β-cell secretory pathway described by Orci and colleagues (Orci et al., 1986). In the absence of an antibody that binds exclusively to insulin and not to proinsulin, this analysis provides a guiding tool for differential binding of interactors to pro and mature forms of insulin along the secretory pathway.
TMEM24, an insulin BIN component enriched in pancreatic islets
Amongst the top proteins with highest mean normalized spectral counts in insulin BIN were the bait protein, insulin, microtubule associated proteins, TMEM24 and IQ motif containing GTPase activating proteins, IQGAPs (Figure 1D and Table S3). The role of microtubular system in insulin secretion has been known for long (Boyd et al., 1982) and more recently IQGAP1, enriched in pancreatic β-cells, has been shown to act as a regulator of insulin secretion acting through exocyst-septin complex and cdc42 (Rittmeyer et al., 2008). However, transmembrane protein 24 TMEM24, is a protein of unknown function apart from its presence in few high throughput studies (Caberoy et al., 2009; Huttlin et al., 2010; Munton et al., 2007) and a single in silico analysis that mapped TMEM24 onto human chromosome 11q23.3, a region frequently deleted or rearranged in neuroblastoma (Katoh, 2004). To validate insulin BIN as a platform for discovery of proinsulin/insulin binding proteins with functional role in insulin maturation and secretion we further characterized TMEM24. TMEM24, found throughout bilaterian animals, consistently showed high level of spectral counts across all replicates (Table S3 and Figure 1D) indicative of protein abundance, likely reflecting strong interaction. This association with proinsulin and insulin was confirmed by Western blotting of immunoprecipitates with specific antibodies against TMEM24 (Figure 2A). Co-immunoprecipitation of exogenously expressed proinsulin-V5 by TMEM24-HA from MIN6 and COS1 cells further confirmed this interaction (Figures 2B and C). Lower expression of proinsulin in COS1 cell extract compared to MIN6 cell extract could be due to increased constitutive secretion in non-insulinoma cells. TMEM24 show high sequence similarity across species and have an N-terminal region with very weak similarity to synaptotagmin, followed by a classic C2 domain and a region suggested to bind specifically to phosphatidylserine (PS) (Figure S3; (Caberoy et al., 2009)), properties typical of synaptic vesicle regulators. Invertebrate homologs also have a C-terminal C1 domain (Colon-Gonzalez and Kazanietz, 2006), which is largely degenerate in vertebrates, and lack the zinc-binding cysteine residues (Figure S3).
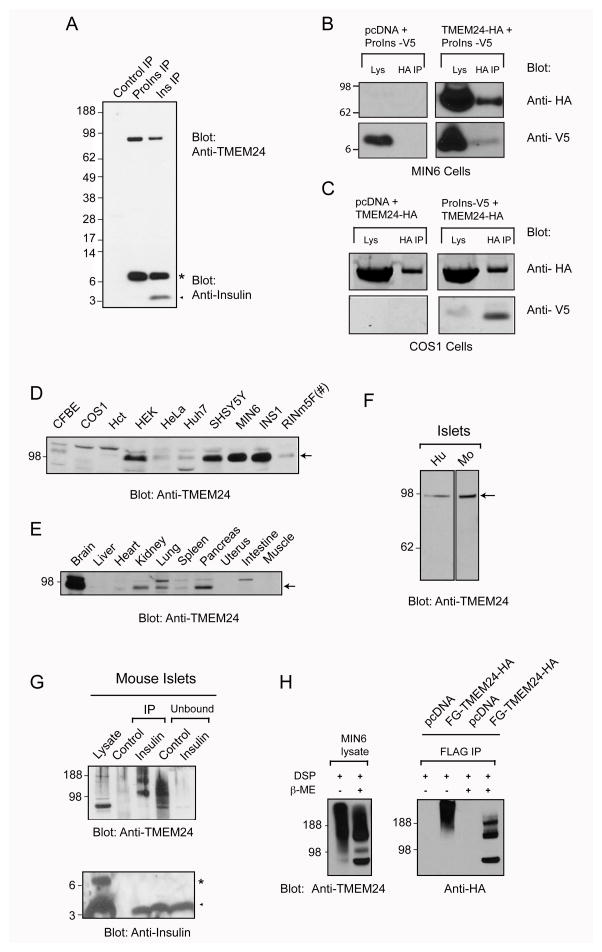
Figure 2. TMEM24 is a pancreatic islet enriched protein that interacts and co-localizes with insulin.
