Abstract
A hallmark of MYC-transformed cells is their aberrant response to antimitogenic signals. Key examples include the inability of MYC-transformed cells to arrest proliferation in response to antimitogenic signals such as TGF-β or DNA damage and their inability to differentiate into adipocytes in response to hormonal stimuli. Given the plethora of antimitogenic signals to which a tumor cell is exposed, it is likely that the ability to confer resistance to these signals is central to the transforming properties of MYC in vivo. At the same time, the inability of MYC-transformed cells to halt cell-cycle progression on stress may establish a dependence on mutations that impair or disable apoptosis. We propose that the interaction of MYC with the zinc finger protein MIZ-1 mediates resistance to antimitogenic signals. In contrast to other interactions of MYC, there is currently little evidence that MIZ-1 associates with MYC in normal, unperturbed cells. The functional interaction of both proteins becomes apparent at oncogenic expression levels of MYC and association with MIZ-1 mediates both oncogenic functions of MYC as well as tumor-suppressive responses to oncogenic levels of MYC.
MYC-transformed cells do not stop dividing in response to antimitogenic signals (e.g., DNA damage). This resistance to antimitogenic signals may involve interactions between MYC and the zinc finger protein MIZ-1.
INTERACTION OF MYC AND MIZ-1
MIZ-1 (Zbtb17) was identified in a two-hybrid screen as a protein interacting with the carboxy-terminus of c-MYC (Peukert et al. 1997). Initial work established that MIZ-1 interacted with both N-MYC and L-MYC, albeit more weakly than with c-MYC. Sequence analysis shows that MIZ-1 is a BTB/POZ-domain zinc finger protein with 13 zinc fingers. MYC interacts with a short helical domain in MIZ-1 that is part of a 50 amino acid stretch interspersed between zinc fingers 12 and 13 (Fig. 1B) (Peukert et al. 1997). BTB/POZ domains are homo- or hetero-oligomerization domains and the POZ domain of MIZ-1 crystallizes as a tetramer (Stead et al. 2007). Deletion of the POZ domain renders the MIZ-1 protein nonfunctional and the truncated protein does not associate with chromatin (Kosan et al. 2010). This finding forms the basis of a mouse model, in which the exons encoding the POZ domain are flanked by loxP-sites and can be deleted by Cre-mediated recombination (Gebhardt et al. 2007). In vivo, MIZ-1 not only forms homo-oligomers, but also forms heterodimers with Bcl-6, another POZ-domain oncoprotein (Phan et al. 2005). The POZ domain is also required for other protein/protein interactions of MIZ-1, but the nature of those interactions is less well understood (Adhikary et al. 2005; Herold et al. 2008).
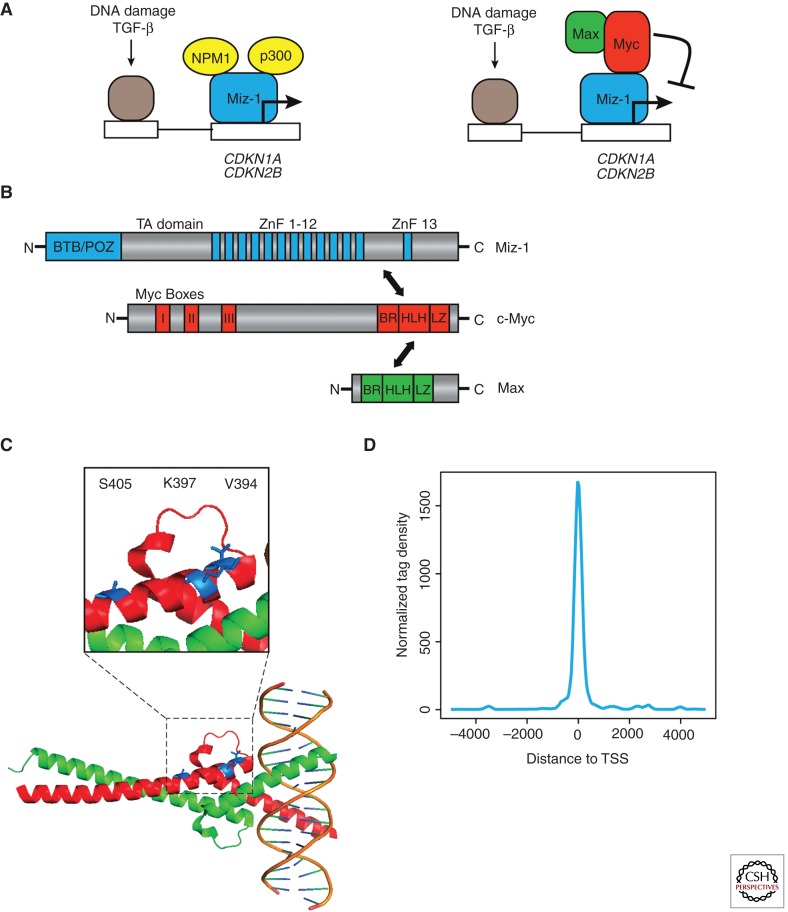
Figure 1.
