Abstract
Campylobacter jejuni is a foodborne bacterial pathogen, which is now considered as a leading cause of human bacterial gastroenteritis. The information regarding ribonucleases in C. jejuni is very scarce but there are hints that they can be instrumental in virulence mechanisms. Namely, PNPase (polynucleotide phosphorylase) was shown to allow survival of C. jejuni in refrigerated conditions, to facilitate bacterial swimming, cell adhesion, colonization and invasion. In several microorganisms PNPase synthesis is auto-controlled in an RNase III (ribonuclease III)-dependent mechanism. Thereby, we have cloned, overexpressed, purified and characterized Cj-RNase III (C. jejuni RNase III). We have demonstrated that Cj-RNase III is able to complement an Escherichia coli rnc-deficient strain in 30S rRNA processing and PNPase regulation. Cj-RNase III was shown to be active in an unexpectedly large range of conditions, and Mn2+ seems to be its preferred co-factor, contrarily to what was described for other RNase III orthologues. The results lead us to speculate that Cj-RNase III may have an important role under a Mn2+-rich environment. Mutational analysis strengthened the function of some residues in the catalytic mechanism of action of RNase III, which was shown to be conserved.
Keywords: Campylobacter jejuni, catalytic activity, manganese, ribonuclease, RNase III
Abbreviations: Cj-RNase III, C. jejuni RNase III; cpm, counts/min; dsRBD, dsRNA binding domain; DSS, disuccinimidyl suberate; DTT, dithiothreitol; Ec-RNase III, E. coli RNase III; LB, Luria–Bertani; PNPase, polynucleotide phosphorylase; RNase III, ribonuclease III; wt, wild-type
INTRODUCTION
Bacteria are able to rapidly adjust their physiology in order to survive in response to environmental demands. This adaptation is accompanied by a fast adjustment of the RNA levels. The cellular concentration of a given RNA is the result of the balance between its synthesis and degradation. RNA degradation involves the concerted action of ribonucleases. Changes in RNA turnover facilitate stress responses, growth phase transitions and production of virulence factors [1,2].
RNase III (ribonuclease III) family of enzymes is a highly conserved group of ds (double-stranded) RNA-specific endoribonucleases widely distributed among prokaryotic and eukaryotic organisms. In bacteria, RNase III has the ability to regulate its own synthesis with a specific cleavage near the 5′end of its mRNA. Moreover, it is also responsible for rRNA operon maturation, processing of cellular, phage and plasmid RNAs, and decay of sRNA/mRNA complexes upon translational silencing [1,3,4].
Bacterial RNase III is the simplest member of the family, containing an endonuclease domain (NucD), characterized by the presence of a set of highly conserved carboxylic acid residues essential for catalytic activity, and a dsRBD (dsRNA binding domain) (Figure 1C). In Escherichia coli, this enzyme is encoded by the rnc gene and it is active as a 52 kDa homodimer [3].
Figure 1. C. jejuni RNase III protein features.
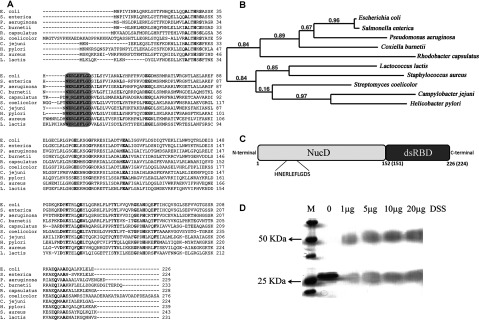
(A) Sequence alignment of RNase III from E. coli (Uniprot ID: P0A7Y0), S. enterica (Uniprot ID: E7V351), P. aeruginosa (Uniprot ID: B7UYX2), C. burnetii (Uniprot ID: P51837), R. capsulatus (Uniprot ID: Q52698), S. coelicolor (Uniprot ID: Q9ZBQ7), C. jejuni (NCBI Reference Sequence: YP_001001278), H. pylori (Uniprot ID: P56118), S. aureus (Uniprot ID: P66668) and L. lactis (Uniprot ID: Q9CHD0). Fully conserved residues are in bold, and the signature sequence of these proteins is highlighted. (B) Phylogenetic tree of RNase III from the species considered in (a). (C) Schematic representation of the domain organization of RNase III, showing the catalytic domain (NucD) (residues 1 to 151 in E. coli protein and 1 to 150 in C. jejuni protein) and the dsRNA binding domain (residues 152–226 in E. coli protein and 151–224 in C. jejuni protein). The signature sequence present in the catalytic domain is emphasized. (D) Cross-linking of RNase III from C. jejuni using DSS. Approximately 0.5 μg of protein was incubated with increasing concentrations of DSS as indicated in the figure. Proteins were visualized by Coomassie Brilliant Blue staining.
Important insights on structural features and mechanism of action of RNase III have been provided by crystallographic studies. Structural analysis of Aquifex aeolicus RNase III revealed a symmetric positioning of the two catalytic domains that form the dimer through hydrophobic interactions, and demonstrated a positional mobility of the dsRBD [5]. A divalent metal ion is required for RNase III activity, with magnesium (Mg2+) as the preferred co-factor. This homodimer uses a two metal mechanism of catalysis, with each active site containing two divalent cations during substrate hydrolysis [6]. The structure of Thermotoga maritime together with the one from A. aeolicus provided the basis for a proposed pathway of dsRNA recognition and cleavage by the bacterial RNase III members [7]. Accordingly, RNase III can affect gene expression in two different ways. When the dsRNA is not bound to the catalytic valley, RNase III binds it without cleaving [8]. In this form, RNase III can affect RNA structures and modulates gene expression [9,10]. In addition, it can also function as a dsRNA-processing enzyme, cleaving both natural and synthetic dsRNA [5]. The RNase III cleavage originates 5′ phosphate and 3′ hydroxyl termini with a two-base overhang at the 3′end [5]. In eukaryotes, homologues of this enzyme have a key role in RNA interference phenomenon that starts with a dsRNA cleavage [3].
Campylobacter jejuni is a Gram-negative microorganism, which remains as one of the major causes of human gastroenteritis worldwide [11]. The main reservoir of C. jejuni is the guts of avian species with up to 109 CFU (colony forming units)/g in faeces [12]. Owing to the spillage of intestinal content that contains a large number of the pathogen, C. jejuni can contaminate cooling water, knives and poultry meat during slaughter. Thus, the consumption of contaminated poultry is the major cause of campylobacteriosis; however, a diverse range of environmental sources are also all recognized causes of infection [13]. Despite the specific microaerobic growth requirements, C. jejuni is ubiquitous in the aerobic environment and is capable to face different stresses during transmission, including temperature changes, starvation, hypo- and hyper-osmotic stress, and desiccation. In addition, during human infection, C. jejuni has to withstand a further range of stresses, including changes in pH and the host innate immune response [14,15]. Although major advances have been made in the understanding of C. jejuni pathobiology and physiology, many unanswered questions remain. A more complete understanding of the regulation of C. jejuni response mechanisms is required to facilitate appropriate intervention strategies in order to reduce C. jejuni-associated diseases [16].
