Abstract
Most pathogens are able to infect multiple hosts but some are highly adapted to a single-host species. A detailed understanding of the basis of host specificity can provide important insights into molecular pathogenesis, the evolution of pathogenic microbes, and the potential for pathogens to cross the species barrier to infect new hosts. Comparative genomics and the development of humanized mouse models have provided important new tools with which to explore the basis of generalism and specialism. This review will examine host specificity of bacterial pathogens with a focus on generalist and specialist serovars of Salmonella enterica.
The host specificity of bacterial pathogens is often a continuum. The genus Salmonella includes a group of generalist and specialist pathogens that are particularly well suited for studying the basis of host specificity. Studying the mechanistic basis of specialism provides insights into coevolution and microbial pathogenesis.
AN EVOLUTIONARY PERSPECTIVE ON HOST SPECIFICITY
Definitions
In host–pathogen interactions, pathogens called generalists are capable of infecting a wide range of host species, whereas others referred to as specialists establish an intimate relationship with only a single-host species. Most pathogens are capable of infecting multiple hosts. Genetic diversity and ecological opportunities for cross-species transmission favor generalism (Woolhouse et al. 2001). The evolutionary dynamic leading to specialism has been long debated. The potential to expand into new niches is implicated as an important driving force in specialism, and some studies suggest that coexistence with other specialists can increase resource exploitation within an ecosystem. Some genetic changes that enhance adaptation to a particular niche may reduce fitness for another niche. This so-called antagonistic pleiotropy can lead to evolutionary trade-offs, which are not necessarily a requirement for specialization but may reinforce tendencies toward specialization. Although often discussed as a dichotomy, host specialization is often a continuum, and many pathogens are able to infect a limited number of hosts (Kirchner and Roy 2002). For example, humans and other primates are susceptible to Shigella, whereas rabbits are resistant but develop inflammation following the injection of Shigella into ileal loops, and mice are resistant to intestinal shigellosis (Phalipon and Sansonetti 2007).
The Host Species Barrier
As pathogens establish defined host ranges, the barrier that must be overcome to make a transition to a new host species is an important determinant of the ecology of infectious diseases. Zoonotic pathogens are able to overcome the species barrier to be transmitted from other vertebrate animals to humans. Crossing the species barrier has important implications for pathogenesis, as pathogens may show increased virulence in hosts to which they are not adapted.
Gene-for-Gene or Multifactorial?
Studies in plants have led to the gene-for-gene model of host pathogen interactions, in which plants that possess a specific resistance gene show resistance to pathogens possessing a corresponding avirulence (avr) gene (Flor 1971). However, the relevance of this model for most animal host–pathogen interactions is limited. Attempts to convert host-specific Salmonella Typhi to a murine pathogen by transduction of DNA from the generalist serovar S. Typhimurium, for example, have led to the conclusion that host specificity in Salmonella is multifactorial (Zahrt 1998). This increases the challenge of understanding the mechanisms underlying host specialization. This review will examine host specificity in bacterial pathogens, with a particular emphasis on the enteric pathogen Salmonella.
EXAMPLES OF SPECIALIST AND GENERALIST BACTERIAL PATHOGENS
Salmonella enterica Serovars
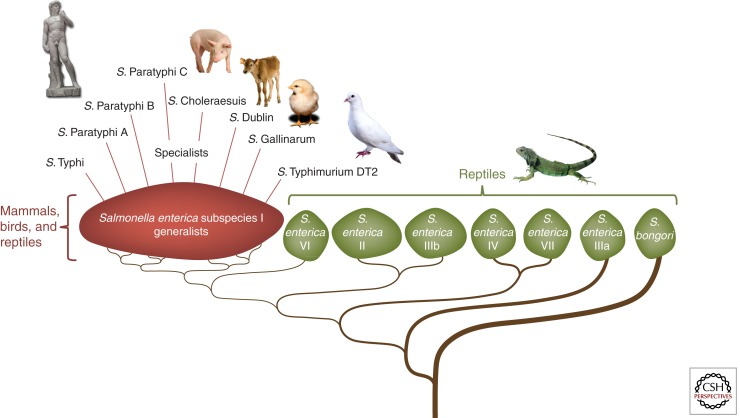
One approach to exploring the genetic basis of host specificity is to compare microbes with a narrow host range (specialists) with close relatives that display broad host specificity (generalists). The genus Salmonella consists of a group of pathogens that are particularly well suited for such a comparison. Salmonella serovars are Gram-negative pathogens belonging to the family Enterobacteriaceae in the phylum Proteobacteria. Most of the >2000 Salmonella serovars are associated with gastroenteritis in humans and have animal reservoirs in a broad range of reptilian, avian, and/or mammalian species (Fig. 1) (Kelterborn 1967). Gastroenteritis is a diarrheal disease that remains localized to the intestine and mesenteric lymph nodes in immunocompetent individuals (Santos et al. 2001). A typical representative generalist serovar associated with gastroenteritis is Salmonella enterica serovar Typhimurium (S. Typhimurium). However, the genus Salmonella also contains a small number of specialists with a narrow host range that are no longer associated with gastroenteritis in humans. Instead, these specialists are associated with disseminated septicemic infections in humans (e.g., typhoid fever) or other animal species (e.g., fowl typhoid). Humans are the only known reservoir for typhoidal Salmonella serovars, which developed in four phylogenetically unrelated clonal lineages within the genus Salmonella, presumably from ancestral organisms that were originally associated with gastroenteritis (Selander et al. 1990). S. enterica serovar Typhi (S. Typhi), the causative agent of typhoid fever, forms one of these clonal lineages, which has been estimated to be between 10,000 and 71,000 years old (Roumagnac et al. 2006). S. enterica serovars Paratyphi C (S. Paratyphi C) and Paratyphi B (S. Paratyphi B) each form single lineages, whereas a fourth lineage is formed by the S. enterica serovars Paratyphi A (S. Paratyphi A) and Sendai (S. Sendai). S. Paratyphi A, S. Paratyphi B, S. Paratyphi C, and S. Sendai cause paratyphoid fever, which is milder in its course but otherwise clinically indistinguishable from typhoid fever.
Figure 1.
Host range of members of the genus Salmonella. The genus Salmonella consists of two species, S. enterica and S. bongori. S. enterica is further subdivided into seven subspecies, designated I, II, IIIa, IIIb, IV, VI, and VII. Serovars of S. bongori and S. enterica subspecies II, IIIa, IIIb, IV, VI, and VII are largely reptile associated and can be occasionally transmitted from this reservoir to humans. The majority of serovars belonging to S. enterica subspecies I are generalists that have reservoirs in mammalian, avian, and reptilian species. Specialists with a more restricted host range have independently evolved from this group several times.
In addition to typhoidal serovars, the genus Salmonella contains a few specialists associated with septicemic illnesses in other animal species. For example, S. enterica serovar Gallinarum (S. Gallinarum) causes pullorum disease (biotype Pullorum) and fowl typhoid (biotype Gallinarum) in poultry, but is avirulent for other animal species (reviewed in Shivaprasad 2000). S. enterica serovars Dublin (S. Dublin) and Choleraesuis (S. Choleraesuis) cause bacteremia in their bovine and porcine hosts, respectively (Hughes et al. 1971; Sojka et al. 1977). However, these pathogens are not fully host restricted, because consumption of unpasteurized milk, undercooked beef, or undercooked pork products can result in invasive bloodstream infections in humans (Saphra and Winter 1957; Fang and Fierer 1991; Threlfall et al. 1992). Closely related specialist/generalist pairs can be found even within a particular Salmonella serovar. For example, whereas the majority of S. Typhimurium isolates are prototypical generalists, a clonal group designated S. Typhimurium phage type DT2 represents a specialist associated with septicemic infections in pigeons (reviewed in Rabsch et al. 2002).
Yersinia Species
A second interesting group of organisms for comparative analysis within the family Enterobacteriaceae is the genus Yersinia, which contains three species pathogenic for humans: Yersinia enterocolitica, Y. pseudotuberculosis, and Y. pestis. The enteropathogenic species Y. enterocolitica and Y. pseudotuberculosis are associated with diarrhea in humans. Both of these pathogens are generalists associated with reservoirs in pigs and wild animals (Kapperud 1991; Fredriksson-Ahomaa 2009) and can also be found in the environment. Y. pestis, the causative agent of plague, branched an estimated 1500 to 20,000 years ago from the closely related Y. pseudotuberculosis serotype O:1b (Achtman et al. 1999). All contemporary Y. pestis isolates arose by clonal diversification from strains associated with the European black death of the 14th century (Bos et al. 2011). Y. pestis circulates in its rodent reservoir by flea-borne transmission, a dramatic change from the fecal–oral transmission of its Y. pseudotuberculosis-like ancestor. The very recent transition from an enteropathogenic lifestyle to that of a vector-borne septicemic infection makes the comparison of the Y. pseudotuberculosis and Y. pestis genomes particularly interesting. Although Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis are able to infect a wide range of mammalian hosts, another Yersinia species called Y. ruckeri is a specialist pathogen of salmonid fish (Sulakvelidze 2000).
