Abstract
Research using ciliates revealed early examples of epigenetic phenomena and continues to provide novel findings. These protozoans maintain separate germline and somatic nuclei that carry transcriptionally silent and active genomes, respectively. Examining the differences in chromatin within distinct nuclei of Tetrahymena identified histone variants and established that transcriptional regulators act by modifying histones. Formation of somatic nuclei requires both transcriptional activation of silent chromatin and large-scale DNA elimination. This somatic genome remodeling is directed by homologous RNAs, acting with an RNA interference (RNAi)-related machinery. Furthermore, the content of the parental somatic genome provides a homologous template to guide this genome restructuring. The mechanisms regulating ciliate DNA rearrangements reveal the surprising power of homologous RNAs to remodel the genome and transmit information transgenerationally.
Ciliates have germline and somatic nuclei, differing in transcriptional activity and genome organization. These nuclei have revealed fascinating epigenetic phenomena (e.g., genome remodeling with RNAi-related machinery).
OVERVIEW
Anyone watching ciliates under a microscope is sure to be fascinated by these complex little animals that use their hair-like cilia to swim, eat, and find a mate. Growing cells duplicate by simple binary fission; yet, periodically, ciliates will mate with a partner or, in some species, undergo self-fertilization, resulting in sexual progeny with a different genotype. What uniquely distinguishes these single-celled eukaryotes is that they maintain two functionally distinct genomes carried in separate nuclei within a common cytoplasm. The smaller of these, the micronucleus, contains the germline genome. It is transcriptionally silent during growth, but stores the genetic information that is passed to progeny at each sexual generation. The larger macronucleus performs somatic functions as it is responsible for all gene expression and, thus, governs the cell’s phenotype. It is discarded at the end of each sexual generation when a new macronucleus differentiates from the germline.
The compartmentalization of gene expression of ciliates implies mechanisms exist that differentially regulate homologous sequences contained within the distinct nuclei. Early studies sought to elucidate the means by which the germline was kept silent and the somatic genome transcriptionally active. Researchers could readily correlate specific histones and their modifications with transcriptional activity or cell cycle stage. For instance, by comparing chromatin proteins from germline and somatic nuclei of Tetrahymena thermophila, some of the first histone variants were identified. Furthermore, new chromatin regulators, such as the first nuclear histone acetyltransferase (HAT), were identified in this ciliate, following up the observation that only the macronucleus contains acetylated histones.
Not only do germline and somatic nuclei have different transcriptional activities, but they have physically different genome organization. During somatic macronuclear development, massive DNA rearrangements generate a streamlined version of the genome. Much of its germline-derived DNA, including most repetitive sequences, is eliminated, whereas all the genes needed for the organism’s survival throughout the life cycle are amplified to achieve a high ploidy level. Moreover, in some ciliate species, protein-encoding sequences that are “scrambled” in the micronucleus are assembled in proper order (unscrambled) in the macronucleus.
Many experiments have shown that DNA rearrangement patterns are not strictly genetically programmed, but are epigenetically controlled, at least in part, by preexisting rearrangements within the parental somatic genome. The implication is that the germline and somatic genomes are compared to each other during nuclear differentiation, a comparison that is likely mediated by homology-dependent interactions between germline and somatic RNAs. Recent studies have indicated that short RNAs produced by an RNA interference (RNAi)-related mechanism are used for the elimination of repetitive DNAs in Paramecium and Tetrahymena, whereas long RNAs play essential roles in the DNA unscrambling of Oxytricha.
In both Paramecium and Tetrahymena, short RNAs are generated from the germline genome during meiosis. The discovery of these small RNAs, together with the demonstration that Argonaute and Dicer homologs are required for DNA rearrangements in these ciliates, have led to the realization that an RNAi-like mechanism is involved. The small RNAs are thought to target homologous sequences for methylation at histone H3 at lysine 9 (H3K9me) and lysine 27 (H3K27me), marking them for elimination. Thus, ciliate DNA rearrangements are mechanistically similar to the more broadly used RNA-directed establishment of heterochromatin. The use of RNAi to eliminate transposable elements further underscores the importance of this pathway as a genome defense mechanism.