It was demonstrated by Haddad et al. [17,18] that the exoribonuclease PNPase (polynucleotide phosphorylase) is involved in low-temperature survival of C. jejuni, and that the lack of PNPase induces swimming limitation, chick colonization delay and decrease of cell adhesion/invasion ability. However, little is known regarding RNases in this pathogen. In E. coli, PNPase synthesis is autocontrolled at a post-transcriptional level in an RNase III-dependent mechanism [19,20]. The functional and evolutionary conservation of the RNase III family in bacteria and higher organisms is indicative of their biological relevance.
We present here the relevant information regarding functionality and biochemical properties of Cj-RNase III (C. jejuni RNase III). We show that this endoribonuclease is able to replace Ec-RNase III (E. coli RNase III) in the 30S processing and in the regulation of PNPase levels. We also demonstrate the dependence of Cj-RNase III catalytic activity on divalent metal ions, and its ability to cleave at different temperatures and pH values that mimic different physiological environments that C. jejuni may face. Finally, a biochemical analysis was assessed on Cj-RNase III mutants with substitutions on two conserved acidic residues in the nuclease domain.
EXPERIMENTAL
Bacterial strains and culture conditions
The strains used in this study are listed in Table 1. C. jejuni 81-176 was routinely cultured on Karmali agar plates (Oxoid) or in BHI (brain heart infusion) broth (Merck). Cultures on plates and broth were incubated at 42°C for 48 and 24h, respectively, under a microaerophilic atmosphere in jars flushed with a gas mixture of 10% (v/v) CO2, 5% (v/v) O2 and 85% (v/v) N2. E. coli strains were cultivated overnight at 37°C in the LB (Luria–Bertani) medium (Sigma). The growth medium was supplemented with ampicillin (100 μg/ml) when appropriate.
Table 1. Strains used in this study.
Construction of the plasmids expressing RNase III from C. jejuni
The C. jejuni 81–176 RNase III gene (rnc) (GenBank accession no. YP_001001278) was amplified from genomic DNA using the primers MO608 (5′-AATTCATATGCAAGGGAAAATGATGAAAAA -3′) and MO612 (5′- GTAGGATCCAATTTCAATCTCGTACCAAAGGTAT -3′) (Table 3) (NdeI and BamHI sites, respectively, are underlined). For rnc amplification by PCR, 1 unit of Phusion High-Fidelity DNA Polymerase (BioLabs) was used. The purified PCR product (734 pb) was cloned into the pGEM-T plasmid (Promega) to generate pGEMT-rnc. To overexpress the Cj-RNase III protein, the rnc gene double digested with NdeI and BamHI was subcloned from pGEMT-rnc into the pET19b expression vector previously digested with the same enzymes (Novagen). Plasmids and primers used are presented in Tables 2 and 3, respectively. The resultant pET19b-rnc was confirmed by DNA sequencing (Beckman Coulter Genomics).
Table 3. Primers used in this study.
| Primers | Sequence (5′-3′) | Restriction site |
|---|---|---|
| MO608 | AATTCATATGCAAGGGAAAATGATGAAAAA | NdeI |
| MO612 | GTAGGATCCAATTTCAATCTCGTACCAAAGGTAT | BamHI |
| Cjej_9 | CATCACAGCAGCGGC | |
| Cjej_10 | GCAGCCGGATCCTAT | |
| D44A_1 | GGTgcaGCTGTGCTTG | Introduction of D44A mutation and PvuII site |
| D44A_2 | CAAGCACAGCtgcACC | |
| E116Q_1 | CAGAcGCGTTAGAtGCCAT | Introduction of E116Q mutation and MluI site |
| E116Q_2 | ATGGCaTCTAACGCgTCTG | |
| E116D_1 | CAGAcGCGTTAcAAGCCAT | Introduction of E116D mutation and MluI site |
| E116D_2 | ATGGCTTgTAACGCgTCTG |
Table 2. Plasmids used in this study.
| Plasmid | Antibiotic resistance | Reference | Comments |
|---|---|---|---|
| pGEM-T | Ampicillin | Promega | Commercial vector |
| pET19b | Ampicillin | Novagen | Commercial expression vector |
| pGEMT:rnc | Ampicillin | This study | Encodes Cj-RNC |
| pET19b-rnc | Ampicillin | This study | Encodes his-Cj-RNC |
| pET19b-rnc_D44A | Ampicillin | This study | Encodes his-Cj-RNC_D44A mutant |
| pET19b-rnc_E116Q | Ampicillin | This study | Encodes his-Cj-RNC_E116Q mutant |
| pET19b-rnc_E116D | Ampicillin | This study | Encodes his-Cj-RNC_E116D mutant |
Construction of RNase III mutants by overlapping PCR
The point mutations D44A, E116Q and E116D were introduced into pET19b-rnc by overlapping PCR [21]. The primers used are listed in Table 3 and base changes are indicated in small letters. All mutant constructs were confirmed by DNA sequencing at STAB Vida, Portugal.
Overexpression and purification of recombinant wt (wild-type) RNase III and mutants from C. jejuni
The plasmids harbouring the histidine-tagged Cj-RNase III wt protein (pET19b-rnc) and the mutants (pET19b-rnc_D44A, pET19b-rnc_E116Q, pET19b-rnc_E116D) were transformed into E. coli BL21(DE3) recArnc105 strain ([22], kindly provided by Professor A. Nicholson to allow the expression of the recombinant proteins. This derivative strain of BL21(DE3), carrying an RNase III mutation, was used since it blocks the regulation of Cj-RNase III by the endogenous E. coli homologue, resulting in a higher yield of the enzyme upon overexpression. Cells were grown at 37°C in 100 ml LB medium supplemented with 100 μg/ml ampicillin to an optical density of 0.5 at 600 nm. At this point, protein expression was induced by addition of 0.5 mM IPTG and left to grow for 2 h. Cells were pelleted by centrifugation and stored at −80°C. Purification was performed by histidine affinity chromatography using HiTrap Chelating HP columns (GE Healthcare) and AKTA FPLC system (GE Healthcare) following a protocol previously described [4]. The purity of the proteins was verified in a SDS/12%PAGE gel followed by Coomassie blue staining. Proteins were quantified using the Bradford Method [23] and 50% (v/v) glycerol was added to the final fractions prior storage at −20°C.
Cross-linking of Cj-RNase III wt and mutants
In order to verify if RNase III wt and mutants are able to dimerize, the purified proteins were incubated with increasing concentrations of DSS (disuccinimidyl suberate). Cross-linking reaction was performed in a final volume of 10 μl, including 0.5 μg of purified protein, 10 mM Hepes pH 7.4, 250 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT (dithiothreitol) and increasing concentrations of DSS (1–20 μg). The reaction was performed at room temperature (~25°C) for 30 min and quenched by adding 1 μl of Tris–HCl 1 M pH 7.5 and SDS loading buffer. The samples were boiled during 5 min and then analysed in a SDS/12% PAGE gel.