Interaction of MYC with MIZ-1. (A) Gene regulation by MYC and MIZ-1. The model proposes that MIZ-1 activates target genes in cooperation with upstream transcription factors that are regulated by specific antimitogenic signals. Association of MYC with MIZ-1 displaces the coactivators, p300 and NPM-1, thereby blocking transcription of MIZ-1-dependent genes. (B) Domains of MIZ-1, MYC, and MAX. The main interaction between MYC and MIZ-1 takes place via the helix-loop-helix domain of MYC and a domain of MIZ-1 that is located between zinc fingers 12 and 13. MAX is a subunit of the trimeric MIZ-1/MYC/MAX complex but does not interact directly with MIZ-1. (C) Structural view of the MIZ-1/MYC interface. The panel shows the carboxy-terminal DNA-binding and dimerization domain of MYC (red) and MAX (green). Indicated in blue are residues of MYC that—when mutated—disrupt or weaken the interaction with MIZ-1. (D) MIZ-1-binding sites are located in core promoters. Shown is a diagram depicting the localization of MIZ-1-binding sites relative to the transcription start sites as deduced from ChIP-sequencing experiments in MDA-MB231 cells.
Mutants in MYC that weaken or disrupt the interaction with MIZ-1 map to the “outside” of the helix-loop-helix (HLH) domain, suggesting that MIZ-1 and MYC can bind adjacent to each other on DNA (Herold et al. 2002). In particular, replacing valine 394 (V394) in MYC with an aspartate residue disrupts the interaction of MYC with MIZ-1. Because this residue points away from MAX and from DNA, the mutation enables the identification of MIZ-1-dependent functions of MYC (Fig. 1C) (Herold et al. 2002). Both transgenic and knockin models for MycV394D have been generated to probe the relevance of the interaction (van Riggelen et al. 2010) (see below). Valine 394 is not conserved in other HLH proteins, arguing that interaction with MIZ-1 is specific for MYC proteins. V394 is conserved in vertebrate MYC proteins, but not, for example, in Drosophila MYC. Correspondingly, homologs of MIZ-1 proteins are found in vertebrate genomes, but not in Drosophila, arguing that the interaction of MYC and MIZ-1 has evolved much later than the core network of MYC, MAX and Mxd proteins (see Conacci-Sorrell et al. 2014).
The consensus DNA sequence to which MIZ-1 binds has not yet been identified. Structural modeling suggests that the 13 zinc fingers of MIZ-1 may wrap around a nucleosome, but this remains to be tested. ChIP-sequencing data identify MIZ-1 as a protein that binds exclusively to core promoters; an example of the results is shown in Figure 1D. Many POZ-domain proteins are transcriptional repressors that localize as insoluble complexes in specific subnuclear compartments. In contrast, free MIZ-1 is a strong transactivator that uses a typical acidic domain for activation of its target genes, using both p300 and Npm1 (nucleophosmin) as coactivators (Staller et al. 2001; Wanzel et al. 2008). Most likely, therefore, the physiological function of MIZ-1 during normal development is to activate a set of target genes; unpublished work from our laboratory shows that direct target genes of MIZ-1 include many genes involved in autophagy and membrane transport.
Transcriptional Repression by MYC via Interaction with MIZ-1
Association with MYC converts MIZ-1 from being a transcriptional activator to a repressor (Staller et al. 2001). Whereas MIZ-1-binding sites are localized in core promoters, their localization relative to the start site of transcription does not support a model of direct steric competition with recruitment of RNA polymerase as a mechanism of repression. Binding of MYC to MIZ-1 induces a change in the biophysical properties of the MIZ-1 protein, renders the complex insoluble and causes it to relocalize within the nucleus (Peukert et al. 1997). Binding of MYC to MIZ-1 displaces both coactivators, p300 and Npm1, from MIZ-1 (Staller et al. 2001; Wanzel et al. 2008). The MYC/MIZ-1 complex in turn may recruit DNA methylases to repress transcription (Brenner et al. 2005; Licchesi et al. 2010). There are several reports that MYC recruits histone deacetylases and polycomb proteins to repress transcription; whether this occurs as part of a MYC/MIZ-1 complex is an open question (Zhang et al. 2012; Corvetta et al. 2013). Association with MIZ-1 also stabilizes MYC and inhibits its proteasomal turnover (Salghetti et al. 1999). This may be either caused by the sequestration described above or simply because of steric masking of carboxy-terminal ubiquitination sites in MYC. Notably, ubiquitination of MYC and proteasomal turnover are critical for transcriptional activation by MYC (Kim et al. 2003; von der Lehr et al. 2003; Adhikary et al. 2005; Farrell and Sears 2014). It is possible, therefore, that inhibition of ubiquitination of MYC by MIZ-1 is critical for blocking its transactivation function.