(A) Immunoprecipitates of MIN6 cells were probed with TMEM24 and insulin antibody; (*) indicates proinsulin band and arrowhead insulin band (B and C) MIN6 or COS1 cell lysates expressing TMEM24-HA and proinsulin-V5 were immunoprecipitated with HA antibodies and western blotted with anti-HA or V5 antibodies (D) tissue extracts (10 μg) from a C57BL/6 mouse or (E) cell lysates (20 μg) of different cell lines were Western blotted using antibodies against TMEM24; (#) Indicates that 10 μg of protein loaded; arrow marks the band corresponding to TMEM24. (F) Human and mouse islets lysates were probed with antibodies against TMEM24 (G) Mouse pancreatic islets were immunoprecipitated with Protein G alone (Control) or insulin antibody; islet lysate, immunoprecipitated proteins and unbound fractions were probed with TMEM24 and Insulin antibody. (H) MIN6 cells untransfected or transfected with either pcDNA or FLAG-TMEM24-HA were in vivo crosslinked with 1 mM DSP. Lysates and FLAG immunoprecipitates were separated in the presence or absence of the reducing agent β-mercaptoethanol followed by Western blotting with antibodies against TMEM24 or HA epitope. See also Figure S3.
To further investigate the tissue specific expression of TMEM24 we analyzed lysates from different cell lines and tissues by Western blot. TMEM24 is highly expressed in kidney and pancreatic tissue as well as in insulinoma cell lines (Figures 2D and E). We also noted a strong enrichment in brain and neuronal cells (Figures 2D and E) suggesting a more general role in secretory pathway function. Islets isolated from mouse and human pancreas showed strong enrichment of TMEM24 (Figure 2F). In islets, TMEM24 co-precipitated under physiological conditions with insulin as a larger protein complex (es) suggesting existence of dimers and/or oligomers (or other protein complexes). A similar result was observed by DSP mediated in vivo cross-linking experiments with endogenous or exogenously transfected TMEM24 in MIN6 cells (Figures 2G and H). Observation of phosphorylated Ser 374 in our mass spectrometry data combined with a tissue specific phosphoproteomic study confirmed phosphorylation of TMEM24 (Huttlin et al., 2010).
Differential extraction of crude membrane fractions from MIN6 cells showed that TMEM24 is an integral membrane protein (Figure S4A). To determine the toplogy of TMEM24 on the membrane, we transfected mouse fibroblasts with FLAG-TMEM24-HA, containing an N-terminal Flag epitope tag and a C-terminal HA epitope tag. Cells were then treated with either digitonin, to selectively permeabilize the plasma membrane, or TX-100 to permeabilize all membranes. As shown in Figure S4B, the HA epitope was accessible when only the plasma membrane was permeabilized suggesting its cytosolic localization whereas FLAG staining was visible only after internal membranes were permeabilized confirming that TMEM24 is a type I transmembrane protein with C-terminal cytosolic region and a short luminal N-terminus. The presence of a large cytosolic region containing a C2 domain and phosphorylation sites suggests that the functional regulation of the TMEM24 could be through its C-terminus.