In vitro transcription
A canonical substrate for RNase III, called R1.1, was constructed using a synthetic DNA template and a promoter oligonucleotide obtained by commercial source (StabVida) for in vitro transcription, using the method described by Milligan et al. [24]. Briefly, the DNA synthetic template (0.5 μM) and the promoter oligonucleotide (0.6 μM) were annealed in 10 mM of Tris–HCl pH 8.0 by heating for 5 min at 70°C, following by incubation for 30 min, at 37°C. In vitro transcription was carried out using ‘Riboprobe in vitro Transcription System’ (Promega) and T7 RNA polymerase with a molar excess of [32P]-α-UTP over non-radioactive UTP. In order to remove DNA template, 1 unit of DNase (Promega) was added and incubated 30 min at 37°C. R1.1 transcript was purified by electrophoresis on an 8.3 M urea/10% PAGE. The gel slice was crushed and the RNA was eluted overnight at room temperature with elution buffer [3 M ammonium acetate pH 5.2, 1 mM EDTA, 2.5% (v/v) phenol pH 4.3]. The RNA was ethanol precipitated and resuspended in RNase-free water. The yield of the labelled substrates [cpm (counts/min)/μl] was determined by scintillation counting.
RNase III activity assays
The activity assays were performed in a final volume of 50 μl containing the RNase III activity buffer (30 mM Tris–HCl pH 8, 160 mM NaCl, 10 mM MgCl2 or MnCl2, 0.1 mM DTT) and 10000 cpm of substrate. As a control, prior to the beginning of each assay an aliquot was taken and was incubated until the end of the assay (without the enzyme). The reactions were started by the addition of the enzyme, and further incubated at the indicated temperature. Aliquots of 5 μl were withdrawn at the time-points indicated in the respective figures, and the reactions were stopped by the addition of formamide containing dye supplemented with 10 mM EDTA. RNase activity assays were carried out at various pH values (pH 5.2, 5.4, 6.1 and 7.3), at different temperatures (4, 30, 37 and 42°C), and at different Mg2+ and Mn2+ concentrations (0.1, 1, 10, 50 and 100 mM). Reaction products were resolved in a 7 M urea/15% PAGE as indicated in the respective figure legends. Signals were visualized by PhosphorImaging and analysed using ImageQuant software (Molecular Dynamics).
Analysis of rRNA patterns on agarose gels
E. coli MG1693 (wt) and SK7622 (Δrnc) alone or carrying the plasmids pGEMT and pGEMT:rnc were grown at 37°C in LB medium to an optical density of 0.5 at 600 nm. Cells were pelleted and stored at −80°C. Total RNA was extracted following the protocol described in Stead et al. [25]. RNA was quantified on a Nanodrop 1000 machine (NanoDrop Technologies), and 15 μg of total RNA were visualized in a 1.5% (w/v) agarose gel.
Western blot
E. coli MG1693 (wt) and SK7622 (Δrnc) alone or carrying the plasmids pGEMT and pGEMT:rnc were grown at 37°C in LB medium to an optical density of 0.5 at 600 nm. Cells were pelleted and lysed by addition of 200 μl of Bug Buster reagent (Merck Millipore). After 10 min of centrifugation at maximum speed, the soluble fraction was transferred to a new tube and protein concentration was measured. 30 ng of soluble protein were separated by SDS–PAGE and transferred to a nitrocellulose membrane (Hybond ECL, GE Healthcare) by electroblotting using the Trans-Blot SD semidry electrophoretic system (Bio-Rad). Membranes were probed with a 1:10000 dilution of anti-PNPase or anti-RNase R antibodies. ECL anti-rabbit IgG-conjugated horseradish peroxidase was used as the secondary reagent in a 1:10000 dilution. Immunodetection was conducted via a chemiluminescence reaction using Amersham ECL Western Blotting Detection Reagents (GE Healthcare).
Sequence alignment and construction of the phylogenetic tree
RNase III sequences of E. coli, Salmonella enterica, Pseudomonas aeruginosa, Coxiella burnetii, Rhodobacter capsulatus, Lactococcus lactis, Staphylococcus aureus, Streptomyces coelicolor, C. jejuni and Helicobacter pylori were aligned using Clustal W2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) [26]. The phylogenetic tree was built using Phylogeny.fr (http://www.phylogeny.fr) [27].
RESULTS AND DISCUSSION
Analysis of C. jejuni RNase III features
RNase III is a specific dsRNA endoribonuclease widely conserved in prokaryotes and eukaryotes. The amino acid alignment of the putative Cj-RNase III with nine different RNase III orthologues from Gram-negative and Gram-positive microorganisms reveals high conservation (Figure 1A). Indeed, Cj-RNase III exhibits the highly conserved amino acid stretch (NERLEFLGDS) in the catalytic domain like in all RNase III members already documented (Figures 1A and 1C) [3]. Furthermore, in Figure 1(B), the phylogenetic tree demonstrates that the sequences can be divided into two groups, which reflects well the phylogenetic relationships of RNase III between the corresponding bacteria. Cj-RNase III is located in a different group than E. coli and S. enterica RNase III. Among the nine microorganisms analysed, the highest similarity is observed with the H. pylori counterpart (48% identity).
Cj-RNase III is a dimeric protein
In order to characterize the activity of Cj-RNase III, we cloned the rnc gene in an expression vector and overexpressed the protein in an E. coli BL21(DE3) recArnc105 strain. Ec-RNase III is able to regulate its own synthesis by cleaving near the 5′end of its own message [28]. The rnc105 mutation changes the highly conserved Gly44 residue in the catalytic domain to aspartic acid abolishing RNase III activity, whereas the recA inactivation increases the stability of the recombinant plasmid. Thus, the E. coli BL21(DE3) recArnc105 strain allowed us to have higher levels of Cj-RNase III expression when compared with the wt strain, by preventing the cleavage of the overexpressed RNase III.
RNase III family members function as a homodimer [29]. To examine whether Cj-RNase III acts like a homodimer we performed a cross-linking assay using the cross-linker DSS. The protein was incubated with different amounts of DSS and the reaction was observed in a SDS/12% PAGE. DSS reacts with primary amino groups forming stable amide bonds. This will ‘fix’ the protein interactions, allowing their identification. In Figure 1(D) is possible to see the formation of a DSS-dependent specie which has two times the size of the monomeric form (~50 kDa). The result is consistent with the formation of a dimer, which led us to conclude that Cj-RNase III also undergoes dimerization.