Target Genes of MYC and MIZ-1
MIZ-1 associates with MYC on a number of target promoters. Best understood are the promoters of two cell-cycle inhibitors, p15Ink4b (CDKN2B) and p21cip1 (CDKN1A) (Seoane et al. 2002). In both cases, the MYC- and MIZ-1-binding sites overlap the transcriptional start site. Binding of the MYC/MIZ-1 complex has little effect on the basal expression of either gene, but blocks their induction by antimitogenic stimuli (Warner et al. 1999; Gebhardt et al. 2006). Association with MIZ-1 is critical for repression of p15Ink4b by MYC in response to TGF-β both in a tissue culture model and in vivo (Gebhardt et al. 2006; van Riggelen et al. 2010). Similarly, MYC inhibits expression of p21Cip1 in response to DNA damage and to inducers of differentiation in culture via MIZ-1 (Seoane et al. 2002; Wu et al. 2003). In vivo, MIZ-1 is critical for suppression of CDKN1A in response to the tumor promoter TPA, during skin carcinogenesis (see below), and for repression of CDKN1A in response to endogenous DNA damage during recombination of T-cell receptors. Similarly, the CDKN1C (p57Kip2) gene is a direct target of MIZ-1 and MYC represses CDKN1C in a MIZ-1-dependent manner during lymphomagenesis (Adhikary et al. 2003; van Riggelen et al. 2010).
Notably, other transcription factors use MIZ-1 as a platform to inhibit expression of cell-cycle inhibitors. For example, the Bcl-6 oncoprotein heterodimerizes with MIZ-1 and suppresses p21Cip1 expression (Phan et al. 2005). This is thought to antagonize the p53-dependent up-regulation of cdkn1a that occurs as a result of DNA damage generated during class switching of immunoglobulin genes in germinal centers. A second example is the Gfi-1 protein, that forms a heterotrimeric complex with MYC and MIZ-1 to repress cdkn2b and cdkn1a, possibly also cdkn1b (p27Kip1), expression during lymphomagenesis (Basu et al. 2009).
A second group of genes targeted by MYC/MIZ-1 complexes are cell-adhesion molecules, most notably integrins (Gebhardt et al. 2006). Repression of integrin expression by MYC has been linked to the exit of hematopoietic and skin stem cells from their niches (Waikel et al. 2001; Wilson et al. 2004), the idea being that the transition from a relatively quiescent cell to a rapidly proliferating transient amplifying cell involves severing of interactions with the stem cell niche. Repression of integrin expression by MYC and MIZ-1 is strongly enhanced by the Arf tumor suppressor protein, suggesting that Arf may antagonize self-renewal of cells that express deregulated MYC (Herkert et al. 2010). Repression of integrin expression by MYC in breast cancer cells has been linked to suppression of cell migration and metastasis (Liu et al. 2012a). Somewhat at odds with this report and the general view of MYC and MIZ-1 as a repressive complex, a recent report suggests that MYC and MIZ-1 cooperate with Skp2 to activate transcription of the RhoA gene and thereby promote metastasis (Chan et al. 2010). We have been unable to reproduce these data and the RhoA promoter is not significantly occupied by MIZ-1 in any cell we analyzed, including breast cancer cells (B von Eyss and M Eilers, unpubl.).
Third, MIZ-1 up-regulates expression of the antiapoptotic BCL-2 gene in several settings; for example, MIZ-1-deficient T lymphocytes fail to up-regulate Bcl-2 in response to stimulation by IL-7, a critical antiapoptotic signaling factor in early lymphocyte development (Saba et al. 2011). Deregulated expression of MYC can repress BCL-2 expression via interaction with MIZ-1 in tissue culture and the ability to bind to MIZ-1 is required for MYC to induce apoptosis in some cell types (Patel and McMahon 2006, 2007). Whether repression of BCL-2 via MIZ-1 is an important proapoptotic mechanism for MYC in vivo remains to be established. Notably, the Bcl-6 oncoprotein can repress transcription of BCL-2 via Miz-1 and large B-cell lymphomas (DLBCL) that have activated Bcl-6 escape repression of Bcl-2 by Bcl-6/MIZ-1 complexes via mutations in the BCL-2 promoter (Saito et al. 2009).