Subcellular localization of TMEM24
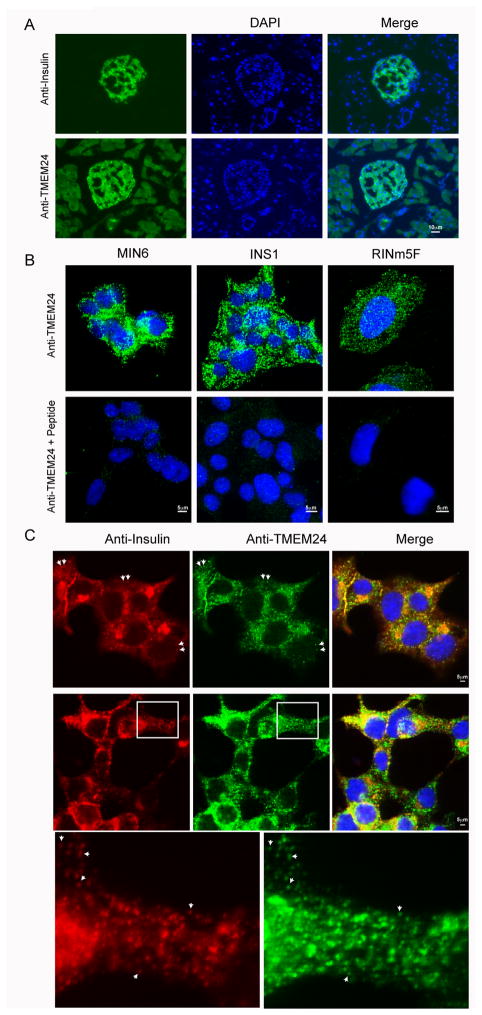
To determine the subcellular localization of TMEM24, consecutive mouse pancreatic cryosections were stained with insulin or TMEM24. As expected, insulin localized specifically to β-cells whereas TMEM24 showed enrichment in islets compared to acinar cells but, was present in all of the secretory cell types within the islets (Figure 3A). Insulinoma cell lines were analyzed by immunofluorescence microscopy to further delineate the localization of TMEM24 within pancreatic β-cells. TMEM24 localized to punctate structures and this staining was lost upon blocking the antibody with the antigenic peptide (Figure 3B) confirming specificity of the antibody. TMEM24 did not show co-localization with several subcellular compartments including trans-Golgi network (TGN38), early endosomes (EEA1), mitochondria (mitotracker) or ERGIC (ERGIC 53) (Figure S5A). Moreover, TMEM24 did not colocalize with plasma membrane-associated SNARE proteins, syntaxin 1 and SNAP25, implicated previously in a readily releasable pool of granules (Figure S5B). However, TMEM24 showed significant colocalization with insulin containing punctate structures (Pearson’s correlation coefficient of 0.60 ± 0.06) (Figure 3C). These results suggest a possibility that TMEM24 plays a universally conserved role in regulated secretion, a conclusion consistent with its abundance in neuronal tissue and co-localization with insulin in pancreatic β-cells.
Figure 3. Subcellular localization of TMEM24.
(A) Consecutive pancreatic cryosections from mouse perfused with 4% paraformaldehyde were immunostained using anti-TMEM24 antibody or anti-insulin antibody followed by Alexafluor conjugated secondary antibody and DAPI staining. Images were obtained at 20X magnification. (B) Insulinoma cell lines MIN6, RINm5F and INS1-E cells were fixed, permeabilized and stained with anti-TMEM24 antibody or anti-TMEM24 antibody incubated with antigenic peptide. Stained cells were visualized by confocal microscopy. (C) INS1-E cells were fixed, permeabilized and co-stained with antibodies against TMEM24 protein (green) and insulin (red). Representative images from two different experiments are shown. Squared region in middle panels are enlarged with white arrowheads indicating punctate structures co-stained by both insulin and TMEM24 antibody. See also Figure S4 and S5.
TMEM24 is not required for insulin processing but plays a role in glucose stimulated insulin secretion
To determine its potential role in insulin maturation, we tested whether TMEM24 is required for the processing of proinsulin to insulin by pulse-chase analysis, following lentivirus-mediated knockdown of TMEM24. MIN6 cells were infected with a lentivirus containing a shRNA targeting TMEM24 and after four days of infection, the extent and rate of processing proinsulin to insulin were unaffected (Figures 4A, B and C), suggesting that TMEM24 is not required for insulin maturation.