IMPLICATIONS FOR VIRULENCE
The Host as Driver of Pathogen Variation
As different hosts represent different environmental niches, the fitness of a pathogen may vary from host to host. Different strains of a pathogen that are highly adapted to specific hosts may evolve in what is referred to as polymorphism. Alternatively, a single pathogen may develop into a generalist with the ability to use different strategies to survive and replicate in different hosts, referred to as polyphenism. In this regard, variation in host phenotypes is an important driver of the evolution of virulence. It has been proposed that pathogens that evolve within a host are more likely to be polymorphic, whereas those evolving outside a host are more likely to be polyphenic (Pfennig 2001). Another potential determinant of pathogen specialization is the strategy used by the host to resist infection (Miller et al. 2005). Theoretical models suggest that attempts by a host to control pathogen replication are more likely to select for polymorphic genotypes than strategies aimed at tolerating pathogen-induced damage.
Effects of Specialization on Virulence
During the coevolution of pathogens and hosts, adaptation is thought to result in the gradual attenuation of virulence, as the survival of the pathogen is dependent on the survival of the host (Dineen 1963; Mims 1975). However, exceptions to this tendency are observed for pathogens in which debility or death of the host are required for transmission. Anderson and May (1982) emphasized that the critical end point of evolution is not necessarily commensalism, but rather maximal transmission of a pathogen, which will tend to be proportional to the pathogen’s reproductive rate. Garnick (1992) further refined this concept by noting that the immune response of the host to a pathogen can limit reproduction, such that optimal transmissibility is dependent on both host and pathogen factors, and the consequences for the host might differ considerably depending on whether the infection is short-lived or chronic. Therefore, the relationship between host specialization and virulence is complex. Human-adapted S. Typhi is generally regarded as more virulent than the generalist serovars, although both specialist and generalist Salmonella serovars can cause a wide range of clinical manifestations ranging from subclinical infection to life-threatening sepsis.
MOLECULAR MECHANISMS OF HOST SPECIFICITY
Microbial Adherence
In some instances, the ability of a pathogen to infect a host has been correlated with its ability to adhere to cells from that species. For example, the enteropathogenic RDEC-1 strain of Escherichia coli is able to cause diarrhea in rabbits but not in rats or guinea pigs and is able to adhere to intestinal brush borders of rabbits but not of the other two species (Cheney et al. 1980). The Opa protein of Neiserria gonorrhoeae is able to bind to the human CEACAM1 glycoprotein but not its canine, bovine, or murine orthologs (Voges et al. 2010), correlating with the specificity of this pathogen for humans. A type-3 secretory system-1 (T3SS-1)-associated lipoprotein called InvH promotes the ability of S. enterica to adhere to and invade cultured epithelial cells (Altmeyer et al. 1993). The effects of InvH on Salmonella interactions with host-intestinal mucosa appear to be host specific, as an invH mutation reduces the severity of Salmonella enteritis in calves but does not affect intestinal colonization or virulence in chicks (Watson et al. 1995, 1998; Porter and Curtiss 1997). The host-specific pathogen S. Typhi also requires InvH for efficient interaction with epithelial cells (Altmeyer et al. 1993) but enters human cells via an interaction between its type-IVB pili and the CFTR (cystic fibrosis transmembrane conductance regulator) protein (Pier et al. 1998; Tsui et al. 2003). Resistance to typhoid has therefore been proposed as a selection pressure that may have maintained the cystic fibrosis mutation at high frequencies in the population (van de Vosse et al. 2005).
Host-specific interactions during entry limit the host range of Listeria monocytogenes (family Listeriaceae, class Bacilli, phylum Firmicutes), an opportunistic food-borne pathogen of humans. The invasion protein internalin (InlA) enables L. monocytogenes to bind E-cadherin on epithelial cells to induce its internalization and crossing of the intestinal barrier. Internalin interacts with E-cadherin from humans but not E-cadherin from mice, owing to a single amino acid difference between the human and murine proteins (Lecuit et al. 1999). Although mice are naturally resistant to oral challenge with L. monocytogenes, transgenic mice expressing human E-cadherin in the intestine become susceptible to infection by this route (Lecuit et al. 2001). The interaction between InlA and human E-cadherin also plays an important role in the ability of L. monocytogenes to cross the placental barrier and cause severe in utero infections (Lecuit et al. 2004). A second invasion protein of L. monocytogenes, InlB, mediates binding to the hepatocyte growth factor receptor (Met) of murine and human cells, but not to Met from rabbits or guinea pigs (Khelef et al. 2006), which represents another example of species specificity. Both InlA and InlB have been implicated in species-specific interactions involved in crossing the placental barrier in vivo (Lecuit et al. 2004; Disson et al. 2008).
Replication in Host Cells
Adherence is only one of the possible mechanisms underlying host specificity. Viruses may show host tropism at the level of adherence and entry or at the level of downstream intracellular events (McFadden 2005). For pathogens able to colonize both cold-blooded and warm-blooded vertebrates, the complex organization of the adaptive immune system in birds and mammals creates unique challenges. In particular, pathogens must confront a broad repertoire of immunoglobulins and highly ordered lymphoid tissues containing mixed populations of lymphocytes, macrophages, and dendritic cells. The ability to survive in macrophages from different types of host has been proposed to be an important factor in Salmonella host specificity (Bäumler et al. 1998). For instance, human-adapted S. Typhi has been reported to display enhanced replication in human macrophages in comparison to the generalist S. Typhimurium, whereas the reverse is true in murine macrophages (Vladoianu et al. 1990; Pascopella et al. 1995; Ishibashi and Arai 1996; Schwan 2000; Xu 2009). The absence of the T3SS effector protein GtgE, a protease that targets the host-cell protein Rab32, has recently been proposed as an explanation for the poor growth of S. Typhi in murine cells (Spanò and Galán 2012).
Toxins
The ability to cause disease in a particular host species can also be affected by a species-specific interaction of toxins with their host-cell targets. Actinobacillus suis and Actinobacillus equuli (family Pasteurellaceae, phylum Proteobacteria) are host-adapted pathogens affecting suckling pigs and newborn foals (sleepy foal disease), respectively. The ability to cause disease in their respective host species is at least, in part, mediated by the elaboration of host-specific pore-forming toxins. Apxl/II, a pore-forming toxin produced by A. suis, shows specific cytotoxicity for porcine leukocytes, whereas Aqx, a pore-forming toxin released by A. equuli, specifically lyses equine leukocytes (reviewed in Frey 2011).
Iron Acquisition and Complement Resistance
Host-specific interactions relating to metal acquisition have been described for the human-adapted pathogens Neisseria gonorrhoeae and Neisseria meningitidis (family Neisseriaceae, phylum Proteobacteria). Like many pathogenic microbes, N. gonorrhoeae and N. meningitidis require iron during extracellular growth in the host. One strategy for iron acquisition is to strip iron from the human iron-binding protein transferrin using transferrin-binding protein B (TbpB) (reviewed in Schryvers and Stojiljkovic 1999). This process is host-specific, because TbpB of N. meningitidis binds human transferrin but not murine transferrin (Renauld-Mongenie et al. 1997). N. meningitidis can persist at least 2 d in intraperitoneally challenged transgenic mice expressing human transferrin, whereas bacteria are rapidly cleared from human transferrin transgenic mice challenged with a N. meningitidis tbpB mutant or from wild-type mice challenged with wild-type N. meningitidis (Zarantonelli et al. 2007). However, mice expressing human transferrin eventually clear even wild-type N. meningitidis, suggesting that additional species-specific interactions may be required for infection. During extracellular growth in the human body, N. meningitidis is exposed to complement. A mechanism that confers complement resistance is the recruitment of a molecule that down-regulates complement activation, complement factor H (fH), to the bacterial surface by factor H-binding protein (fHbp) (Schneider et al. 2006; Ngampasutadol et al. 2008). N. meningitidis fHbp binds fH from humans, but not fH from primates or mice (Granoff et al. 2009). Interestingly, differences between human and primate fH sequences cluster to the precise site of interaction with fHbp (Schneider et al. 2009), which likely represents another interaction contributing to the host specificity of N. meningitidis (Everett et al. 2011).
IMPLICATIONS OF HOST SPECIFICITY FOR TRANSMISSION
Host restriction limits the number of susceptible hosts available for transmission. Although this is rarely a limiting factor for transmission of human-adapted microbes in urban settings, it must have posed a formidable challenge for the persistence of pathogens within the small groups of human hunters and gatherers that existed before the agricultural revolution of the Neolithic age. One way for a microbe to meet this challenge is the development of a chronic persistent state. The transition from a broad-host-range pathogen (generalist) that is cleared from the body within a few weeks to a host-restricted pathogen (specialist) that chronically persists within a host for decades is illustrated by comparison of the lifestyles of broad-host-range and host-restricted Salmonella serovars.
Generalist Salmonella Serovars
Upon ingestion, the broad-host-range pathogen S. Typhimurium uses its virulence factors, two type-III secretion systems (T3SS-1 and T3SS-2), to invade epithelial cells (Galán and Curtiss III 1989) and survive in macrophages (Ochman et al. 1996), which elicits acute intestinal inflammation (Tsolis et al. 1999), producing symptoms of gastroenteritis within 24 hours of ingestion (Glynn and Palmer 1992). During the ensuing host response, neutrophils migrate into the intestinal lumen and produce reactive oxygen species in an attempt to kill bacteria. A by-product of oxyradical production is the generation of a new respiratory electron acceptor, tetrathionate, which enables S. Typhimurium to out-compete obligate anaerobic microbes in the gut lumen (Winter et al. 2010a) by utilizing carbon sources that cannot be fermented (Thiennimitr et al. 2011). The resulting bloom of S. Typhimurium in the intestinal lumen enhances its transmission to the next host by the fecal–oral route (Lawley et al. 2008). With the onset of adaptive immunity, the pathogen is cleared from intestinal tissues, and bacterial shedding in the feces subsides within a few weeks.