MOUSE MODELS
The biochemical analyses described above suggest that MIZ-1 can act both as an activator and as a repressor (because it forms repressive complexes with MYC and Bcl-6) and that levels of MYC may be a critical determinant that distinguishes the two forms of gene regulation. The mouse models described below provide in vivo evidence for this model. A summary of the results is shown in Table 1.
Table 1.
Mouse models to interrogate the physiological roles of MIZ-1
| Model | Tissue | Phenotype | Molecular analysis |
|---|---|---|---|
| Normal development | |||
| MizΔ (constitutive deletion of MIZ-1) | – | Early embryonic lethality and apoptosis of ectodermal cells during gastrulation | Deregulation of Cdkn1c expression |
| Miz-1ΔPOZ (fl/fl) (inducible deletion of the POZ domain of Miz-1) | T lymphocytes | Cell-cycle arrest and apoptosis during early T-cell development | Up-regulation of Cdkn1a and of proapoptotic target genes of p53 |
| B lymphocytes | Apoptosis of early B-cell progenitors | Reduced expression of Bcl-2 and inhibition of IL-7-dependent signal transduction | |
| Keratinocytes | Disruption of hair growth cycle and premature senescence | Reduced expression of Cdkn2b, Itga6, Itgb4 et al. | |
| Myc V394D knockin | – | Reduction in body weight | – |
| Tumor models | |||
| Miz-1ΔPOZ (fl/fl) | TPA/DMBA-induced skin carcinogenesis | Delay in tumorigenesis | Derepression of Cdkn1a Phenotype rescued by deletion of Cdkn1a |
| MycV394D transgene | T lymphomagenesis | Delay in lymphoma formation and TGF-β-dependent senescence | Derepression of cdkn2b and Cdkn1c expression |
| MycV394D knockin | APC-driven colon carcinogenesis | Delay in tumorigenesis | Not analyzed |
Normal Development
The murine miz-1 gene is essential during early development; homozygous deletion of miz-1 causes massive apoptosis of ectodermal cells at E7.5 during gastrulation (Adhikary et al. 2003). The phenotype of miz-1ΔPOZ mice is less severe, but mice still die late in embryogenesis (Kosan et al. 2010). Mice that carry a conditional deletion of the MIZ-1/POZ domain have been analyzed in several tissues. The strongest effects occur in B and T lymphocytes. The effects of MIZ-1 deletion in hematopoietic cells have been described in detail in a recent review (Moroy et al. 2011). Comparison with mice that carry a conditional deletion in c-myc or with mycv394d knockin mice shows that these effects are unlikely to be because of disruption of MYC/MIZ-1 complexes. The identity of partner proteins that mediate MIZ-1-dependent repressive effects in hematopoietic cells has not been firmly established. As mentioned above, candidates include the Bcl-6 and Gfi-1 transcriptional repressor proteins, but the in vivo function of both Bcl-6/MIZ-1 and Gfi-1/MIZ-1 complexes has not been determined. Currently, no effects of MIZ-1 deletion during unperturbed development can be ascribed to an interaction with MYC.
Tumor Models
The data described above suggest that association with MIZ-1 may be critical for MYC-dependent proliferation and escape from senescence (via suppression of cell-cycle inhibitors), apoptosis (via suppression of Bcl-2), migration and metastasis as well as exit from the stem cell compartment (via suppression of integrin expression). The data raise the possibility that MYC-dependent repression via MIZ-1 is relevant at multiple stages of MYC-dependent tumor development. Data from several mouse tumor models begin to provide experimental support for this notion.
Lymphomagenesis
Deregulated expression of MYC driven by an immunoglobulin heavy chain promoter causes lymphomagenesis and is used as a model for the development of human Burkitt’s lymphoma. Using an MYCV394D allele, instead of wild-type MYC, in a transgenic model strongly delays lymphomagenesis but does not abolish it (van Riggelen et al. 2010). Surprisingly, prelymphomagenic states in both mice are virtually identical and the difference between both genotypes becomes apparent only during lymphoma formation. Lymphomas expressing V394D MYC show a moderately decreased percentage of apoptotic cells, but a strongly enhanced percentage of senescent cells characterized by trimethylation of H3K9. Molecularly, they are characterized by enhanced expression of cdkn2b and cdkn1c mRNAs. Expression of both cell-cycle inhibitors is driven by high levels of TGF-β, which is synthesized by lymphoma cells themselves (but not by normal lymphocytes) and by invading tumor macrophages (Reimann et al. 2010). Blockade of TGF-β signaling abolishes MIZ-1-dependent senescence (van Riggelen et al. 2010). Together, the data show that the MYC/MIZ-1 complex has a specific role in antagonizing TGF-β-induced senescence during lymphomagenesis.