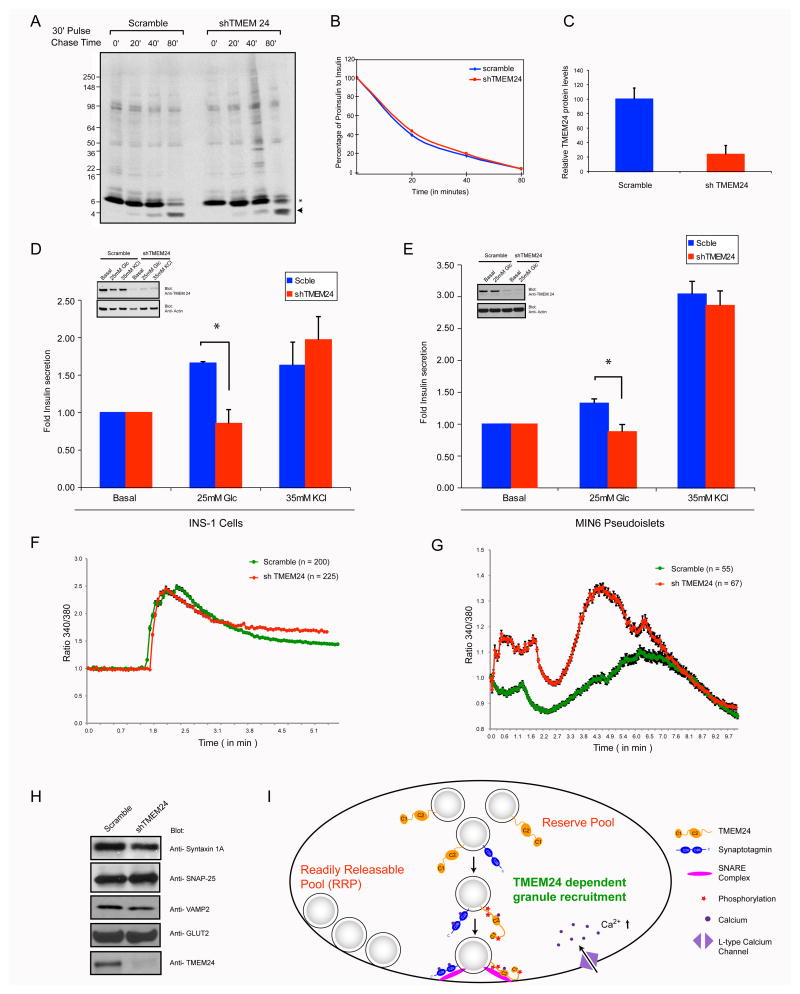
Figure 4. TMEM24 has a role in glucose stimulated insulin secretion.
MIN6 cells transduced with scrambled or shTMEM24 were (A) pulse labeled with 35S and then chased with complete media. Cell lysates were immunoprecipitated with antibodies against insulin and samples were analyzed by autoradiography (B) Ratio of proinsulin to insulin at different time points were quantified from the pulse chase analysis by setting the zero time point to a 100. (C) Amount of TMEM24 in lysates were quantified, normalized to actin and plotted. (D) INS1-E cells or (E) MIN6 pseudoislets were transduced with scrambled or TMEM24 shRNA. Amount of insulin secreted under basal, 25 mM glucose or 35 mM KCl were assayed by ELISA. Relative insulin secretion was calculated by normalizing to insulin secretion under basal conditions. Average ± SEM of three independent knockdown and secretion assays shown (* p< 0.05). MIN6 cells transduced with scrambled or shTMEM24 loaded with Fura 2 and changes in [Ca2+]i were measured in the presence of (F) 35 mM KCl or (G) 25 mM glucose. Data represented as Average + SEM from indicated number of cells for each condition (H) Levels of SNARE proteins and GLUT2 were assessed by Western blotting upon TMEM24 knockdown (I) Model for proposed function of TMEM24 indicates possible role for TMEM24 in the recruitment of the reserved pool to the membrane for release. Figure illustrates a Ca2+-dependent and perhaps phosphorylation dependent recruitment of the reserve pool to manage a response to increased blood glucose.
We focused on the role of TMEM24 in insulin secretion using INS-1 derived 832/13 clonal cells. All of the secretagogues including KCl can evoke rapid release of the RRP responsible for the first phase insulin secretion while only glucose and nutrient metabolites can cause first phase and a sustained second phase secretion by mobilization of the reserve pool (Wang and Thurmond 2009). Even with 70% reduction in protein levels of TMEM24 in cells, the basal and KCl-induced insulin secretion from granules docked to the cell surface remain unchanged, emphasizing that TMEM24 is unlikely to be a regulator of the RRP. In contrast, glucose induced secretion resulting from recruitment of granules from the cell interior to its periphery (Ohara-Imaizumi et al., 2007; Wang and Thurmond, 2009) was consistently reduced by 50% or greater compared to the control cells (Figure 4D). Similar to INS1-E cells, glucose stimulated secretion was reduced upon partial TMEM24 knockdown in MIN6 pseudoislets ((Kelly et al., 2011), Figure 4E) without affecting the pseudoislet structure. The decrease in TMEM24 did not cause significant destabilization or reduced expression of SNARE proteins, considered important for insulin secretion, or GLUT2 (the β-cell glucose transporter), confirming that the reduced insulin secretion is not due to an overall decrease in fusion components or decreased uptake of glucose (Figure 4H).