Host-Specific Salmonella Serovars
Similar to S. Typhimurium, the host-restricted S. Typhi uses its two type-III secretion systems (T3SS-1 and T3SS-2), to invade epithelial cells (Elsinghorst et al. 1989) and survive in macrophages (Forest et al. 2010). However, S. Typhi does not elicit overt host responses during the initial invasion of the intestinal mucosa, as indicated by a 2-wk incubation period of typhoid fever (Olsen et al. 2003). S. Typhi avoids the generation of responses during the initial phase of infection because it evades innate immunity by expressing a capsular polysaccharide (Raffatellu et al. 2005; Wilson et al. 2008; Haneda et al. 2009) and by rapidly repressing expression of flagella and T3SS-1 during the transition through the epithelial lining (Winter et al. 2010b). These mechanisms enable S. Typhi to slip past the host’s defenses to cause an invasive bloodstream infection. Systemic dissemination results in colonization of internal organs and enables the pathogen to establish chronic carriage in the gall bladder in a fraction of infected individuals (e.g., Typhoid Mary). Chronic carriers are important reservoirs for transmission within the human population, intermittently releasing bacteria through the bile duct into the intestine (Putnam 1927; Leavitt 1992).
Other host-restricted Salmonella serovars also cause systemic infections that provide access to new routes of transmission. For example, the most important route of transmission for poultry-adapted S. Gallinarum is infection of the ovaries, which results in vertical spread via the egg to the chick or poult (Shivaprasad 2000). In cattle, long-term S. Dublin carrier animals contribute to transmission of the pathogen within infected herds by periodically shedding bacteria into milk (Nielsen et al. 2004). These examples help to illustrate that within the genus Salmonella, the transition from a pathogen with a broad host range to a pathogen with a restricted host range commonly involves a change in the route of transmission.
Yersinia Species and the Importance of Vector-Borne Transmission
The transition from free-living Y. pseudotuberculosis to host-dependent Y. pestis also involved a dramatic change in the route of transmission. Although Y. pseudotuberculosis is an enteric pathogen that is transmitted via the fecal–oral route, the vector-borne transmission of Y. pestis exchanges a transition through the mammalian intestine for a transition through the flea gut. However, the selective forces that selected for this change in the route of transmission during the emergence of Y. pestis from an ancestral Y. pseudotuberculosis-like organism remain unknown.
Dependence on an arthropod vector for transmission may restrict host availability and therefore impose constraints on the opportunity for a pathogen to encounter novel hosts. For example, Lyme disease is an important zoonotic infection in the Northeastern United States, where a tick vector transmits the Borellia burgdorferi spirochete to Peromyscus leucopus, a highly competent and ubiquitous rodent reservoir host (LoGiudice et al. 2003). In contrast, Lyme disease is nonendemic in Northern Colorado where the primary rodent reservoir is Neotoma mexacana, a solitary species with little human contact (Maupin et al. 1994).
GENOMIC SIGNATURES OF HOST SPECIFICITY
Genomic Decay
Host specificity is associated with a number of genomic signatures, including genomic decay, genomic rearrangements, and gene acquisition by lateral gene transfer. The genomes of specialists commonly show signs of genomic decay, as indicated by gene deletion or gene inactivation through point mutation (pseudogene formation). For example, 80 genes have been deleted from the genome of Y. pestis since this pathogen diverged from Y. pseudotuberculosis and became host dependent (Chain et al. 2004). Furthermore, the genome of Y. pestis contains ∼208 pseudogenes, compared with only 62 in Y. pseudotuberculosis (Parkhill et al. 2001; Chain et al. 2004). The specialist Y. ruckeri has the smallest genome of any Yersinia species, although this does not appear to be a result of reductive evolution, as its genome contains relatively few pseudogenes. Both Y. pestis and Y. ruckeri lack genes for urease, vitamin B12 metabolism, and the methionine salvage pathway, perhaps because these pathogens do not colonize the mammalian intestine (Chen et al. 2010). The genome of the generalist S. Typhimurium (strain LT2) contains 25 pseudogenes (McClelland et al. 2001), far fewer than the genomes of host-restricted pathogens such as S. Typhi (204 pseudogenes), S. Paratyphi A (173 pseudogenes), S. Paratyphi C (152 pseudogenes), S. Gallinarum (309 pseudogenes), S. Dublin (177 pseudogenes), or S. Choleraesuis (151 pseudogenes) (Parkhill et al. 2001; McClelland et al. 2004; Chiu et al. 2005; Thomson et al. 2008; Liu et al. 2009; Betancor et al. 2012). The pigeon-associated S. Typhimurium DT2 strain 94-213 has 84 pseudogenes, including 21 loci that are intact in other closely related S. Typhimurium strains, and shows distinctive patterns of gene expression (Kingsley et al. 2013). Similarly, the presence of 77 pseudogenes in the multidrug-resistant S. Typhimurium ST313 genotype has led to the suggestion that this lineage is becoming host-adapted to humans (Kingsley et al. 2009; Feasey et al. 2012).
One reason for genomic decay in specialists is related to their divergence from a generalist ancestor along with a change in their route of transmission. When genes necessary for transmission of the ancestral organism are no longer under selection, their functions become dispensable. As a result, such genes accumulate mutations or are deleted. For example, the shdA and misL genes encode nonfimbrial adhesins that enhance intestinal colonization of S. Typhimurium (Kingsley et al. 2000, 2002; Dorsey et al. 2005). Pseudogene formation in host-restricted Salmonella serovars has inactivated both shdA (S. Typhi, S. Paratyphi A, S. Paratyphi C, S. Dublin, and S. Gallinarum) and misL (S. Typhi) (Parkhill et al. 2001; McClelland et al. 2004; Thomson et al. 2008; Liu et al. 2009; Betancor et al. 2012). Another group of intestinal colonization factors of S. Typhimurium is encoded by fimbrial operons of the chaperone usher assembly class (Weening et al. 2005). Genomic decay of genes encoding these intestinal colonization factors is evident in host-restricted Salmonella serovars, as pseudogenes are present in 7 of 11 fimbrial operons in S. Typhi strain CT18 (Townsend et al. 2001), 8 of 12 fimbrial operons in S. Gallinarium (Thomson et al. 2008), and 3 of 12 fimbrial operons in S. Paratyphi A (McClelland et al. 2004). It has been postulated that mutations that alter adhesion-binding specificity or affinity can also contribute to adaptation to different host environments (Kisiela et al. 2012). The generalist S. Typhimurium uses tetrathionate respiration encoded by the ttrRS/ttrBCA gene cluster to out-compete microbes in the gut during its fecal–oral transmission cycle (Winter et al. 2010a). Pseudogenes in this pathway are found in S. Typhi strain CT18 (ttrS) and S. Gallinarum (ttrB, ttrC) (Parkhill et al. 2001; Thomson et al. 2008). Tetrathionate respiration supports growth of S. Typhimurium on ethanolamine as a carbon source in the gut lumen during gastroenteritis (Thiennimitr et al. 2011). This pathway is degraded in specialists S. Paratyphi C (eutA, eutC, eutK, and eutN) and S. Choleraesuis (eutN) (Liu et al. 2009). Growth on ethanolamine under anaerobic conditions necessitates biosynthesis of vitamin B12 by the cob/cbi gene cluster (Roof and Roth 1988), which contains pseudogenes in S. Typhi (cbiM, cbiK, cbiJ, and cbiC), S. Paratyphi A (cbiA), and S. Gallinarum (cobD, cbiD, and cbiC) (Parkhill et al. 2001; McClelland et al. 2004; Thomson et al. 2008).
Many of the pseudogenes found in the genomes of host-restricted Salmonella serovars encode functions that aid intestinal colonization and competition with the intrinsic microbiota during gastroenteritis. These genes are intact in broad-host-range Salmonella serovars because they confer properties that enhance transmission of these pathogens by the fecal–oral route during gastroenteritis (Lawley et al. 2008), which imposes selective constraints that prevent loss of the encoded functions by mutation. However, these genes are no longer under selection in host-restricted pathogens that cause septicemic infections and transmit from reservoirs in the gall bladder (e.g., S. Typhi), the udder (e.g., S. Dublin), or the ovaries (e.g., S. Gallinarum). Thus, one driving force responsible for pseudogene formation in host-restricted Salmonella serovars is that functions required for transmission from an intestinal reservoir are no longer under selection. The resulting genomic decay of these pathways in host-restricted Salmonella serovars represent an experiment of nature, giving rise to a nonsaturation mutagenesis of genes necessary for fecal–oral transmission of generalists associated with gastroenteritis.
Genomic Rearrangements
Genomic rearrangements are another genomic signature associated with specialists but absent from generalists. The chromosomes of E. coli and S. Typhimurium display a high degree of conservation with regard to their gene order (Riley and Anilionis 1978), suggesting that this feature may be under selection. Large rearrangements that alter chromosomal gene order are commonly associated with a lower growth rate, because inversions may alter gene dosage, replication-transcription conflicts, or chromosome symmetry (Hill and Gray 1988; Rebollo et al. 1988; Campo et al. 2004). The generalist S. Typhimurium needs to multiply at a maximum growth rate in the intestine because successful transmission requires bacterial numbers in the feces to reach a sufficient threshold (Lawley et al. 2008). Thus, the maximum growth rate needed for transmission might place the gene order on the chromosomes of broad-host-range Salmonella serovars under selection.