Skin Papilloma
Two very distinct descriptions of mouse phenotypes lacking c-myc in skin have been reported. The more severe phenotype reports a complete loss of skin architecture (Zanet et al. 2005). A second report argues that deletion of c-MYC has mild phenotypes during normal development, but that MYC is critical for suppression of cdkn1a during papilloma formation (Oskarsson et al. 2006). Similarly, deletion of the MIZ-1/POZ domain has a moderate effect on normal homeostasis, but MIZ-1 is required to suppress cdkn1a during the rapid proliferation of keratinocytes induced by the tumor promoter TPA, in a skin carcinogenesis model (Gebhardt et al. 2007; Honnemann et al. 2012). In the absence of functional MIZ-1, keratinocytes do not expand and they undergo rapid differentiation in response to TPA. Importantly, the phenotypes of miz-1ΔPOZ mice are fully rescued by codeletion of cdkn1a, but not by deletion of cdkn2b, arguing that MIZ-1 has a critical function as a repressor of cdkn1a during skin carcinogenesis (Honnemann et al. 2012). Recently, deletion of the MYC-associated HectH9/Huwe1 ubiquitin ligase has been shown to enhance transcriptional repression of cdkn2b and cdkn1a by MYC/MIZ-1 and promote papilloma formation, further supporting the view that repression by MYC/MIZ-1 is critical in this tumor model (Inoue et al. 2013).
Colon Carcinoma
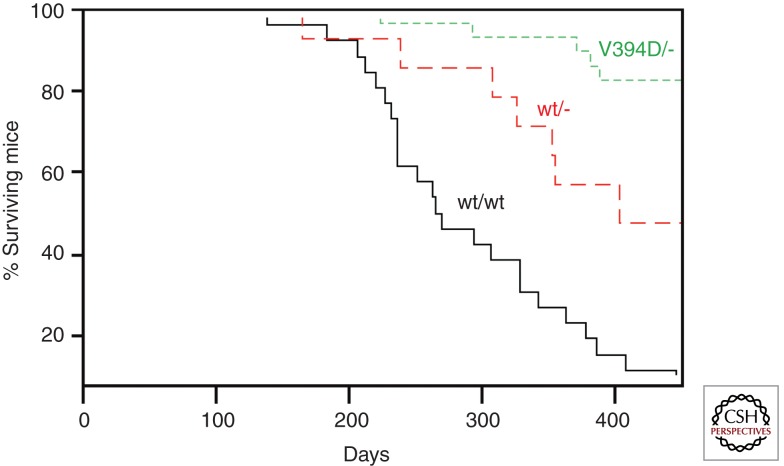
Deregulated expression of Myc as a consequence of mutations in the APC gene (as well as other mutations in the Wnt pathway) has been unequivocally established as a key driver of colon carcinogenesis (Sansom et al. 2007). V394D knockin mice develop normally and are fertile (M Eilers, unpubl.). This has allowed a test of whether the interaction of an endogenous Myc protein with Miz-1 is important for colorectal carcinogenesis. No difference in tumor formation was observed in homozygous wild-type or V394D knockin mice. Because MycV394D is reduced but not abolished in its affinity to Miz-1, one possible explanation for this observation is that that the very high levels of Myc found in APC-deficient tumors provide sufficient Myc/Miz-1-dependent repression even in V394D knockin mice. Consistent with this interpretation, mice that are hemizygous for mycv394d show a much-retarded rate of colorectal carcinogenesis relative to mice that carry one copy of wild-type myc (Fig. 2). The data argue that the interaction of endogenous deregulated Myc with Miz-1 is required for intestinal tumorigenesis.
Figure 2.
Role of interaction of endogenous MYC with MIZ-1 for APC-driven colorectal carcinogenesis. The panel shows Kaplan-Meier survival curves of animals conditionally deleted for APC at the start of the experiment. Survival curves are shown for animals expressing either two alleles of wild-type (wt) MYC or one allele of wtMYC or MYCV394D, respectively. Tumorigenesis in animals with two alleles of MYCV394D was not significantly different from wild-type animals.