Granule release in first and second phases of secretion is tightly coupled to elevated Ca2+ concentration; KCl mediated insulin secretion occurs by closure of K+ channels that causes Ca2+ influx through L-type Ca2+ channels whereas glucose stimulation induces various metabolic signals which in turn leads to an increase in cytoplasmic Ca2+ concentration (Pedersen and Sherman, 2009). To determine whether knockdown of TMEM24 altered Ca2+ influx, scramble and shTMEM24 treated cells were loaded with the Ca2+ binding dye, Fura 2, and stimulated with KCl or glucose. As shown in Figure 4F, TMEM24 knockdown in cells did not show any difference in [Ca2+]i concentration when stimulated with KCl, suggesting that TMEM24 does not affect the activity of L-type Ca2+ channels, responsible for localized Ca2+ influx in proximity to docked RRP. However, cells lacking TMEM24 did show a slight increase in intracellular [Ca2+]i upon stimulation with glucose, possibly due to downstream events reflecting loss of TMEM24 function (Figure 4G). Together, these results suggests that impairment of the second phase of insulin secretion in the absence of TMEM24 is not due to reduced Ca2+ levels but possibly because of Ca2+ mediated, TMEM24 dependent downstream events leading to the targeting of RP to plasma membrane for release.
We suggest that TMEM24, a late-acting insulin BIN component, is a membrane-associated synaptotagmin-like protein that associates with proinsulin and insulin. TMEM24 functions in the sustained glucose induced insulin secretion, possibly by acting as a missing link in the recruitment of reserve pool of granules (newcomers) to late events leading to secretion. The identification and characterization of TMEM24 suggests that other components of the insulin BIN are likely to provide, in an unbiased way, important clues to the largely uncharacterized molecular features of the insulin processing and granule maturation pathways required to sustain normal glucose homeostasis.
DISCUSSION
The work reported herein describes the molecular description of insulin maturation (the insulin BIN) using a conformational intermediate based mass spectrometry approach. This approach allowed us to get at the molecular events differentiating early from late steps in the genesis of storage granules, a pathway that heretofore has been largely limited to morphological descriptions (Orci et al., 1986; Orci et al., 1985). Our results illustrate that proinsulin maturation evolves by an unanticipated and surprisingly complex set of protein-protein interactions that reflects the dynamic folding compartments dictating protein trafficking through the exocytic pathway (Balch et al., 2008; Hutt and Balch, 2013; Powers and Balch, 2013). The insulin BIN provides a critical resource to characterize previously unknown molecular features of the β-cell secretory pathway.
Comparison of our interactome dataset with whole cell human and mouse islet proteomic and gene expression studies (Mahdi et al., 2012; Marselli et al., 2010; Schrimpe-Rutledge et al., 2012; Waanders et al., 2009) highlights the physiological relevance of the interactome to intact islets. Of the 230 proteins that we identified in the secretory pathway of the MIN6 interactome, 168 were represented in the mouse islet proteome giving an overlap of 73% (Waanders et al., 2009). Comparison of changes in the MIN6 interactome in proteomic abundance with human islets after 24 h of 5 mM or 15 mM glucose incubation (Schrimpe-Rutledge et al., 2012) revealed an overlap of 67.8% (156 proteins out of 230) where ~67% of the proteins in our interactome were represented to be up (31%), down (27%) or unchanged (9%) (Table S3) suggesting the interactome components are largely very dynamic in response to glucose challenge.
Interestingly, only a limited set of proteins comprising 21 out of 230 (9%), involved in membrane trafficking, were found to interact with proinsulin at early steps of synthesis and folding. This likely reflects its normally transient residence in the ER during nascent synthesis followed by efficient folding and export as shown by pulse chase and electron microscope autoradiography experiments (Orci et al., 1986; Wang et al., 2011). Early interactors confirmed previously known functions as well as revealed other interactions including the ER oxidase, Erp44. In contrast to paucity of ER interactors, we discovered a large number of interactors (147 out of 230; 64%) to likely facilitate post-ER, Golgi and post-Golgi maturation events directing insulin to secretory granules and involved in regulated release. Proteins including the Rab GTPase Rab27a, fell into group 3 and reflect interactions with mature insulin responsible for secretion (62 out of 230; 27%). The absence of any plasma membrane associated SNARE family members in our dataset is in agreement with the idea that only a small pool of granules are docked to the plasma membrane (~7% of total granules (Henquin, 2011), with the majority of insulin immunoprecipitated coming from the reserve pool. Because insulin is the major cargo in a β-cell, our results demonstrate that the multiple direct or indirect protein interactions (the insulin BIN) shown here may play a major role in biogenesis and management of the β-cell.