In contrast, host-restricted Salmonella serovars no longer depend on multiplication at a maximal rate to successfully compete with other microbes in the intestine because their transmission success is aided by chronic carriage, a state of relative dormancy that is likely associated with slow growth. Therefore, selective constraints that prevent chromosomal rearrangements may no longer apply to host-restricted Salmonella serovars. This might explain why major genomic rearrangements owing to homologous recombination between the rrn operons are a characteristic feature of the chromosomes of S. Typhi isolates (Liu and Sanderson 1995, 1996; Kothapalli et al. 2005; Matthews et al. 2010). Similar genome rearrangements are also found in other host-restricted lineages within the genus Salmonella, including S. Paratyphi C, S. Gallinarum, and S. Typhimurium phage type DT2 (Liu and Sanderson 1998; Helm et al. 2004; Wu et al. 2005; Liu et al. 2007). Thus, rearrangements are a genomic signature of host-restricted members of the genus Salmonella that rely on chronic persistence in the host for transmission.
Lateral Gene Transfer
A third genomic signature associated with specialists is the acquisition of genetic material by lateral gene transfer, but this genomic signature is also commonly found in genomes of generalists. In some cases, laterally acquired genomic regions help explain how host-restricted pathogens adapted to their new lifestyle as they diverged from their generalist ancestors. For example, the generalist S. Typhimurium triggers innate inflammatory responses that prevent bacterial dissemination from the intestine (reviewed in Santos et al. 2009). In contrast, the specialist S. Typhi evades innate immunity through a rapid change in virulence gene expression when the pathogen enters the intestinal mucosa (Winter et al. 2010b). These changes in virulence gene expression are mediated by the TviA protein, which activates expression of a capsular polysaccharide, the Vi antigen, and represses expression of flagella and T3SS-1 (Winter et al. 2009). The tviA gene and genes involved in the biosynthesis of the capsular polysaccharide (tviBCDE vexABCDE) are encoded by a horizontally acquired DNA region designated Salmonella pathogenicity island (SPI)-7 (Virlogeux et al. 1995; Parkhill et al. 2001). SPI-7 is present in two other host-restricted serovars associated with disseminated septicemic infections, S. Dublin and S. Paratyphi C, suggesting that convergence by horizontal transfer of this DNA region occurred in their evolutionary history (Hashimoto and Khan 1997; Morris et al. 2003; Pickard et al. 2003). S. Typhi also produces a cytolethal distending toxin known as the “typhoid toxin,” which is encoded by a pathogenicity islet (Spanò et al. 2008; Hodak and Galán 2013), although recent genomic studies have shown the presence of the islet in some nontyphoidal serovars as well (den Bakker et al. 2011).
Similarly, acquisition of the 10-kb pPst plasmid and the 100-kb pFra plasmid by lateral gene transfer enabled Y. pestis to adapt to the vector-borne route of transmission after its divergence from a Y. pseudotuberculosis-like ancestor. For instance, the ymt gene located on the pFra plasmid encodes phospholipase D, which enhances the ability of Y. pestis to colonize the flea midgut (Hinnebusch et al. 2002). The pFra plasmid also carries the caf1M1A1 operon, which encodes a protein capsule that is required for infection of mice by the flea-borne route (Sebbane et al. 2009). The pla gene carried on the pPst plasmid encodes a plasminogen activator that is specifically required for inducing the bubonic form of plague in the mouse after a flea bite (Sebbane et al. 2006). Thus, acquisition of two Y. pestis-specific plasmids by lateral gene transfer facilitated the rapid evolutionary transition of Y. pestis to flea-borne transmission.
MODELS TO STUDY HOST SPECIFICITY
Cell Culture Models
Comparing infectivity in cultured cells from permissive and nonpermissive hosts has been a useful approach to understanding the molecular mechanisms of host specificity shown by viral pathogens, but has been generally less useful for bacteria. One exception has been the avian-specific S. enterica serovar Gallinarum biotypes Gallinarum and Pullorum, which have lost their ability to show mannose-sensitive hemagglutination and also show reduced adherence to and invasion of human HEp-2 epithelial cells owing to changes in their type-1 fimbriae. Expression of type-1 fimbriae from generalist S. Typhimurium restores hemagglutination and dramatically enhances HEp-2 invasion by S. Gallinarum and S. Pullorum. As mentioned previously, comparative survival in human and murine macrophages correlates with host specificity in S. Typhi and S. Typhimurium (Vladoianu et al. 1990; Pascopella et al. 1995; Ishibashi and Arai 1996; Schwan 2000; Xu 2009). However, comparative survival in chicken macrophages does not correlate with the avian virulence of S. Gallinarum (Chadfield et al. 2003). Cell culture has been useful for understanding the basis of host specificity in hemotropic Bartonella spp. The ability to adhere to and invade erythrocytes from humans, cats, mice, or rats recapitulates the host specificity of B. quintana, B. henseleae, B. tribocorum, and B. birtlesii observed in vivo (Vayssier-Taussat et al. 2010). The ability of Bartonella to infect specific hosts appears to be encoded within the Trw type-IV secretory system of these pathogens. In contrast, the host specificity of rabbit enteropathogenic E. coli strains is not mirrored by in vitro studies of ileal brush border adherence or erythrocyte agglutination (Robins-Browne et al. 1994). In summary, cell culture can be useful for exploring the mechanistic basis of host specificity but in many cases fails to recapitulate the complexity of in vivo host-bacterial interactions.
Comparative Zoology
Where permissive and nonpermissive animal hosts have been available for study, the comparative zoology of host–pathogen interactions has been informative. It has been reported, for instance, that type-1 fimbriae are not the sole factor responsible for the avian host specificity of S. Gallinarum, because S. Typhimurium is better able to proliferate in deep tissues and cause lethal infection in mice after intravenous inoculation, which bypasses the mucosal stage of infection, whereas the opposite is true in chickens (Barrow et al. 1994). The investigators making these observations concluded that host specificity for Salmonella is primarily expressed at the level of the reticuloendothelial system. Both bovine-adapted Salmonella serovar Dublin and swine-adapted S. Choleraesuis are virulent when administered orally to cattle, whereas avian S. Gallinarum and ovine S. Abortusovis are not. The initial interactions of these serovars with the intestinal mucosa do not appear to be appreciably different, but only S. Dublin is able to persist within the cattle intestine and translocate to mesenteric lymph nodes (Paulin et al. 2002).
Mice fail to develop diarrhea following oral Salmonella administration, representing an important difference between murine and human hosts. However, the administration of streptomycin in drinking water disrupts the endogenous intestinal flora and renders mice susceptible to Salmonella colitis (Bohnhoff and Miller 1962; Barthel et al. 2003). Specialist and generalist Salmonella strains differ in their ability to cause enteritis in the streptomycin-treated mouse model, although substantial strain-to-strain differences within serovars are observed (Suar et al. 2006). The human-adapted serovars S. Typhi and S. Paratyphi do not elicit inflammation in this model (Suar et al. 2006; Raffatellu et al. 2007; Haneda et al. 2009), correlating with their reduced propensity to cause diarrhea in humans. Introduction of the viaB capsule locus into S. Typhimurium suppresses the inflammatory pathology elicited by this generalist serovar, indicating that S. Typhi uses a specific mechanism to reduce intestinal inflammation.
Humanized Mice
Human-adapted pathogens present a special challenge for experimentalists. Until recently, researchers have had to extrapolate from observations in related animal-adapted pathogens, an approach with obvious limitations. For example, S. Typhimurium infection in mice is often touted as a model to understand S. Typhi infection in humans, but “murine typhoid” and human typhoid are different in a number of fundamental respects. As discussed earlier, typhoidal and nontyphoidal Salmonella serovars differ with regard to virulence factors, pseudogene content, and interactions with the innate immune system. Studies in mice have shown that IFN-γ/IL-12 signaling and the Nox2 phagocyte oxidase are critically important for host resistance to Salmonella (Hess et al. 1996; Mastroeni et al. 1996, 2000). Yet, although humans deficient in these host defenses show enhanced susceptibility to nontyphoidal Salmonella infections, they do not appear to be appreciably more susceptible to S. Typhi (de Jong et al. 1998; Winkelstein et al. 2000; Ottenhoff et al. 2002). Similarly, mice lacking CD4+ T cells are highly susceptible to S. Typhimurium, as are humans with low CD4+ T-cell counts owing to HIV infection (Fischl et al. 1986; Hess et al. 1996; Gordon et al. 2002), but HIV infection does not confer an increased susceptibility to typhoid fever (Gordon 2008). To render mice susceptible to an S. Typhi challenge, hog gastric mucin must be coadministered to incapacitate host phagocytes, or massive bacterial inocula must be given (Collins and Carter 1978; Hone et al. 1991). Such models provide limited insight into the pathogenesis of actual human typhoid.