Human Tumors
The human MIZ-1 gene is localized at 1p36.13. Point mutations in MIZ-1 in human tumors are rare and no evidence exists that they alter the function of the protein. In contrast, deletions of variable size and location within 1p36 are frequent in multiple human tumors, leading to the suggestion that the region contains several tumor suppressor genes (Bagchi and Mills 2008). One example is neuroblastoma and MIZ-1 has tumor-suppressor properties in this neoplasm (Ikegaki et al. 2007). Mutations and SNPs close to the MIZ-1 gene have been associated with blindness and congenital heart failure.
SIGNALING VIA MYC AND MIZ-1
The signals that control MIZ-1 expression and function and the assembly of MYC/MIZ-1 complexes remain poorly defined. MIZ-1 is phosphorylated by Akt in the DNA-binding domain at S428, leading to association with a 14-3-3 protein and inhibition of DNA binding (Wanzel et al. 2005). Potentially most revealing is the finding that MIZ-1 is activated when a ribosomal protein, Rpl23, is depleted in cells; this induces a stress response during which a coactivator of MIZ-1, nucleophosmin (Npm1), is released from the nucleolus, in which it normally resides, into the nucleus (Wanzel et al. 2008). Npm1 is a multifunctional protein that has, among others, a rate-limiting function in ribosome biogenesis; expression levels of Npm1 can be rate limiting for cell growth (Colombo et al. 2011). Npm1 interacts with the Arf tumor suppressor protein and Arf and MIZ-1 antagonize each other’s function at multiple levels (Itahana et al. 2003). Consequently, several proteins, including both MYC and MIZ-1, interact with both Npm1 and Arf (Qi et al. 2004; Li et al. 2008; Herkert et al. 2010). Arf promotes the assembly of MYC/MIZ-1 complexes, enhances repression by MYC/MIZ-1 complexes, and antagonizes transcriptional activation by MYC (Boone et al. 2011). The data suggest, in line with the observations from the mouse models, that assembly of MYC/MIZ-1 complexes has features of a stress response that occurs predominantly in response to supraphysiological levels of MYC, which induce expression of Arf (Zindy et al. 1998).
Transcription-independent and cytosolic functions of MIZ-1 have been reported that are linked to association of MIZ-1 with microtubules and to its ability to inhibit the ubiquitin ligase Huwe1/HectH9; Huwe1 in turn has been implicated in regulating Jnk activation by the TNF receptor (Ziegelbauer et al. 2001; Liu et al. 2009, 2012b). Currently unknown is whether these functions are regulated by association with MYC and, if so, whether either activation of TNF or association with microtubules is an upstream regulator of MIZ-1-dependent transcription.
CONCLUDING REMARKS
We believe that there are three central open questions:
Currently available data are compatible with two mechanistic models. The original model (Fig. 1A) suggested that MAX/MYC/MIZ-1 complexes assemble on a specific subset of MYC target genes, whereas other genes bind MYC/MAX complexes, but not MIZ-1. This model originated from the idea that there are separate classes of E-Box and core-promoter-binding sites for MYC. However, the finding that the majority of MYC/MAX-binding sites, including E-boxes, are localized at core promoters suggests a more provocative model in which MYC/MIZ-1 complexes constitute a fraction of MYC complexes on all MYC-bound core promoters. If this model were correct, it would have significant implications for models that view MYC as a general amplifier of transcription, because it suggests that both activating and repressive complexes of MYC coexist at core promoters. Therefore, identification of the full spectrum of MYC/MIZ-1 target genes by ChIP sequencing will be necessary to clarify whether the association with MIZ-1 affects general models of MYC function.
Most currently known functions of the MYC/MIZ-1 complex become apparent during tumorigenesis, suggesting that the tumor-promoting role of the MYC/MIZ-1 complex may be a gain-of-function of supraphysiological levels of MYC. However, the interface of the MYC protein that binds MIZ-1 appears to have been positively selected for during evolution, raising the question of whether there are physiological conditions under which the complex assembles. One possibility is that suppression of antiproliferative signals is critical during physiological conditions of rapid cell growth (e.g., during tissue regeneration after injury). Alternatively, the promotion or facilitation of MYC-induced apoptosis by the MYC/MIZ-1 complex balances the ability to counteract senescence and therefore the sum of tumor-suppressive functions of the complex outweighs its oncogenic functions.
Finally, the question arises as to how far human tumors depend on MIZ-1-mediated repression by MYC and whether targeting the complex will have therapeutic benefit. Recent data suggest that the OmoMyc-dominant-negative allele of MYC differentially affects MYC/MAX and MYC/MIZ-1 complexes (Soucek et al. 2008; Savino et al. 2011). If these can be extended to human tumors, proof-of-principle experiments will be possible that allow us to decipher the contribution of MYC/MIZ-1 complexes to MYC-dependent transformation of human tumors.