As a validation of this concept and based on the predictive value of the insulin BIN, we identified and characterized a type I transmembrane protein, TMEM24 that is enriched in pancreatic islets, strongly associated with proinsulin and insulin with high spectral coverage and suggested a functional role in reserve pool granule maturation for release. Intriguingly, TMEM24 is an ancient protein, which could reflect a key role in the evolution of regulated secretory pathway function across diverse phyla. We demonstrated that TMEM24 knockdown specifically reduced glucose-induced insulin secretion, but not the readily releasable KCl induced secretion, providing critical insight into the molecular components dictating reserve pool release. Strikingly, all homologs of TMEM24 contain an ~130 amino acid C2 domain that folds into an eight stranded β-sandwich found in over 100 proteins involved in vesicle trafficking and signal transduction. C2 domains bind phospholipids in a Ca2+-dependent manner and mediate protein-membrane associations that facilitate exocytosis of synaptic vesicles and insulin granules (Friedrich et al., 2010). Different pools of granules in β-cells have different Ca2+ sensitivities. Thus, availability of different Ca2+ sensors may be crucial in generating the dynamic responses critical for glucose homeostasis managed by insulin release (Pedersen and Sherman, 2009). While the precise mechanism by which TMEM24 functions in glucose/Ca2+-sensitive reserve pool granule maturation remains to be determined, one speculative possibility is that TMEM24 could undergo glucose-induced phosphorylation/dephosphorylation changes. These changes could mediate its interaction with other proteins to facilitate the recruitment of newcomer granules to phospholipid bilayers in a Ca2+-dependent fashion, a model consistent with the similarity of TMEM24 to synaptotagmin family members and their role(s) in synchronous vesicle release (Figure 4I).
Our quantitative description of the molecular pathway defining the β–cell secretory axis opens the door for unprecedented level of insight into the complex environment controlling β-cell secretory axis function. The insulin BIN sets the stage for discovery and mechanistic analyses of proteins in pancreatic β-cells likely contributing to glucose homeostasis in health and disease.
EXPERIMENTAL PROCEDURES
Antibodies, plasmids and cell lines
Detailed description of all antibodies, DNA constructs, cell lines and shRNA lentiviral purification and concentration is described in online Supplemental Material.
Immunoprecipitation of insulin
MIN6 cell pellets were resuspended in 50 mM Tris pH 7.5, 150 mM NaCl and 1% TX-100 with protease inhibitor cocktail (Roche) lysed on ice for 30 min, clarified by centrifugation and precleared with Protein G beads (Amersham) for 1 h at 4°C. Supernatant was incubated either with Protein G beads alone (control) or with control IgG, insulin or proinsulin antibodies crosslinked to beads for 3–4 h at 4°C. The beads were pelleted, washed with lysis buffer twice and with buffer without detergent once. Bound proteins were eluted with 100 μl of 50 mM Tris pH 7.5 and 1% SDS at 100°C for 5 min. Eluted samples (1/10th total) were analyzed by silver stain before analysis by mass spectrometry. Rest of the eluate was precipitated with 4 volumes of acetone at −20°C for overnight, spun at maximum speed and pellets were dried and stored at −80°C.
Pulse chase analysis
MIN6 cells were plated at 8×105 cells in 35 mm dishes and infected with 20 μl of concentrated scrambled or TMEM24 shRNA virus. After 72 h of infection, cells were washed with PBS twice and pulse chase analysis was carried out as described in online Supplemental Material.
ELISA
ELISA was performed using Rat/mouse ELISA kit (Millipore, EZRMI-13K) according to the manufacturer’s instructions and as described in detail in online Supplemental Material.
Supplementary Material
HIGHLIGHTS.