It is generally believed that one of the reasons for the host restriction of S. Typhi is that mice possess some host defenses that are absent from humans. Consistent with this idea, a recent study suggests that inactivation of the murine Toll-like receptor 11 (TLR11), which is absent in humans, renders mice susceptible to S. Typhi infection (Mathur et al. 2012). TLR11-deficient mice thus represent a potential model to study aspects of typhoid fever pathogenesis (Fang and Bäumler 2012). Conversely, it is thought that L. monocytogenes does not infect mice by the oral route because receptors for epithelial invasion are not expressed in this species. The finding that transgenic expression in mice of the InlA receptor human E-cadherin renders these animals susceptible to invasion of enterocytes and crossing of the intestinal barrier supports this idea (Lecuit et al. 2001). Similarly, Streptococcus pyogenes secretes an enzyme that specifically activates human plasminogen and increases virulence in transgenic mice expressing human plasminogen (Sun et al. 2004). N. meningitidis expresses an iron acquisition system (TbpA and TbpB) that specifically strips iron from human transferrin and enhances growth in transgenic mice expressing human transferrin (Zarantonelli et al. 2007). Collectively, these studies show that the targeted deletion or addition of genes can increase the susceptibility of mice to selected human-specific pathogens.
Recent technological advances have created “humanized” chimeric mice that reconstitute specific anatomic and functional elements of human biology in laboratory mice (Mosier et al. 1988; Traggiai et al. 2004). Although studies of infection in humanized mice have only recently begun, this approach promises to provide unprecedented insights into mechanisms of host interaction with human pathogens including hepatitis viruses, HIV, CMV, Epstein-Barr virus, dengue virus, N. meningitidis, and Plasmodium falciparum (e.g., Bente 2005; Jiménez-Díaz 2009; Smith et al. 2010; Berges and Rowan 2011; Sato et al. 2011; Zeisel et al. 2011; Melican et al. 2013).
The generation of humanized mice requires genetic modifications that render mice receptive to the engraftment of human tissues, hematopoietic stem cells, or peripheral blood mononuclear cells. These may include a scid (severe combined immunodeficiency) mutation, which results in sensitivity to radiation and impaired B- and T-cell maturation, a NOD (nonobese diabetic) mutation, which confers reduced natural killer (NK) cell activity and additional innate immune defects, and targeted mutation of the Il2rg (interleukin-2 receptor γ-chain) locus, which results in severe impairment of T, B, and NK cell development (Shultz et al. 2007; Bernard et al. 2008; Brehm et al. 2010). A combination of mutations is required to support high levels of foreign cell engraftment, and transgenic human cytokine expression may enhance the subsequent survival, development, and differentiation of human cells in the murine host (Manz 2007). Following γ-irradiation of immunodeficient mice at birth, human cells or tissues may be introduced at various sites, such as the blood, liver, heart, peritoneal cavity, bone marrow, or kidney.
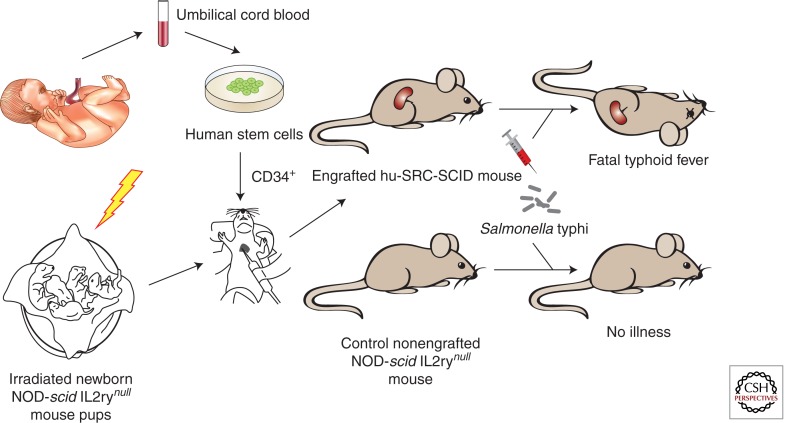
Despite their severe deficiencies in both innate and adaptive immunity, NOD-scid IL2rγnull mice are completely resistant to S. Typhi administered by intraperitoneal inoculation. However, engraftment with human hematopoietic stem cells, to create hu-SRC-SCID mice, renders the animals susceptible to progressive lethal infection (Fig. 2) (Libby et al. 2010). This suggests that S. Typhi requires human hematopoietic cells to proliferate and cause progressive infection in vivo. The pathology observed in hu-SRC-SCID mice infected with S. Typhi resembles human typhoid in a number of respects, including hepatic Kupffer cell swelling and splenic granulomatous inflammation with epithelioid macrophages and multinucleated giant cells (Mallory 1898; Ayhan et al. 1973; Bharadwaj et al. 2009). Elevated serum levels of the cytokines IL-6, IFN-γ, and TNF-α are also observed, as in patients with typhoid fever (Butler et al. 1993; Keuter et al. 1994). Engraftment of human hematopoietic stem cells into a different mouse background, Rag2 IL2rγnull mice, has also been shown to render mice susceptible to S. Typhi infection. However, although the humanized Rag2 IL2rγnull mice show high organism burdens indicative of bacterial replication, their infections do not appear to be as lethal as those observed in hu-SRC-SCID mice (Song et al. 2010; Firoz Mian et al. 2011). It is not clear whether this is owing to different levels or types of human cell engraftment, or perhaps to contributions of the NOD mutation to typhoid susceptibility. Preliminary observations indicate that at least some S. Typhi virulence determinants are required for virulence in humanized mice (Libby et al. 2010; Song et al. 2010), and the production of human antibodies to S. Typhi was observed in some infected Rag2 IL2rγnull mice, suggesting that humanized mice may become useful for the preclinical evaluation of typhoid vaccine candidates (Song et al. 2010). Collectively, these initial observations indicate that humanized mice can recapitulate certain aspects of human typhoid and will be useful to obtain new insights into typhoid pathogenesis, host specificity, and immunity.
Figure 2.
Humanized mouse model of typhoid fever. Newborn immunodeficient NOD-scid IL2rγnull mice are irradiated before transplantation of human CD34+ hematopoietic stem cells from T-cell-depleted umbilical cord blood. Engrafted mice that have been repopulated with human SCID repopulating cells (hu-SRC) are now susceptible to lethal systemic infection with the human-adapted pathogen Salmonella Typhi.
Despite their considerable promise, humanized mice at the present time have a number of significant limitations for the study of infectious diseases, including cost, various defects in reconstituted immunity (including mucosal immunity in the intestinal tract), mouse-to-mouse variability, and graft-versus-host disease. Further refinements to optimize engraftment can be anticipated as humanized mouse models continue to be developed and refined.
CONCLUSIONS
Importance of Understanding Pathogen Host Specificity
Host specialism represents an avenue of evolutionary development that takes a pathogen down an irreversible path of adaptation to a specific host. Specialism may be viewed as a gambit by the pathogen, in which exploitation of a specific niche is achieved at the potential expense of a trade-off in versatility (Bäumler et al. 1998). The extinction of the host can now result in the elimination of the pathogen as well. Yet the substantial number of host-specific pathogens attests to the fact that the benefits of this strategy can outweigh the risks. For the researcher, host specificity provides a valuable opportunity to study the complexities of coevolution between two organisms whose fates have become inextricably intertwined.
Crossing the Species Barrier
Another important reason to understand the basis of host specificity is that host barriers are relative, not absolute. Zoonotic pathogens, which are able to overcome the species barrier to infect humans, comprise a majority of the emerging and reemerging microbial challenges that afflict mankind (Taylor et al. 2001; Woolhouse et al. 2001). By exploring the mechanistic basis of specialism, one can gain essential insights into the pathogenesis of human-specific infections and anticipate future threats from pathogens waiting to make the species jump. As the playwright Arthur Miller wrote, “You specialize in something until one day you find it is specializing in you” (Miller 1968).
ACKNOWLEDGMENTS
The authors acknowledge research support from the National Institutes of Health (AI39557, AI40124, AI44170, AI44486, AI76246, AI77629, AI88122, AI91966, and AI96528).