Functional complementation of Cj-RNase III in E. coli
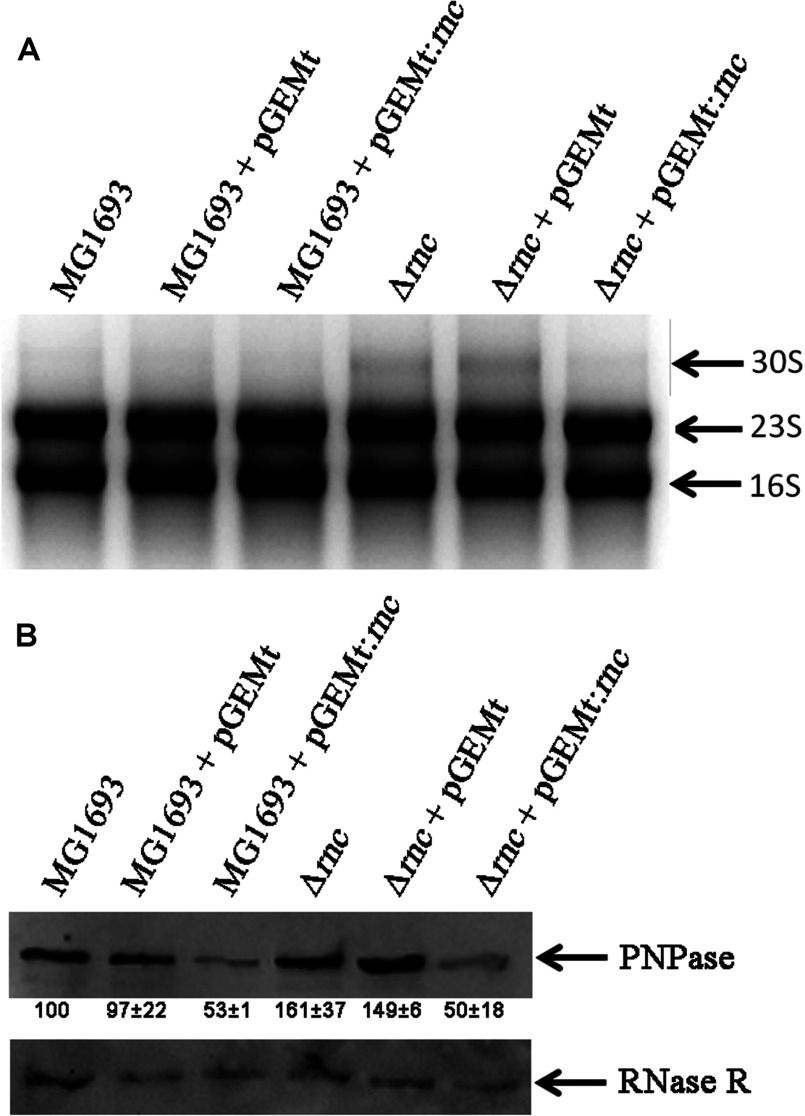
RNase III is involved in the processing of 30S rRNA. In the E. coli strain SK7622 that has a deletion in the rnc gene, is possible to detect the unprocessed 30S rRNA due to RNase III absence [30] (Figure 2A). To see whether the Cj-RNase III was able to complement the loss of endogenous RNase III in E. coli, we have transformed the SK7622 (Δrnc) strain with the plasmid pGEMT:rnc that carries the rnc gene from C. jejuni and analysed the rRNA pattern. SK7622 does not express T7 RNA polymerase, and the expression of the Cj-RNase III is this strain is derived from the weak expression of the plasmid. The results obtained revealed that in the presence of pGEMT:rnc the 30S rRNA species is no longer observed (Figure 2A). We can then conclude that Cj-RNase III is able to functionally replace E. coli RNase III in the 30S processing and that this may be also its role in C. jejuni.
Figure 2. E. coli complementation by Cj-RNase III.
(A) Processing of 30S rRNA. Total RNA was extracted from E. coli MG1693 (wt) and SK7622 (Δrnc) empty or containing pGEMt and pGEMt:rnc plasmids, and analysed in a 1.5% agarose gel. Bands were visualized by staining with ethidium bromide. The RNA species are indicated in the figure. This experiment was performed in quadruplicate. (B) PNPase regulation. Total soluble proteins were extracted from E. coli MG1693 (wt) and SK7622 (Δrnc) empty or containing pGEMt and pGEMt:rnc plasmids and 30 ng of protein were applied in a SDS/Page gel. A Western blot was performed using anti-PNPase and anti-RNase R antibodies. This experiment was performed in quadruplicate. PNPase levels were quantified and the values are indicated in the figure.
It is also known that RNase III is involved in the posttranscriptional control of pnp message, which codes for the exoribonuclease PNPase. Strains deficient in RNase III present higher levels of PNPase when compared with the wt [31,32]. Since we showed that Cj-RNase III is able to complement an E. coli mutant at the level of 30S processing, we checked if the same is valid for PNPase regulation. For that purpose, we extracted the total soluble proteins and performed a Western blot using anti-PNPase antibodies. By comparing the E. coli parental and mutant strains, we can see that the PNPase levels are ~1.5-fold higher in Δrnc and Δrnc+pGEMt strains (Figure 2B). The presence of pGEMT:rnc plasmid that harbors Cj-RNase III in both parental and mutant strains, causes a substantial decrease in PNPase levels due to the presence of higher levels of this protein (Figure 2B). As a control, we used antibodies against another ribonuclease, RNase R, which regulation does not involve RNase III. We can see that for RNase R there are no changes at the protein level (Figure 2B), indicating that the modifications observed for PNPase are specific.
These complementation assays indicate that Cj-RNase III is efficient in the processing of E. coli rRNA and in the cleavage of pnp message, demonstrating that this enzyme is able to substitute its E. coli counterpart in rRNA processing and in the control of mRNA levels. This is an interesting result considering the phylogenetic distance between RNase III from C. jejuni and E. coli (Figure 1B). Similar results were previously obtained for the Lactococcus lactis RNase III [33], strengthening the high conservation of this family of enzymes.
Divalent metal ion dependence of Cj-RNase III catalytic activity
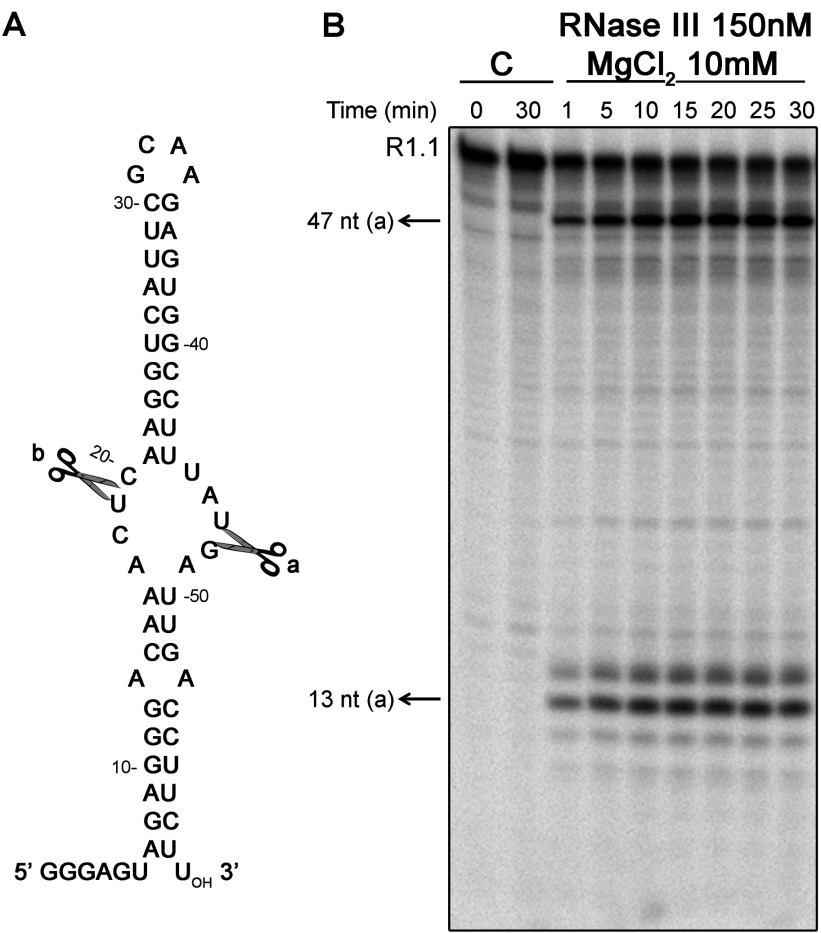
R1.1 RNA is a well-known 60 nt (nucleotides) substrate for E. coli RNase III, which contains a single primary cleavage site (Figure 3A, (a)) that is recognized in vivo and in vitro, and a secondary site [Figure 3A, (b)] that is cleaved only under specific conditions [34]. This canonical substrate was used to check the activity of Cj-RNase III purified in this work. It is known that the cleavage of dsRNA by RNase III is dependent of divalent cations [7]. Ec-RNase III can use different divalent ions with Mg2+ as the preferred co-factor [35]. A cleavage assay using Mg2+ is shown in Figure 3(B), which reveals that Cj-RNase III is catalytically active and cleaves R1.1.