Defined insulin biosynthetic interaction network (insulin BIN)
Describes the molecular components involved in insulin trafficking and secretion
Identified C2 domain containing transmembrane protein, TMEM24
TMEM24 manages glucose stimulated insulin secretion from the reserve pool of granules
Acknowledgments
We would like to thank Harish Pillai for help with Log2 graph, heat map and SAINT analysis, Drs. Sherry Niessen, Cristian Ruse, Patricia Szajner, Atanas Koulov, Sandra Pankow and Patricia Tu for help with aspects of Mass spectrometry runs, Drs.Tao Xu and Robin Park for assistance with SEQUEST analysis, Dr. Hyungwon Choi for insight on SAINT analysis, Drs. Lesley Page, Mathew Wheeler and Patricia Szajner for critical reading and suggestions on the manuscript, Dr. Mathew Wheeler, Dr. Maria Cecilia Marcondes and Dr.Justin Hassler for help with mouse islet isolation, Dr.Ya-Wen Liu for mouse fibroblasts, Dr.Jiansong Tong for help with mouse perfusion, Helen Plutner and Brooke Haddock for the characterization of 18D2 antibody. These studies were supported by NIH grant GM033301 and GM42336 to WEB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002;21:835–844. doi: 10.1093/emboj/21.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft FM, Rorsman P. Diabetes mellitus and the beta cell: the last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back SH, Kaufman RJ. Endoplasmic reticulum stress and type 2 diabetes. Annual review of biochemistry. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Boyd AE, 3rd, Bolton WE, Brinkley BR. Microtubules and beta cell function: effect of colchicine on microtubules and insulin secretion in vitro by mouse beta cells. J Cell Biol. 1982;92:425–434. doi: 10.1083/jcb.92.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner Y, Coute Y, Iezzi M, Foti M, Fukuda M, Hochstrasser DF, Wollheim CB, Sanchez JC. Proteomics analysis of insulin secretory granules. Mol Cell Proteomics. 2007;6:1007–1017. doi: 10.1074/mcp.M600443-MCP200. [DOI] [PubMed] [Google Scholar]
- Caberoy NB, Zhou Y, Alvarado G, Fan X, Li W. Efficient identification of phosphatidylserine-binding proteins by ORF phage display. Biochem Biophys Res Commun. 2009;386:197–201. doi: 10.1016/j.bbrc.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Gonzalez F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim Biophys Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Friedrich R, Yeheskel A, Ashery U. DOC2B, C2 domains, and calcium: A tale of intricate interactions. Mol Neurobiol. 2010;41:42–51. doi: 10.1007/s12035-009-8094-8. [DOI] [PubMed] [Google Scholar]
- Furuta M, Carroll R, Martin S, Swift HH, Ravazzola M, Orci L, Steiner DF. Incomplete processing of proinsulin to insulin accompanied by elevation of Des-31,32 proinsulin intermediates in islets of mice lacking active PC2. J Biol Chem. 1998;273:3431–3437. doi: 10.1074/jbc.273.6.3431. [DOI] [PubMed] [Google Scholar]
- Henquin JC. The dual control of insulin secretion by glucose involves triggering and amplifying pathways in beta-cells. Diabetes Res Clin Pract. 2011;93(Suppl 1):S27–31. doi: 10.1016/S0168-8227(11)70010-9. [DOI] [PubMed] [Google Scholar]
- Hickey AJ, Bradley JW, Skea GL, Middleditch MJ, Buchanan CM, Phillips AR, Cooper GJ. Proteins associated with immunopurified granules from a model pancreatic islet beta-cell system: proteomic snapshot of an endocrine secretory granule. J Proteome Res. 2009;8:178–186. doi: 10.1021/pr800675k. [DOI] [PubMed] [Google Scholar]
- Hosaka M, Watanabe T, Yamauchi Y, Sakai Y, Suda M, Mizutani S, Takeuchi T, Isobe T, Izumi T. A subset of p23 localized on secretory granules in pancreatic beta-cells. J Histochem Cytochem. 2007;55:235–245. doi: 10.1369/jhc.6A7093.2006. [DOI] [PubMed] [Google Scholar]
- Huang XF, Arvan P. Intracellular transport of proinsulin in pancreatic beta-cells. Structural maturation probed by disulfide accessibility. J Biol Chem. 1995;270:20417–20423. doi: 10.1074/jbc.270.35.20417. [DOI] [PubMed] [Google Scholar]
- Hutt DM, Balch WE. Cold Spring Harbor perspectives in biology. 2013. Expanding Proteostasis by Membrane Trafficking Networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K, Ohara-Imaizumi M, Takahashi N, Mizutani S, Zhao S, Kikuta T, Kasai H, Nagamatsu S, Gomi H, Izumi T. Rab27a mediates the tight docking of insulin granules onto the plasma membrane during glucose stimulation. The Journal of clinical investigation. 2005;115:388–396. doi: 10.1172/JCI22955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of TMEM24 family genes in silico. Int J Oncol. 2004;25:759–764. [PubMed] [Google Scholar]