Footnotes
Editors: Pascale Cossart and Stanley Maloy
Additional Perspectives on Bacterial Pathogenesis available at www.perspectivesinmedicine.org
REFERENCES
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci 96: 14043–14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer RM, McNern JK, Bossio JC, Rosenshine I, Finlay BB, Galan JE 1993. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol Microbiol 7: 89–98 [DOI] [PubMed] [Google Scholar]
- Anderson RM, May RM 1982. Coevolution of hosts and parasites. Parasitology 85: 411–426 [DOI] [PubMed] [Google Scholar]
- Ayhan A, Gokoz A, Karacadag S, Telatar H 1973. The liver in typhoid fever. Am J Gastroenterol 59: 141–146 [PubMed] [Google Scholar]
- Barrow PA, Huggins MB, Lovell MA 1994. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect Immun 62: 4602–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71: 2839–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Tsolis RM, Ficht TA, Adams LG 1998. Evolution of host adaptation in Salmonella enterica. Infect Immun 66: 4579–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bente DA, Melkus MW, Garcia JV, Rico-Hesse R 2005. Dengue fever in humanized NOD/SCID mice. J Virol 79: 13797–13799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Rowan MR 2011. The utility of the new generation of humanized mice to study HIV-1 infection: Transmission, prevention, pathogenesis, and treatment. Retrovirology 8: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Peakman M, Hayday AC 2008. Establishing humanized mice using stem cells: Maximizing the potential. Clin Exp Immunol 152: 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancor L, Yim L, Martínez A, Fookes M, Sasias S, Schelotto F, Thomson N, Maskell D, Chabalgoity JA 2012. Genomic comparison of the closely related Salmonella enterica serovars Enteritidis and Dublin. Open Microbiol J 6: 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj S, Anim JT, Ebrahim F, Aldahham A 2009. Granulomatous inflammatory response in a case of typhoid fever. Med Princ Pract 18: 239–241 [DOI] [PubMed] [Google Scholar]
- Bohnhoff M, Miller CP 1962. Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J Infect Dis 111: 117–127 [DOI] [PubMed] [Google Scholar]
- Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, Coombes BK, McPhee JB, DeWitte SN, Meyer M, Schmedes S, et al. 2011. A draft genome of Yersinia pestis from victims of the Black Death. Nature 478: 506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm MA, Shultz LD, Greiner DL 2010. Humanized mouse models to study human diseases. Curr Opin Endocrinol Diabetes Obes 17: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T, Ho M, Acharya G, Tiwari M, Gallati H 1993. Interleukin-6, γ interferon, and tumor necrosis factor receptors in typhoid fever related to outcome of antimicrobial therapy. Antimicrob Agents Chemother 37: 2418–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo N, Dias MJ, Daveran-Mingot ML, Ritzenthaler P, Le Bourgeois P 2004. Chromosomal constraints in Gram-positive bacteria revealed by artificial inversions. Mol Microbiol 51: 511–522 [DOI] [PubMed] [Google Scholar]
- Chadfield MS, Brown DJ, Aabo S, Christensen JP, Olsen JE 2003. Comparison of intestinal invasion and macrophage response of Salmonella Gallinarum and other host-adapted Salmonella enterica serovars in the avian host. Vet Microbiol 92: 49–64 [DOI] [PubMed] [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, et al. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci 101: 13826–13831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PE, Cook C, Stewart AC, Nagarajan N, Sommer DD, Pop M, Thomason B, Kiley Thomason MP, Lentz S, Nolan N, et al. 2010. Genomic characterization of the Yersinia genus. Genome Biol 11: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney CP, Schad PA, Formal SB, Boedeker EC 1980. Species specificity of in vitro Escherichia coli adherence to host intestinal cell membranes and its correlation with in vivo colonization and infectivity. Infect Immun 28: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, Tang P, Chu C, Hu S, Bao Q, Yu J, Chou YY, Wang HS, Lee YS 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res 33: 1690–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FM, Carter PB 1978. Growth of salmonellae in orally infected germfree mice. Infect Immun 21: 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong R, Altare F, Haagen IA, Elferink DG, Boer T, van Breda Vriesman PJ, Kabel PJ, Draaisma JM, van Dissel JT, Kroon FP, et al. 1998. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 280: 1435–1438 [DOI] [PubMed] [Google Scholar]
- den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L, Hoelzer K, Rodriguez-Rivera LD, Brown S, Bolchakova E, et al. 2011. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12: 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen JK 1963. Immunological aspects of parasitism. Nature 197: 268–269 [DOI] [PubMed] [Google Scholar]
- Disson O, Grayo S, Huillet E, Nikitas G, Langa-Vives F, Dussurget O, Ragon M, Le Monnier A, Babinet C, Cossart P, et al. 2008. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455: 1114–1118 [DOI] [PubMed] [Google Scholar]
- Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Bäumler AJ 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 57: 196–211 [DOI] [PubMed] [Google Scholar]
- Elsinghorst EA, Baron LS, Kopecko DJ 1989. Penetration of human intestinal epithelial cells by Salmonella: Molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli. Proc Natl Acad Sci 86: 5173–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett J, Lea SM, Caesar JJE, Johnson S, Tang CM 2011. Investigating host specificity of Neisseria meningitidis. Mol Immunol 48: 1673 [Google Scholar]
- Fang FC, Bäumler AJ 2012. A tollgate for typhoid. Cell 151: 473–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC, Fierer J 1991. Human infection with Salmonella dublin. Medicine 70: 198–207 [DOI] [PubMed] [Google Scholar]
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA 2012. Invasive non-typhoidal Salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 379: 2489–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoz Mian M, Pek EA, Chenoweth MJ, Ashkar AA 2011. Humanized mice are susceptible to Salmonella typhi infection. Cell Mol Immunol 8: 83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl MA, Dickinson GM, Sinave C, Pitchenik AE, Cleary TJ 1986. Salmonella bacteremia as manifestation of acquired immunodeficiency syndrome. Arch Intern Med 146: 113–115 [PubMed] [Google Scholar]
- Flor HH 1971. Current status of the gene-for-gene concept. Ann Rev Phytopathol 9: 275–296 [Google Scholar]
- Forest CG, Ferraro E, Sabbagh SC, Daigle F 2010. Intracellular survival of Salmonella enterica serovar Typhi in human macrophages is independent of Salmonella pathogenicity island (SPI)-2. Microbiology 156: 3689–3698 [DOI] [PubMed] [Google Scholar]
- Fredriksson-Ahomaa M 2009. Epidemiology of human Yersinia pseudotuberculosis infection. Archiv Fur Lebensmittelhygiene 60: 82–87 [Google Scholar]
- Frey J 2011. The role of RTX toxins in host specificity of animal pathogenic Pasteurellaceae. Vet Microbiol 153: 51–58 [DOI] [PubMed] [Google Scholar]
- Galán JE, Curtiss R III 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci 86: 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnick E 1992. Parasite virulence and parasite-host coevolution: A reappraisal. J Parasitol 78: 381–386 [PubMed] [Google Scholar]
- Glynn JR, Palmer SR 1992. Incubation period, severity of disease, and infecting dose: Evidence from a Salmonella outbreak. Am J Epidemiol 136: 1369–1377 [DOI] [PubMed] [Google Scholar]
- Gordon MA 2008. Salmonella infections in immunocompromised adults. J Infect 56: 413–422 [DOI] [PubMed] [Google Scholar]
- Gordon MA, Banda HT, Gondwe M, Gordon SB, Boeree MJ, Walsh AL, Corkill JE, Hart CA, Gilks CF, Molyneux ME 2002. Non-typhoidal Salmonella bacteraemia among HIV-infected Malawian adults: High mortality and frequent recrudescence. AIDS 16: 1633–1641 [DOI] [PubMed] [Google Scholar]
- Granoff DM, Welsch JA, Ram S 2009. Binding of complement factor H (fH) to Neisseria meningitidis is specific for human fH and inhibits complement activation by rat and rabbit sera. Infect Immun 77: 764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneda T, Winter SE, Butler BP, Wilson RP, Tukel C, Winter MG, Godinez I, Tsolis RM, Bäumler AJ 2009. The capsule-encoding viaB locus reduces intestinal inflammation by a Salmonella pathogenicity island 1-independent mechanism. Infect Immun 77: 2932–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Khan AQ 1997. Comparison of ViaB regions of Vi-positive organisms. FEMS Microbiol Lett 157: 55–57 [DOI] [PubMed] [Google Scholar]
- Helm RA, Porwollik S, Stanley AE, Maloy S, McClelland M, Rabsch W, Eisenstark A 2004. Pigeon-associated strains of Salmonella enterica serovar Typhimurium phage type DT2 have genomic rearrangements at rRNA operons. Infect Immun 72: 7338–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Ladel C, Miko D, Kaufmann SH 1996. Salmonella typhimurium aroA-infection in gene-targeted immunodeficient mice: Major role of CD4+ TCR-αβ cells and IFN-γ in bacterial clearance independent of intracellular location. J Immunol 156: 3321–3326 [PubMed] [Google Scholar]
- Hill CW, Gray JA 1988. Effects of chromosomal inversion on cell fitness in Escherichia coli K-12. Genetics 119: 771–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296: 733–735 [DOI] [PubMed] [Google Scholar]
- Hodak H, Galán JE 2013. A Salmonella Typhi homologue of bacteriophage muramidases controls typhoid toxin secretion. EMBO Rep 14: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hone DM, Harris AM, Chatfield S, Dougan G, Levine MM 1991. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine 9: 810–816 [DOI] [PubMed] [Google Scholar]
- Hughes LE, Gibson EA, Roberts HE, Davies ET, Davies G, Sojka WJ 1971. Bovine salmonellosis in England and Wales. Br Vet J 127: 225–238 [DOI] [PubMed] [Google Scholar]
- Ishibashi Y, Arai T 1996. A possible mechanism for host-specific pathogenesis of Salmonella serovars. Microb Pathog 21: 435–446 [DOI] [PubMed] [Google Scholar]