ACKNOWLEDGMENTS
Work in M.E.’s laboratory on MIZ-1 is supported by grants from the Deutsche Forschungsgemeinschaft, the Thyssen Foundation, and the Deutsche Krebshilfe (German Cancer Aid).
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Adhikary S, Peukert K, Karsunky H, Beuger V, Lutz W, Elsasser HP, Moroy T, Eilers M 2003. Miz1 is required for early embryonic development during gastrulation. Mol Cell Biol 23: 7648–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. 2005. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123: 409–421 [DOI] [PubMed] [Google Scholar]
- Bagchi A, Mills AA 2008. The quest for the 1p36 tumor suppressor. Cancer Res 68: 2551–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Liu Q, Qiu Y, Dong F 2009. Gfi-1 represses CDKN2B encoding p15INK4B through interaction with Miz1. Proc Natl Acad Sci 106: 1433–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DN, Qi Y, Li Z, Hann SR 2011. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci 108: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, et al. 2005. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J 24: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, Wu J, Nakayama KI, Kang HY, Huang HY, et al. 2010. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol 12: 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Alcalay M, Pelicci PG 2011. Nucleophosmin and its complex network: A possible therapeutic target in hematological diseases. Oncogene 30: 2595–2609 [DOI] [PubMed] [Google Scholar]
- *.Conacci-Sorrell M, McFerrin L, Eisenman RN 2014. An overview of MYC and its interactome. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvetta D, Chayka O, Gherardi S, D’Acunto CW, Cantilena S, Valli E, Piotrowska I, Perini G, Sala A 2013. Physical interaction between MYCN oncogene and polycomb repressive complex 2 (PRC2) in neuroblastoma: Functional and therapeutic implications. J Biol Chem 288: 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Farrell AS, Sears RC 2014. MYC degradation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Frye M, Herold S, Benitah SA, Braun K, Samans B, Watt FM, Elsasser HP, Eilers M 2006. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J Cell Biol 172: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt A, Kosan C, Herkert B, Moroy T, Lutz W, Eilers M, Elsasser HP 2007. Miz1 is required for hair follicle structure and hair morphogenesis. J Cell Sci 120: 2586–2593 [DOI] [PubMed] [Google Scholar]
- Herkert B, Dwertmann A, Herold S, Abed M, Naud JF, Finkernagel F, Harms GS, Orian A, Wanzel M, Eilers M 2010. The Arf tumor suppressor protein inhibits Miz1 to suppress cell adhesion and induce apoptosis. J Cell Biol 188: 905–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell 10: 509–521 [DOI] [PubMed] [Google Scholar]
- Herold S, Hock A, Herkert B, Berns K, Mullenders J, Beijersbergen R, Bernards R, Eilers M 2008. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO Jl 27: 2851–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnemann J, Sanz-Moreno A, Wolf E, Eilers M, Elsasser HP 2012. Miz1 is a critical repressor of cdkn1a during skin tumorigenesis. PLoS ONE 7: e34885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegaki N, Gotoh T, Kung B, Riceberg JS, Kim DY, Zhao H, Rappaport EF, Hicks SL, Seeger RC, Tang XX 2007. De novo identification of MIZ-1 (ZBTB17) encoding a MYC-interacting zinc-finger protein as a new favorable neuroblastoma gene. Clin Cancer Res 13: 6001–6009 [DOI] [PubMed] [Google Scholar]
- Inoue S, Hao Z, Elia AJ, Cescon D, Zhou L, Silvester J, Snow B, Harris IS, Sasaki M, Li WY, et al. 2013. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev 27: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell 12: 1151–1164 [DOI] [PubMed] [Google Scholar]
- Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP 2003. Skp2 regulates myc protein stability and activity. Mol Cell 11: 1177–1188 [DOI] [PubMed] [Google Scholar]
- Kosan C, Saba I, Godmann M, Herold S, Herkert B, Eilers M, Moroy T 2010. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity 33: 917–928 [DOI] [PubMed] [Google Scholar]
- Li Z, Boone D, Hann SR 2008. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci 105: 18794–18799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licchesi JD, Van Neste L, Tiwari VK, Cope L, Lin X, Baylin SB, Herman JG 2010. Transcriptional regulation of Wnt inhibitory factor-1 by Miz-1/c-Myc. Oncogene 29: 5923–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhao Y, Eilers M, Lin A 2009. Miz1 is a signal- and pathway-specific modulator or regulator (SMOR) that suppresses TNF-α-induced JNK1 activation. Proc Natl Acad Sci 106: 18279–18284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Radisky DC, Yang D, Xu R, Radisky ES, Bissell MJ, Bishop JM 2012a. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol 14: 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yan J, Jiang S, Wen J, Chen L, Zhao Y, Lin A 2012b. Site-specific ubiquitination is required for relieving the transcription factor Miz1-mediated suppression on TNF-α-induced JNK activation and inflammation. Proc Natl Acad Sci 109: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroy T, Saba I, Kosan C 2011. The role of the transcription factor Miz-1 in lymphocyte development and lymphomagenesis-binding Myc makes the difference. Seminars Immunol 23: 379–387 [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Essers MA, Dubois N, Offner S, Dubey C, Roger C, Metzger D, Chambon P, Hummler E, Beard P, et al. 2006. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev 20: 2024–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH, McMahon SB 2006. Targeting of Miz-1 is essential for Myc-mediated apoptosis. J Biol Chem 281: 3283–3289 [DOI] [PubMed] [Google Scholar]
- Patel JH, McMahon SB 2007. BCL2 is a downstream effector of MIZ-1 essential for blocking c-MYC-induced apoptosis. J Biol Chem 282: 5–13 [DOI] [PubMed] [Google Scholar]
- Peukert K, Staller P, Schneider A, Carmichael G, Hanel F, Eilers M 1997. An alternative pathway for gene regulation by Myc. EMBO J 16: 5672–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol 6: 1054–1060 [DOI] [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431: 712–717 [DOI] [PubMed] [Google Scholar]
- Reimann M, Lee S, Loddenkemper C, Dörr JR, Tabor V, Aichele P, Stein H, Dörken B, Jenuwein T, Schmitt CA 2010. Tumor stroma-derived TGF-β limits Myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17: 262–272 [DOI] [PubMed] [Google Scholar]
- Saba I, Kosan C, Vassen L, Moroy T 2011. IL-7R-dependent survival and differentiation of early T-lineage progenitors is regulated by the BTB/POZ domain transcription factor Miz-1. Blood 117: 3370–3381 [DOI] [PubMed] [Google Scholar]
- Saito M, Novak U, Piovan E, Basso K, Sumazin P, Schneider C, Crespo M, Shen Q, Bhagat G, Califano A, et al. 2009. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci 106: 11294–11299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti SE, Kim SY, Tansey WP 1999. Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J 18: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Savino M, Annibali D, Carucci N, Favuzzi E, Cole MD, Evan GI, Soucek L, Nasi S 2011. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS ONE 6: e22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J 2002. Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419: 729–734 [DOI] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, et al. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol 3: 392–399 [DOI] [PubMed] [Google Scholar]
- Stead MA, Trinh CH, Garnett JA, Carr SB, Baron AJ, Edwards TA, Wright SC 2007. A β-sheet interaction interface directs the tetramerisation of the Miz-1 POZ domain. J Mol Biol 373: 820–826 [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Muller J, Otto T, Beuger V, Yetil A, Choi PS, Kosan C, Moroy T, Felsher DW, Eilers M 2010. The interaction between Myc and Miz1 is required to antagonize TGF-β-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev 24: 1281–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, et al. 2003. The F-Box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 11: 1189–1200 [DOI] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet 28: 165–168 [DOI] [PubMed] [Google Scholar]
- Wanzel M, Kleine-Kohlbrecher D, Herold S, Hock A, Berns K, Park J, Hemmings B, Eilers M 2005. Akt and 14-3-3η regulate Miz1 to control cell-cycle arrest after DNA damage. Nat Cell Biol 7: 30–41 [DOI] [PubMed] [Google Scholar]
- Wanzel M, Russ AC, Kleine-Kohlbrecher D, Colombo E, Pelicci PG, Eilers M 2008. A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nat Cell Biol 10: 1051–1061 [DOI] [PubMed] [Google Scholar]
- Warner BJ, Blain SW, Seoane J, Massague J 1999. Myc downregulation by transforming growth factor β required for activation of the p15Ink4b G1 arrest pathway. Mol Cell Biol 19: 5913–5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev 18: 2747–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG 2003. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22: 351–360 [DOI] [PubMed] [Google Scholar]
- Zanet J, Pibre S, Jacquet C, Ramirez A, de Alboran IM, Gandarillas A 2005. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J Cell Sci 118: 1693–1704 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao X, Fiskus W, Lin J, Lwin T, Rao R, Zhang Y, Chan JC, Fu K, Marquez VE, et al. 2012. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell 22: 506–523 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ziegelbauer J, Shan B, Yager D, Larabell C, Hoffmann B, Tjian R 2001. Transcription factor MIZ-1 is regulated via microtubule association. Mol Cell 8: 339–349 [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]