Figure 3. Substrate cleavage in vitro by Cj-RNase III.
(A) Sequence and secondary structure of R1.1 RNA substrate. The cleavage sites are indicated. (B) In vitro cleavage of R1.1 RNA by RNase III from C. jejuni. The activity assay was performed at 37°C as described in the Experimental section using Mg2+ as co-factor. Protein concentration is indicated in the figure. Samples were taken during the reaction at the time points indicated and reaction products were analysed in a 15% PAGE/7 M urea gel. Control reactions with no enzyme added were incubated at the maximum reaction time. Length of substrates and degradation products are indicated. These experiments were performed at least in triplicate.
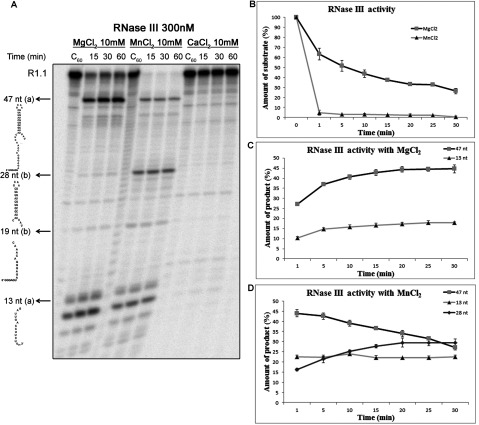
We also tested the activity of Cj-RNase III in the presence of other two divalent ions, Mn2+ (manganese) and Ca2+ (calcium). It was described that Mn2+ is also able to support catalysis, while Ca2+ functions as an inhibitor [29,35]. As expected, Ca2+ at 10 mM does not support Cj-RNase III catalysis, but Mn2+ does (Figure 4A). However, the RNA cleavage pattern is different according to the divalent ion used. With Mg2+, RNase III cleaves R1.1 into two major products (47 and 13 nt) resulting from cleavage in the primary site (a) (Figure 3A), and a fraction of the substrate is not digested (Figures 3 and 4). When higher amount of enzyme is used (1200 nM) and the reaction occurs for longer periods of time (2 h), we can observe that cleavage at the secondary site (b) can also occur (results not shown). However, in the presence of 10 mM of Mn2+ it is possible to observe immediately the formation of the four degradation products (47, 28, 19 and 13 nt), which results from cleavage at both sites (a and b) (Figure 4A).
Figure 4. Cj-RNase III activity with different divalent ions.
(A) 300 nM of recombinant protein were incubated with 10000 cpm of R1.1 substrate at 37°C in a reaction buffer with MgCl2, MnCl2 and CaCl2. This experiment was performed at least in triplicate. (B) Determination of the amount of substrate degraded in the presence of 300 nM of Cj-RNase III with MgCl2 and MnCl2. (C) Determination of the amount of degradation products (47 and 13 nt) in the presence of MgCl2. (D) Determination of the amount of degradation products (47, 28 and 13 nt) in the presence of MnCl2.
We have quantified the amount of substrate present at each time point of the reaction with Mg2+ and Mn2+ at the same concentration and using the same amount of protein. The graphic in Figure 4(B) shows that, after 1 min of reaction, Cj-RNase III degraded>95% of the substrate in the presence of 10 mM of Mn2+, while only ~35% is degraded in the presence of 10 mM of Mg2+. Moreover, in those conditions, the degradation obtained in the presence of Mg2+ did not reach more than 70% during the 30 min of reaction (Figure 4(B)). We also quantified the degradation products of the reaction in both conditions. In the presence of Mg2+, we can observe the formation of the 47 and 13 nt fragments resulting of cleavage in (a) site (Figure 4(C)). In the presence of Mn2+, we see a decreased in the 47 nt fragment and an increased in the 28 nt, which results at cleavage at (b) site, while the 13 nt remains constant (Figure 4D). These results support the conclusion that cleavage at (b) site is only promoted in the presence of Mn2+.
In E. coli, it is known that Mn2+ can support catalysis when used in a concentration range of 0.1–1 mM. For higher concentrations (> 5mM), Mn2+ was shown to have an inhibitory effect [35]. In addition, Ec-RNase III cleaves R1.1 substrate with the same efficiency at the optimal concentrations of Mg2+ and Mn2+ [36]. Thus, we decided to test the activity of Cj-RNase III in a broad range of Mg2+ and Mn2+ concentrations from 0.1 to 100 mM. The results presented in Figure 5 indicate that this protein is active over a wide range of Mg2+ concentrations. However, it seems to prefer values of the order of 10–50 mM (Figure 5A) since we can observe higher consumption of the substrate. Moreover, in the presence of Mn2+ the protein is most active between 1 and 10 mM (Figure 5B). When higher concentrations are used (above 20 mM), the reaction is inhibited. Overall, our experiments show that in contrary to what was reported for E. coli RNase III, the Mn2+ seems to be the preferred co-factor of Cj-RNase III. Also, Mn2+ shows to have an inhibitory effect when increased concentrations are used, however the enzyme supports a higher concentration of this metal ion when compared with E. coli counterpart.
Figure 5. Cj-RNase III activity with different concentrations of divalent ions.
300 nM of recombinant protein were incubated with 10000 cpm of R1.1 substrate at 37°C in a reaction buffer with different MgCl2 (A) or MnCl2 (B) concentrations, ranging from 0.1 to 100 mM. These experiments were performed at least in triplicate.
Our study strengthen the idea that Mn2+ and Mg2+ ions are responsible to control or adapt ribonuclease activity. Very interestingly, a recent study suggests that RNase HII from Pyrococcus furiosus digests dsRNA in the presence of Mn2+ ions, whereas there is no dsRNA digestion activity using Mg2+ in the reaction [37].