- Jiménez-Díaz MB, Mulet T, Viera S, Gómez V, Garuti H, Ibáñez J, Alvarez-Doval A, Shultz LD, Martínez A, Gargallo-Viola D, Angulo-Barturen I 2009. Improved murine model of malaria using Plasmodium falciparum competent strains and non-myelodepleted NOD-scid IL2Rγnull mice engrafted with human erythrocytes. Antimicrob Agents Chemother 53: 4533–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapperud G 1991. Yersinia enterocolitica in food hygiene. Int J Food Microbiol 12: 53–65 [DOI] [PubMed] [Google Scholar]
- Kelterborn E 1967. Salmonella-species. First isolations, names and occurrence. Springer-Verlag, New York [Google Scholar]
- Keuter M, Dharmana E, Gasem MH, van der Ven-Jongekrijg J, Djokomoeljanto R, Dolmans WM, Demacker P, Sauerwein R, Gallati H, van der Meer JW 1994. Patterns of proinflammatory cytokines and inhibitors during typhoid fever. J Infect Dis 169: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Khelef N, Lecuit M, Bierne H, Cossart P 2006. Species specificity of the Listeria monocytogenes InlB protein. Cell Microbiol 8: 457–470 [DOI] [PubMed] [Google Scholar]
- Kingsley RA, van Amsterdam K, Kramer N, Bäumler AJ 2000. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect Immun 68: 2720–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Santos RL, Keestra AM, Adams LG, Bäumler AJ 2002. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol Microbiol 43: 895–905 [DOI] [PubMed] [Google Scholar]
- Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V, et al. 2009. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res 19: 2279–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Kay S, Connor T, Barquist L, Salt L, Holt KE, Sivaraman K, Wileman T, Goulding D, Clare S, et al. 2013. Genome and transcriptome adaptation accompanying emergence of the definitive type 2 host-restricted Salmonella enterica serovar Typhimurium pathovar. mBio 4: e00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner JW, Roy BA 2002. Evolutionary implications of host-pathogen specificity: Fitness consequences of pathogen virulence traits. Evol Ecol Res 4: 27–48 [Google Scholar]
- Kisiela DI, Chattopadhyay S, Libby SJ, Karlinsey JE, Fang FC, Tchesnokova V, Kramer JJ, Beskhlebnaya V, Samadpour M, Grzymajlo K, et al. 2012. Evolution of Salmonella enterica virulence via point mutations in the fimbrial adhesin. PLoS Pathog 8: e1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli S, Nair S, Alokam S, Pang T, Khakhria R, Woodward D, Johnson W, Stocker BA, Sanderson KE, Liu SL 2005. Diversity of genome structure in Salmonella enterica serovar Typhi populations. J Bacteriol 187: 2638–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM 2008. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun 76: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt JW 1992. “Typhoid Mary” strikes back. Bacteriological theory and practice in early twentieth-century public health. Isis 83: 608–629 [DOI] [PubMed] [Google Scholar]
- Lecuit M, Dramsi S, Gottardi C, Fedor-Chaiken M, Gumbiner B, Cossart P 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J 18: 3956–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P 2001. A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science 292: 1722–1725 [DOI] [PubMed] [Google Scholar]
- Lecuit M, Nelson DM, Smith SD, Khun H, Huerre M, Vacher-Lavenu MC, Gordon JI, Cossart P 2004. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: Role of internalin interaction with trophoblast E-cadherin. Proc Natl Acad Sci 101: 6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby SJ, Brehm MA, Greiner DL, Shultz LD, McClelland M, Smith KD, Cookson BT, Karlinsey JE, Kinkel TL, Porwollik S, et al. 2010. Humanized nonobese diabetic-scid IL2rγnull mice are susceptible to lethal Salmonella Typhi infection. Proc Natl Acad Sci 107: 15589–15594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Sanderson KE 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc Natl Acad Sci 92: 1018–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Sanderson KE 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc Natl Acad Sci 93: 10303–10308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Sanderson KE 1998. Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS Microbiol Lett 164: 275–281 [DOI] [PubMed] [Google Scholar]
- Liu WQ, Liu GR, Li JQ, Xu GM, Qi D, He XY, Deng J, Zhang FM, Johnston RN, Liu SL 2007. Diverse genome structures of Salmonella paratyphi C. BMC Genomics 8: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WQ, Feng Y, Wang Y, Zou QH, Chen F, Guo JT, Peng YH, Jin Y, Li YG, Hu SN, et al. 2009. Salmonella paratyphi C: Genetic divergence from Salmonella choleraesuis and pathogenic convergence with Salmonella typhi. PLoS ONE 4: e4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice K, Ostfield RS, Schmidt KA, Keesing F 2003. The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proc Natl Acad Sci 100: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory FB 1898. A histological study of typhoid fever. J Exp Med 3: 611–638 [PMC free article] [PubMed] [Google Scholar]
- Manz MG 2007. Human-hemato-lymphoid-system mice: Opportunities and challenges. Immunity 26: 537–541 [DOI] [PubMed] [Google Scholar]
- Mastroeni P, Harrison JA, Chabalgoity JA, Hormaeche CE 1996. Effect of interleukin 12 neutralization on host resistance and γ interferon production in mouse typhoid. Infect Immun 64: 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni P, Vazquez-Torres A, Fang FC, Xu Y, Khan S, Hormaeche CE, Dougan G 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J Exp Med 192: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Oh H, Zhang D, Park SG, Seo J, Koblansky A, Hayden MS, Ghosh S 2012. A mouse model of Salmonella Typhi infection. Cell 151: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews TD, Edwards R, Maloy S 2010. Chromosomal rearrangements formed by rrn recombination do not improve replichore balance in host-specific Salmonella enterica serovars. PLoS ONE 5: e13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin GO, Gage KL, Piesman J, Montenieri J, Sviat SL, VanderZanden L, Happ CM, Dolan M, Johnson BJ 1994. Discovery of an enzootic cycle of Borrelia burgdorferi in Neotoma mexicana and Ixodes spinipalpis from northern Colorado, an area where Lyme disease is nonendemic. J Infect Dis 170: 636–643 [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413: 852–856 [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, et al. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet 36: 1268–1274 [DOI] [PubMed] [Google Scholar]
- McFadden G 2005. Poxvirus tropism. Nat Rev Microbiol 3: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melican K, Michea Veloso P, Martin T, Bruneval P, Duménil G 2013. Adhesion of Neisseria meningitidis to dermal vessels leads to local vascular damage and purpura in a humanized mouse model. PLoS Pathog 9: e1003139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A 1968. The price: A play. Secker and Warburg, London [Google Scholar]
- Miller MR, White A, Boots M 2005. The evolution of host resistance: Tolerance and control as distinct strategies. J Theor Biol 236: 198–207 [DOI] [PubMed] [Google Scholar]
- Mims CA 1975. The meaning of persistent infections in nature. Bull World Health Org 52: 745. [PMC free article] [PubMed] [Google Scholar]
- Morris C, Tam CK, Wallis TS, Jones PW, Hackett J 2003. Salmonella enterica serovar Dublin strains, which are Vi antigen-positive use type IVB pili for bacterial self-association and human intestinal cell entry. Microb Pathog 35: 279–284 [DOI] [PubMed] [Google Scholar]
- Mosier DE, Gulizia RJ, Baird SM, Wilson DB 1988. Transfer of a functional human immune system to mice with severe combined immunodeficiency. Nature 335: 256–259 [DOI] [PubMed] [Google Scholar]
- Ngampasutadol J, Ram S, Gulati S, Agarwal S, Li C, Visintin A, Monks B, Madico G, Rice PA 2008. Human factor H interacts selectively with Neisseria gonorrhoeae and results in species-specific complement evasion. J Immunol 180: 3426–3435 [DOI] [PubMed] [Google Scholar]
- Nielsen LR, Schukken YH, Grohn YT, Ersboll AK 2004. Salmonella Dublin infection in dairy cattle: Risk factors for becoming a carrier. Prev Vet Med 65: 47–62 [DOI] [PubMed] [Google Scholar]
- Ochman H, Soncini FC, Solomon F, Groisman EA 1996. Identification of a pathogenicity island for Salmonella survival in host cells. Proc Natl Acad Sci 93: 7800–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SJ, Bleasdale SC, Magnano AR, Landrigan C, Holland BH, Tauxe RV, Mintz ED, Luby S 2003. Outbreaks of typhoid fever in the United States, 1960–99. Epidemiol Infect 130: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenhoff TH, Verreck FA, Lichtenauer-Kaligis EG, Hoeve MA, Sanal O, van Dissel JT 2002. Genetics, cytokines and human infectious disease: Lessons from weakly pathogenic mycobacteria and salmonellae. Nat Genet 32: 97–105 [DOI] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413: 848–852 [DOI] [PubMed] [Google Scholar]
- Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small PL 1995. Host restriction phenotypes of Salmonella typhi and Salmonella gallinarum. Infect Immun 63: 4329–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin SM, Watson PR, Benmore AR, Stevens MP, Jones PW, Villarreal-Ramos B, Wallis TS 2002. Analysis of Salmonella enterica serotype-host specificity in calves: Avirulence of S. enterica serotype gallinarum correlates with bacterial dissemination from mesenteric lymph nodes and persistence in vivo. Infect Immun 70: 6788–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfennig KS 2001. Evolution of pathogen virulence: The role of variation in host phenotype. Proc Biol Sci 268: 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalipon A, Sansonetti PJ 2007. Shigella’s ways of manipulating the host intestinal innate and adaptive immune system: A tool box for survival? Immunol Cell Biol 85: 119–129 [DOI] [PubMed] [Google Scholar]
- Pickard D, Wain J, Baker S, Line A, Chohan S, Fookes M, Barron A, Gaora PO, Chabalgoity JA, Thanky N, et al. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J Bacteriol 185: 5055–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier GB, Grout M, Zaidi T, Meluleni G, Mueschenborn SS, Banting G, Ratcliff R, Evans MJ, Colledge WH 1998. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature 393: 79–82 [DOI] [PubMed] [Google Scholar]
- Porter SB, Curtiss RR 1997. Effect of inv mutations on Salmonella virulence and colonization in 1-day-old White Leghorn chicks. Avian Dis 41: 45–57 [PubMed] [Google Scholar]