- Kelly C, McClenaghan NH, Flatt PR. Role of islet structure and cellular interactions in the control of insulin secretion. Islets. 2011;3:41–47. doi: 10.4161/isl.3.2.14805. [DOI] [PubMed] [Google Scholar]
- Mahdi T, Hanzelmann S, Salehi A, Muhammed SJ, Reinbothe TM, Tang Y, Axelsson AS, Zhou Y, Jing X, Almgren P, et al. Secreted frizzled-related protein 4 reduces insulin secretion and is overexpressed in type 2 diabetes. Cell metabolism. 2012;16:625–633. doi: 10.1016/j.cmet.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Marselli L, Thorne J, Dahiya S, Sgroi DC, Sharma A, Bonner-Weir S, Marchetti P, Weir GC. Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PloS one. 2010;5:e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, Gehrig P, Potthast F, Rutishauser D, Gerrits B, et al. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol Cell Proteomics. 2007;6:283–293. doi: 10.1074/mcp.M600046-MCP200. [DOI] [PubMed] [Google Scholar]
- Ohara-Imaizumi M, Fujiwara T, Nakamichi Y, Okamura T, Akimoto Y, Kawai J, Matsushima S, Kawakami H, Watanabe T, Akagawa K, et al. Imaging analysis reveals mechanistic differences between first- and second-phase insulin exocytosis. J Cell Biol. 2007;177:695–705. doi: 10.1083/jcb.200608132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara-Imaizumi M, Nakamichi Y, Tanaka T, Ishida H, Nagamatsu S. Imaging exocytosis of single insulin secretory granules with evanescent wave microscopy: distinct behavior of granule motion in biphasic insulin release. J Biol Chem. 2002;277:3805–3808. doi: 10.1074/jbc.C100712200. [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Amherdt M, Madsen O, Perrelet A, Vassalli JD, Anderson RG. Conversion of proinsulin to insulin occurs coordinately with acidification of maturing secretory vesicles. J Cell Biol. 1986;103:2273–2281. doi: 10.1083/jcb.103.6.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Amherdt M, Madsen O, Vassalli JD, Perrelet A. Direct identification of prohormone conversion site in insulin-secreting cells. Cell. 1985;42:671–681. doi: 10.1016/0092-8674(85)90124-2. [DOI] [PubMed] [Google Scholar]
- Pedersen MG, Sherman A. Newcomer insulin secretory granules as a highly calcium-sensitive pool. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7432–7436. doi: 10.1073/pnas.0901202106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Balch WE. Diversity in the origins of proteostasis networks - a driver for protein function in evolution. Nature reviews. Molecular cell biology. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittmeyer EN, Daniel S, Hsu SC, Osman MA. A dual role for IQGAP1 in regulating exocytosis. J Cell Sci. 2008;121:391–403. doi: 10.1242/jcs.016881. [DOI] [PubMed] [Google Scholar]
- Schrimpe-Rutledge AC, Fontes G, Gritsenko MA, Norbeck AD, Anderson DJ, Waters KM, Adkins JN, Smith RD, Poitout V, Metz TO. Discovery of Novel Glucose-Regulated Proteins in Isolated Human Pancreatic Islets Using LC-MS/MS-Based Proteomics. J Proteome Res. 2012;11:3520–3532. doi: 10.1021/pr3002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino S, Shibasaki T, Minami K. Dynamics of insulin secretion and the clinical implications for obesity and diabetes. The Journal of clinical investigation. 2011;121:2118–2125. doi: 10.1172/JCI45680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckale J, Solimena M. The insulin secretory granule as a signaling hub. Trends Endocrinol Metab. 2010;21:599–609. doi: 10.1016/j.tem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen Y, Yuan Q, Tang W, Zhang X, Osei K. Control of precursor maturation and disposal is an early regulative mechanism in the normal insulin production of pancreatic beta-cells. PloS one. 2011;6:e19446. doi: 10.1371/journal.pone.0019446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Thurmond DC. Mechanisms of biphasic insulin-granule exocytosis - roles of the cytoskeleton, small GTPases and SNARE proteins. J Cell Sci. 2009;122:893–903. doi: 10.1242/jcs.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark P, Andersson A, Westermark GT. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiological reviews. 2011;91:795–826. doi: 10.1152/physrev.00042.2009. [DOI] [PubMed] [Google Scholar]
- Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6:75–77. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- Zhu X, Orci L, Carroll R, Norrbom C, Ravazzola M, Steiner DF. Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10299–10304. doi: 10.1073/pnas.162352799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.