The maintenance of metal homoeostasis is essential for proper functioning of the cell, and has significant implications for microbial adaptations in changing environments. Bacteria have evolved several mechanisms to acquire metals and to sustain growth in environments with limited amounts of free trace metals [38]. Manganese is an example of a cofactor that could replace the role of Mg2+ in certain proteins that has emerged as a very important metal in virulence [39]. It is known that macrophages have a poor magnesium, acidic and low oxygen environment [39]. Curiously, divalent manganese is thermodynamically favored in acidic environments and in the absence of oxygen. Campylobacter is able to survive within macrophages for a period of 24–30 h. It is within the macrophages that Mn2+ homoeostasis is thought to be important for C. jejuni; however, the specific cellular role of this metal is not yet established [40]. It raises the question if a possible shift of the metal cofactor could be related with an adaptation of the enzyme activities to new conditions. The results obtained in this work lead us to speculate that Cj-RNase III may have an important role under a Mn2+-rich environment, namely during invasion and infection.
pH dependence and thermostability of Cj-RNase III catalytic activity
C. jejuni faces different pH values during the infection process, and is able to survive during the passage from food or water source into the host gastrointestinal tract. Thus, this pathogen is prepared to sense and respond to a rapid decrease of pH (normally below the threshold for growth) [41]. It is known that the pH of a human stomach depends on physiological variables. Following transit through the stomach, bacteria encounter a variety of environments within the intestine, ranging from mildly acidic (pH 5.5) to moderately alkaline (pH 7.4) [42,43]. Taking this into account, we have tested the activity of Cj-RNase III at different pH values between 5.2 and 7.3, in the presence of 10 mM Mg2+ or Mn2+. As showed in Figure 6, Cj-RNase III is active in all pH tested, and it seems that the protein was efficient in cleaving R1.1 RNA even in the mildly acidic comparing with the standard conditions (Figure 3).
Figure 6. Cj-RNase III activity using different pH values.
300 nM of recombinant protein were incubated with 10000 cpm of R1.1 substrate at 37°C in a reaction buffer with MgCl2 (A) or MnCl2 (B) and different pH, ranging from 5.2 to 7.3. These experiments were performed at least in triplicate.
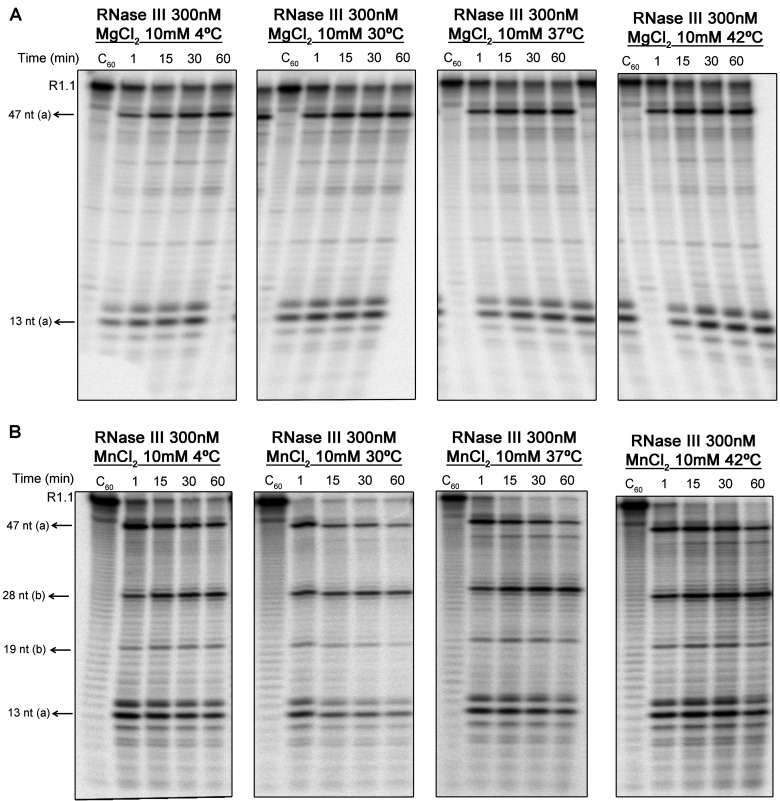
Differences of temperature constitute another key factor during C. jejuni survival. Although this microorganism is able to grow at 37°C, its optimal temperature is about 42°C, the avian body temperature [44]. Moreover, it has the potential to survive under refrigerated temperatures [45]. The ability of C. jejuni to survive refrigeration is of major interest to food safety and public health, since it is a typically intervention used in the control of bacterial growth in food. For this reason, we decided to analyse the activity of Cj-RNase III at different temperatures: 4, 30, 37 and 42°C. The results obtained show that Cj-RNase III is active in an unexpectedly large range of temperatures from 4 to 42°C (Figure 7).
Figure 7. Cj-RNase III activity at different temperatures.
300 nM of recombinant protein were incubated with 10000 cpm of R1.1 substrate at 4, 30, 37 and 42°C in a reaction buffer with MgCl2 (A) or MnCl2 (B). These experiments were performed at least in triplicate.
Altogether, these experiments showed that Cj-RNase III is active in a wide range of conditions. The pH tolerance of Cj-RNase III and its capability to cleave over different temperatures, including 4°C could be indicative of a possible role of RNase III during the acid shock and temperature response of Campylobacter during the infection process.
Analysis of the effect of point mutations in the activity of Cj-RNase III in the presence of Mg2+ and Mn2+
Structural advances combined with directed mutagenesis provide powerful tools to uncover important issues of RNase III catalytic activity. RNase III orthologues have eight highly conserved aspartate (D) and glutamate (E) residues in the catalytic domain (E38, E41, D45, E65, E100, D114, E117 and D126 in E. coli), which were suggested to have an important role in activity. In E. coli the metal binding is coordinated by four of these residues: E41 and D45 that belong to the signature box (NERLEFLGDS), and D114 and E117 [5]. Substitution of D45 for an alanine (Ec-D45A) had a severe impact on the catalytic function of Ec-RNase III both in vitro [46] and in vivo [5]. The replacement of E117 by an alanine or lysine also blocked the phosphodiester hydrolysis, and was also reported to suppress RNase III activity in vivo [29]. Additionally, two other substitutions of E117 residue were performed, in which this amino acid was substituted for an aspartic acid (Ec-E117D) and for a glutamine (Ec-E117Q) [36]. The introduction of these mutations in Ec-RNase III induced minimal structural changes but affected the activity of the enzyme in the presence of Mg2+. Curiously, the presence of Mn2+ in the reaction rescued the activity of the Ec-D45A and Ec-E117D mutant [36,46]. Considering the importance that Mn2+ plays in the activity of Cj-RNase III, we decided to alter the equivalent E117 in C. jejuni protein (Cj-E116) for aspartic acid and glutamine and see the effect of these mutations in the activity of the enzyme. We also substituted the equivalent residue D45 (Cj-D44) for an alanine and checked its effect on catalysis.
As already mentioned, in order to be active, the nuclease domains of RNase III undergoes dimerization. After purification of the mutant proteins, their ability to form dimers was tested by incubation the DSS cross-linker. The mutants exhibited normal homodimeric behavior, comparable with the wt protein (Figure 8A).
Figure 8. Characterization of Cj-RNase III mutants.