- Putnam P 1927. The trend of typhoid fever mortality in the United States. Am J Hyg 7: 762–781 [Google Scholar]
- Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschape H, Adams LG, Bäumler AJ 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun 70: 2249–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Bäumler AJ 2005. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun 73: 3367–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, Rossetti CA, Lawhon SD, Chu H, Lau T, Bevins CL, et al. 2007. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect Immun 75: 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo JE, Francois V, Louarn JM 1988. Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc Natl Acad Sci 85: 9391–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld-Mongenie G, Poncet D, von Olleschik-Elbheim L, Cournez T, Mignon M, Schmidt MA, Quentin-Millet MJ 1997. Identification of human transferrin-binding sites within meningococcal transferrin-binding protein B. J Bacteriol 179: 6400–6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Anilionis A 1978. Evolution of the bacterial genome. Ann Rev Microbiol 32: 519–560 [DOI] [PubMed] [Google Scholar]
- Robins-Browne RM, Tokhi AM, Adams LM, Bennett-Wood V 1994. Host specificity of enteropathogenic Escherichia coli from rabbits: Lack of correlation between adherence in vitro and pathogenicity for laboratory animals. Infect Immun 62: 3329–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Roth JR 1988. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol 170: 3855–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TA, Acosta CJ, Farrar J, Dougan G, et al. 2006. Evolutionary history of Salmonella typhi. Science 314: 1301–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Bäumler AJ 2001. Animal models of Salmonella infections: Enteritis vs. typhoid fever. Microb Infect 3: 1335–1344 [DOI] [PubMed] [Google Scholar]
- Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, Tsolis RM, Baumler AJ 2009. Life in the inflamed intestine, Salmonella style. Trends Microbiol 17: 498–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphra I, Winter JW 1957. Clinical manifestations of salmonellosis in man—An evaluation of 7779 human infections identified at the New York Salmonella Center. N Engl J Med 256: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Sato K, Misawa N, Nie C, Satou Y, Iwakiri D, Matsuoka M, Takahashi R, Kuzushima K, Ito M, Takada K, et al. 2011. A novel animal model of Epstein–Barr virus-associated hemophagocytic lymphohistiocytosis in humanized mice. Blood 117: 5663–5673 [DOI] [PubMed] [Google Scholar]
- Schneider MC, Exley RM, Chan H, Feavers I, Kang YH, Sim RB, Tang CM 2006. Functional significance of factor H binding to Neisseria meningitidis. J Immunol 176: 7566–7575 [DOI] [PubMed] [Google Scholar]
- Schneider MC, Prosser BE, Caesar JJE, Kugelberg E, Li S, Zhang Q, Quoraishi S, Lovett JE, Deane JE, Sim RB, et al. 2009. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature 458: 890–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schryvers AB, Stojiljkovic I 1999. Iron acquisition systems in the pathogenic Neisseria. Mol Microbiol 32: 1117–1123 [DOI] [PubMed] [Google Scholar]
- Schwan WR, Huang XZ, Hu L, Kopecko DJ 2000. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect Immun 68: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci 103: 5526–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbane F, Jarrett C, Gardner D, Long D, Hinnebusch BJ 2009. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect Immun 77: 1222–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander RK, Beltran P, Smith NH, Helmuth R, Rubin FA, Kopecko DJ, Ferris K, Tall BD, Cravioto A, Musser JM 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect Immun 58: 2262–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad HL 2000. Fowl typhoid and pullorum disease. Rev Sci Tech 19: 405–424 [DOI] [PubMed] [Google Scholar]
- Shultz LD, Ishikawa F, Greiner DL 2007. Humanized mice in translational biomedical research. Nat Rev Immunol 7: 118–130 [DOI] [PubMed] [Google Scholar]
- Smith MS, Goldman DC, Bailey AS, Pfaffle DL, Kreklywich CN, Spencer DB, Othieno FA, Streblow DN, Garcia JV, Fleming WH, et al. 2010. Granulocyte-colony stimulating factor reactivates human cytomegalovirus in a latently infected humanized mouse model. Cell Host Microbe 8: 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka WJ, Wray C, Shreeve J, Benson AJ 1977. Incidence of Salmonella infection in animals in England and Wales 1968–1974. J Hyg 78: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Willinger T, Rongvaux A, Eynon EE, Stevens S, Manz MG, Flavell RA, Galan JE 2010. A mouse model for the human pathogen Salmonella Typhi. Cell Host Microbe 8: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Galán JE 2012. A Rab32-dependent pathway contributes to Salmonella Typhi host restriction. Science 338: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Ugalde JE, Galán JE 2008. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3: 30–38 [DOI] [PubMed] [Google Scholar]
- Suar M, Jantsch J, Hapfelmeier S, Kremer M, Stallmach T, Barrow PA, Hardt WD 2006. Virulence of broad- and narrow-host-range Salmonella enterica serovars in the streptomycin-pretreated mouse model. Infect Immun 74: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulakvelidze A 2000. Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: The ignored species. Microbes Infect 2: 297–513 [DOI] [PubMed] [Google Scholar]
- Sun H, Ringdahl U, Homeister JW, Fay WP, Engelberg NC, Yang AY, Rozek LS, Wang X, Sjöbring U, Ginsburg D 2004. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305: 1283–1286 [DOI] [PubMed] [Google Scholar]
- Taylor LH, Latham SM, Woolhouse ME 2001. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356: 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci 108: 17480–17485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, et al. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res 18: 1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall EJ, Hall ML, Rowe B 1992. Salmonella bacteraemia in England and Wales, 1981–1990. J Clin Pathol 45: 34–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J, Dougan G, et al. 2001. Salmonella enterica serotype Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun 69: 2894–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E, Chicha L, Mazzucchelli L, Bronz L, Piffaretti JC, Lanzavecchia A, Manz MG 2004. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science 304: 104–107 [DOI] [PubMed] [Google Scholar]
- Tsolis RM, Adams LG, Ficht TA, Bäumler AJ 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67: 4879–4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui IS, Yip CM, Hackett J, Morris C 2003. The type IVB pili of Salmonella enterica serovar Typhi bind to the cystic fibrosis transmembrane conductance regulator. Infect Immun 71: 6049–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vosse E, Ali S, de Visser AW, Surjadi C, Widjaja S, Vollaard AM, van Dissel JT 2005. Susceptibility to typhoid fever is associated with a polymorphism in the cystic fibrosis transmembrane conductance regulator (CFTR). Hum Genet 118: 138–140 [DOI] [PubMed] [Google Scholar]
- Vayssier-Taussat M, Le Rhun D, Deng HK, Biville F, Cescau S, Danchin A, Marignac G, Lenaour E, Boulouis HJ, Mavris M, et al. 2010. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog 6: e1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlogeux I, Waxin H, Ecobichon C, Popoff MY 1995. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology 141: 3039–3047 [DOI] [PubMed] [Google Scholar]
- Vladoianu IR, Chang HR, Pechere JC 1990. Expression of host resistance to Salmonella typhi and Salmonella typhimurium: Bacterial survival within macrophages of murine and human origin. Microb Pathog 8: 83–90 [DOI] [PubMed] [Google Scholar]
- Voges M, Bachmann V, Kammerer R, Gophna U, Hauck CR 2010. CEACAM1 recognition by bacterial pathogens is species specific. BMC Microbiol 10: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PR, Paulin SM, Bland AP, Jones PW, Wallis TS 1995. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect Immun 63: 2743–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PR, Galyov EE, Paulin SM, Jones PW, Wallis TS 1998. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect Immun 66: 1432–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun 73: 3358–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, Bäumler AJ 2008. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol 10: 876–890 [DOI] [PubMed] [Google Scholar]
- Winkelstein JA, Marino MC, Johnston RBJ, Boyle J, Curnutte J, Gallin JI, Malech HL, Holland SM, Ochs H, Quie P, et al. 2000. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 79: 155–169 [DOI] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Russmann H, Bäumler AJ 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol 74: 175–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. 2010a. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467: 426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Godinez I, Yang H-J, Russmann H, Andrews-Polymenis HL, Bäumler AJ 2010b. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog 6: e1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME, Taylor LH, Haydon DT 2001. Population biology of multihost pathogens. Science 292: 1109–1112 [DOI] [PubMed] [Google Scholar]
- Wu KY, Liu GR, Liu WQ, Wang AQ, Zhan S, Sanderson KE, Johnston RN, Liu SL 2005. The genome of Salmonella enterica serovar Gallinarum: Distinct insertions/deletions and rare rearrangements. J Bacteriol 187: 4720–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Maloy S, McGuire KL 2009. Macrophages influence Salmonella host-specificity in vivo. Microb Pathog 47: 212–222 [DOI] [PubMed] [Google Scholar]
- Zahrt TC 1998. “A genetic analysis of the Salmonella typhi host range”. PhD thesis, University of Illinois, Urbana-Champaign [Google Scholar]
- Zarantonelli ML, Szatanik M, Giorgini D, Hong E, Huerre M, Guillou F, Alonso JM, Taha MK 2007. Transgenic mice expressing human transferrin as a model for meningococcal infection. Infect Immun 75: 5609–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel MB, Da Costa D, Baumert TF 2011. Opening the door for hepatitis C virus infection in genetically humanized mice. Hepatology 54: 1873–1875 [DOI] [PubMed] [Google Scholar]