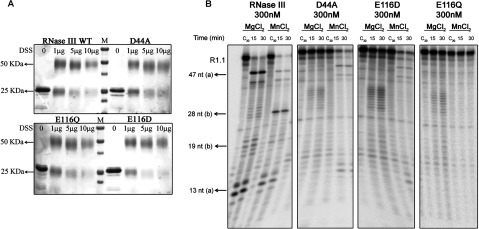
(A) Cross-linking of RNase III mutants from C. jejuni using DSS. Roughly 0.5 μg of protein was incubated with increasing concentrations of DSS as indicated in the figure. Proteins were visualized by Coomassie Brilliant Blue staining. (B) Cj-RNase III mutants activity in the presence of Mg2+ and Mn2+. Roughly 300 nM of protein were incubated with 10000 cpm of R1.1 substrate at 37°C in a reaction buffer with MgCl2 or MnCl2. These experiments were performed at least in triplicate.
We also analysed the capacity of the mutant proteins to support catalysis. For this purpose, we tested the activity of the enzymes in the conditions where the wt Cj-RNase III carries out efficient degradation of the RNA substrate, in the presence of Mg2+ or Mn2+. Using Mg2+ as cofactor, the results reveal that the mutants do not cleave efficiently R1.1 RNA (Figure 8B). However, for the three mutants, it is possible to observe a residual activity. The same was verified for the equivalent mutants in E. coli when extended reaction times and high enzyme concentrations were used [36,46]. These data suggest a conservation of the role of these residues in the catalytic mechanism of action of RNase III.
Regarding the use of Mn2+ in the reactions, we have shown that this metal ion was able to rescue the activity of Cj-D44A and Cj-E116D mutants. As previously discussed, in the presence of Mn2+, the wt Cj-RNase III is able to recognize the secondary site (b) of R1.1 RNA (Figures 3 and 4). Nevertheless, Cj-D44A and Cj-E116D mutants are no longer able to cleave at this site (Figure 8b), even when higher amounts of protein are used (results not shown). When dsRNA is cleaved, two hydrolysis events occur at each active center of RNase III. Moreover, it is known that the residues D44 and E110 in A. aeolicus (D45 and E116 in C. jejuni, respectively) are responsible for a second RNA-cutting site [5]. This is in good agreement with the lost of Cj-D44A and Cj-E116D mutants to recognize and cleave the R1.1 substrate at the secondary site.
Regarding the Cj-E116Q mutant, we demonstrated that Mn2+ was not able to restore its catalytic activity (Figure 8B). Our findings are consistent with the weak affinity of the carboxamide group for divalent metal ions as previously discussed by Sun and Nicholson, which explains the inability of Mg2+ and Mn2+ ions to restore the activity of this mutant [36].
Overall, these results show that D44 and E116 residues are important for the catalytic activity of Cj-RNase III by providing a metal-binding site, which is recognized by Mg2+ and Mn2+. The mutants are not able to cleave in the presence of Mg2+, but a different behaviour is seen in the presence of Mn2+. Moreover, our data strengthen that in fact RNase III family of enzymes contains a conserved mechanism of action.
In this paper, we have undertaken a functional and biochemical analysis of Cj-RNase III. We demonstrated that Cj-RNase III is able to complement an E. coli rnc deficient strain in 30S rRNA processing and in regulation of PNPase levels. These observations reinforce the conservation of these functions in bacteria, since similar results were previously obtained for other microorganisms [33]. We were also able to show that Cj-RNase III is active in a wide range of conditions. Such plasticity may be important for the adaptation of this pathogen to different conditions during infection. The data obtained lead us to speculate that Cj-RNase III may have an important role under a Mn2+-rich environment, contrarily to the consensus that Mg2+ is the physiologic relevant cofactor for RNase III orthologues.
PNPase has an impact on cell biology of C. jejuni, namely in survival at refrigerated conditions, bacterial swimming, colonization, cell adhesion and invasion. Moreover, its levels are controlled in an RNase III-dependent manner. Taking these into account, RNase III might be also a main player in the adaptation of this pathogen to environmental demands. Hence, it is crucial to better understand the mechanism of action of this endoribonuclease.
ACKNOWLEDGMENTS
We want to thank Professor Allen W. Nicholson for providing BL21 (DE3) rnc105 recA strain. We also thank Andreia Aires for technical support in the laboratory.
AUTHOR CONTRIBUTION
Nabila Haddad, Margarida Saramago and Rute G. Matos designed and conducted the experiments; Nabila Haddad, Margarida Saramago, Rute G. Matos, Hervé Prévost and Cecília M. Arraiano analysed the data; Nabila Haddad, Margarida Saramago, Rute G. Matos, Hervé Prévost and Cecília M. Arraiano wrote the paper.
FUNDING
RNA research in UMR1014 SECALIM was funded by the Région Pays de la Loire through the programme grant GENICAMP. N.H. was supported by the GENICAMP programme and a Ph.D. scholarship awarded by the Institut National de la Recherche Agronomique and Région des Pays de la Loire. Work at ITQB was supported by grants from Fundação para a Ciência e Tecnologia (FCT), including grant number PEst-OE/EQB/LA0004/2011 and project number PTDC/QUI-BIQ/111757/2009. M.S. was a recipient of Doctoral fellowship [grant number SFRH/BD/65607/2009] and R.G.M. was a recipient of a Post-Doctoral fellowship [grant number SFRH/BPD/75887/2011], both funded by FCT.
References
- 1.Arraiano C. M., Mauxion F., Viegas S. C., Matos R. G., Seraphin B. Intracellular ribonucleases involved in transcript processing and decay: Precision tools for RNA. Biochim. Biophys. Acta. 2013;1829:491–513. doi: 10.1016/j.bbagrm.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Silva I. J., Saramago M., Dressaire C., Domingues S., Viegas S. C., Arraiano C. M. Importance and key events of prokaryotic RNA decay: the ultimate fate of an RNA molecule. Wiley Interdiscip. Rev. RNA. 2011;2:818–836. doi: 10.1002/wrna.94. [DOI] [PubMed] [Google Scholar]
- 3.Arraiano C. M., Andrade J. M., Domingues S., Guinote I. B., Malecki M., Matos R. G., Moreira R. N., Pobre V., Reis F. P., Saramago M., et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 4.Viegas S. C., Silva I. J., Saramago M., Domingues S., Arraiano C. M. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 2011;39:2918–2930. doi: 10.1093/nar/gkq1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaszczyk J., Tropea J. E., Bubunenko M., Routzahn K. M., Waugh D. S., Court D. L., Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 6.Meng W., Nicholson A. W. Heterodimer-based analysis of subunit and domain contributions to double-stranded RNA processing by Escherichia coli RNase III in vitro. Biochem. J. 2008;410:39–48. doi: 10.1042/BJ20071047. [DOI] [PubMed] [Google Scholar]
- 7.Gan J., Shaw G., Tropea J. E., Waugh D. S., Court D. L., Ji X. A stepwise model for double-stranded RNA processing by ribonuclease III. Mol. Microbiol. 2008;67:143–154. doi: 10.1111/j.1365-2958.2007.06032.x. [DOI] [PubMed] [Google Scholar]
- 8.Blaszczyk J., Gan J., Tropea J. E., Court D. L., Waugh D. S., Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure. 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Dasgupta S., Fernandez L., Kameyama L., Inada T., Nakamura Y., Pappas A., Court D. L. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III – the effect of dsRNA binding on gene expression. Mol. Microbiol. 1998;28:629–640. doi: 10.1046/j.1365-2958.1998.00828.x. [DOI] [PubMed] [Google Scholar]
- 10.Oppenheim A. B., Kornitzer D., Altuvia S., Court D. L. Posttranscriptional control of the lysogenic pathway in bacteriophage lambda. Progr. Nucleic Acid Res. Mol. Biol. 1993;46:37–49. doi: 10.1016/s0079-6603(08)61017-x. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Palacios G. M. The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 2007;44:701–703. doi: 10.1086/509936. [DOI] [PubMed] [Google Scholar]
- 12.Newell D. G., Fearnley C. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 2003;69:4343–4351. doi: 10.1128/AEM.69.8.4343-4351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson C. K., Ethelberg S., Van Pelt W., Tauxe R. V. Epidemiology of Campylobacter jejuni infections in industrialized nations. In: Nachamkin I., Szymanski C. M., Blaser M. J., editors. Campylobacter. Washington, DC: ASM Press; 2008. [Google Scholar]
- 14.Murphy C., Carroll C., Jordan K. N. Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J. Appl. Microbiol. 2006;100:623–632. doi: 10.1111/j.1365-2672.2006.02903.x. [DOI] [PubMed] [Google Scholar]
- 15.Park S. F. Campylobacter jejuni stress responses during survival in the food chain and colonization. In: Ketley J. M., Konkel M. E., editors. Campylobacter Molecular and Cellular Biology. Norfolk, U.K.: Horizon Bioscience; 2005. [Google Scholar]
- 16.Pittman M. S., Elvers K. T., Lee L., Jones M. A., Poole R. K., Park S. F., Kelly D. J. Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 2007;63:575–590. doi: 10.1111/j.1365-2958.2006.05532.x. [DOI] [PubMed] [Google Scholar]
- 17.Haddad N., Burns C. M., Bolla J. M., Prévost H., Fédérighi M., Drider D., Cappelier J. M. Long-term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity. Appl. Environ. Microbiol. 2009;75:7310–7318. doi: 10.1128/AEM.01366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haddad N., Tresse O., Rivoal K., Chevret D., Nonglaton Q., Burns C. M., Prevost H., Cappelier J. M. Polynucleotide phosphorylase has an impact on cell biology of Campylobacter jejuni. Front. Cell Infect. Microbiol. 2012;2:30. doi: 10.3389/fcimb.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarrige A. C., Mathy N., Portier C. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 2001;20:6845–6855. doi: 10.1093/emboj/20.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robert-Le Meur M., Portier C. E. coli polynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 1992;11:2633–2641. doi: 10.1002/j.1460-2075.1992.tb05329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi R. Recombinant PCR. In: Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., editors. PCR Protocols: a Guide to Methods and Applications. San Diego, CA: Academic Press; 1990. pp. 171–183. [Google Scholar]
- 22.Amarasinghe A. K., Calin-Jageman I., Harmouch A., Sun W., Nicholson A. W. Escherichia coli ribonuclease III: affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 2001;342:143–158. doi: 10.1016/s0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- 23.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stead M. B., Agrawal A., Bowden K. E., Nasir R., Mohanty B. K., Meagher R. B., Kushner S. R. RNAsnap: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 2012;40:e156. doi: 10.1093/nar/gks680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Dereeper A., Audic S., Claverie J. M., Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsunaga J., Simons E. L., Simons R. W. RNase III autoregulation: structure and function of rncO, the posttranscriptional ‘operator’. RNA. 1996;2:1228–1240. [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Nicholson A. W. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical footprinting using catalytically inactive RNase III mutants. EMBO J. 1996;15:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 30.Babitzke P., Granger L., Olszewski J., Kushner S. R. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J. Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′ end. EMBO J. 1987;6:2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zilhao R., Cairrao F., Régnier P., Arraiano C. M. PNPase modulates RNase II expression in Escherichia coli: implications for mRNA decay and cell metabolism. Mol. Microbiol. 1996;20:1033–1042. doi: 10.1111/j.1365-2958.1996.tb02544.x. [DOI] [PubMed] [Google Scholar]
- 33.Amblar M., Viegas S. C., Lopez P., Arraiano C. M. Homologous and heterologous expression of RNase III from Lactococcus lactis. Biochem. Biophys. Res. Commun. 2004;323:884–890. doi: 10.1016/j.bbrc.2004.08.167. [DOI] [PubMed] [Google Scholar]
- 34.Gross G., Dunn J. J. Structure of secondary cleavage sites of E. coli RNAase III in A3t RNA from bacteriophage T7. Nucleic Acids Res. 1987;15:431–442. doi: 10.1093/nar/15.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. L., Chelladurai B. S., Zhang K., Nicholson A. W. Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res. 1993;21:1919–1925. doi: 10.1093/nar/21.8.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun W., Nicholson A. W. Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant. Biochemistry. 2001;40:5102–5110. doi: 10.1021/bi010022d. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura S., Fujishima K., Sato A., Tsuchiya D., Tomita M., Kanai A. Characterization of RNase HII substrate recognition using RNase HII-argonaute chimaeric enzymes from Pyrococcus furiosus. Biochem. J. 2010;426:337–344. doi: 10.1042/BJ20091553. [DOI] [PubMed] [Google Scholar]
- 38.Kovacs-Simon A., Titball R. W., Michell S. L. Review Lipoproteins of bacterial pathogens. Infect. Immun. 2011;79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papp-Wallace K. M., Maguire M. E. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 40.Pogacar M. S., Mihaljevic R. R., Klancnik A., Brumuini G., Abram M., Mozina S. S. Survival of stress exposed Campylobacter jejuni in the murine macrophage J774 cell line. Int. J. Food Microbiol. 2009;129:68–73. doi: 10.1016/j.ijfoodmicro.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Reid A. N., Pandey R., Palyada K., Naikare H., Stintzi A. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl. Environ. Microbiol. 2008;74:1583–1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holzer P. Taste receptors in the gastrointestinal tract. V. Acid sensing in the gastrointestinal tract. American J. Physiol. Gastrointest. Liver Physiol. 2007;292:G699–G705. doi: 10.1152/ajpgi.00517.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao K. A., Yazaki E., Evans D. F., Carbon R. Objective evaluation of small bowel and colonic transit time using pH telemetry in athletes with gastrointestinal symptoms. Br. J. Sports Med. 2004;38:482–487. doi: 10.1136/bjsm.2003.006825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee A., Smith S. C., Coloe P. J. Survival and growth of Campylobacter jejuni after artificial inoculation onto chicken skin as a function of temperature and packaging conditions. J. Food Protect. 1998;61:1609–1614. doi: 10.4315/0362-028x-61.12.1609. [DOI] [PubMed] [Google Scholar]
- 45.Chan K. F., Tran H. L., Kanenaka R. Y., Kathariou S. Survival of clinical and poultry-derived isolates of Campylobacter jejuni at a low temperature (4°C) Appl. Environ. Microbiol. 2001;67:4186–4191. doi: 10.1128/AEM.67.9.4186-4191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun W., Li G., Nicholson A. W. Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry. 2004;43:13054–13062. doi: 10.1021/bi049258i. [DOI] [PubMed] [Google Scholar]
- 47.Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol. Rev. 1980;44:1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C., Robinson R. A. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]