In contrast, another type of noncoding RNA, long macronuclear RNAs, is used to guide the DNA unscrambling event in Oxytricha, which is a ciliate distantly related to Paramecium and Tetrahymena. Transcription of the parental macronuclear genome occurs during early conjugation in Oxytricha, and RNAi knockdown of specific parental macronuclear long RNAs inhibits unscrambling in the corresponding loci in the new macronucleus. Moreover, the injection of artificial RNAs reprograms the unscrambling order. Therefore, long macronuclear RNAs epigenetically regulate DNA unscrambling probably by acting as templates to guide the rearrangements.
Fully understanding these two curious DNA rearrangement processes, small RNA-directed DNA elimination in Paramecium and Tetrahymena and long RNA-directed DNA unscrambling in Oxytricha, will undoubtedly provide new insights into the roles of RNA in the epigenetic programming of the genome.
1. CILIATES: SINGLE CELLS WITH TWO DIFFERENT GENOMES
Ciliates, which comprise a monophyletic lineage that is an estimated 1 billion years old (Philippe et al. 2000), were among the first unicellular eukaryotes to be used as genetic models. In the late 1930s, when T.M. Sonneborn discovered the mating types of Paramecium aurelia (Sonneborn 1937), the chromosome theory of inheritance elaborated by T.H. Morgan was still unsatisfying to many researchers, in particular embryologists (see Felsenfeld 2014 for historical detail). Unable to envision how such static entities as genes could be the sole basis of heredity, they believed that the cytoplasm had to be involved, if only to coordinate gene action (see Harwood 1985). Although mainstream geneticists largely focused on gene action, Sonneborn’s early genetic analyses showed that the transmission of many heritable characteristics could not be fully explained by Mendel’s laws. The study of ciliates has revealed some of the first examples of cytoplasmic inheritance and continues to provide new insights into epigenetic mechanisms.
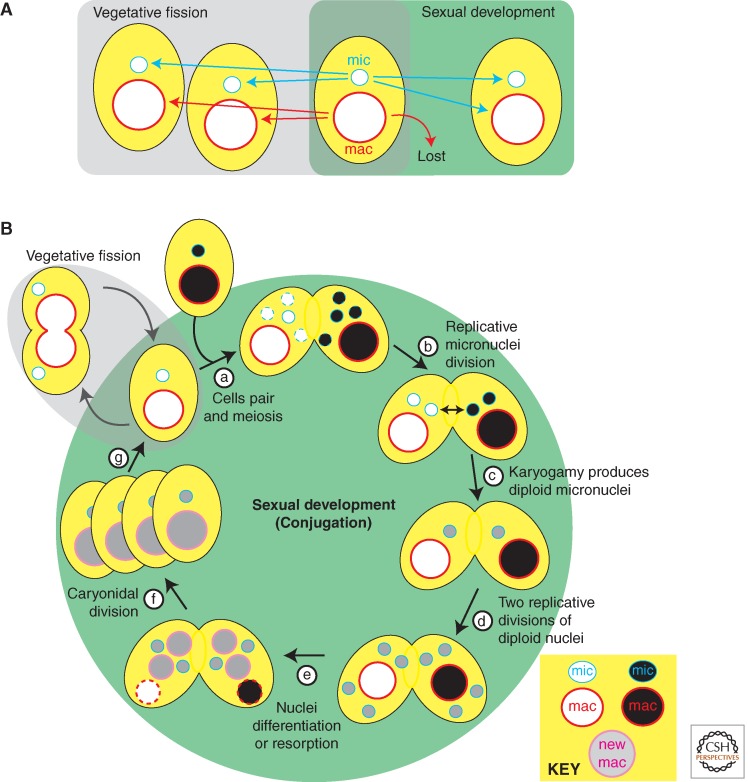
A key biological attribute of ciliates that provides insight into epigenetic mechanisms is nuclear dimorphism: Each cell contains two kinds of nuclei that differ in structure and function. The diploid micronuclei are transcriptionally silent during vegetative growth, but contain the germline genome. These nuclei undergo meiosis to produce gametic nuclei that transmit the germline genome to the next sexual generation (Fig. 1). In contrast, the highly polyploid macronuclei are responsible for gene expression during vegetative growth and thus govern the cell’s phenotype, but they are lost during sexual development and can therefore be considered the equivalent of the soma (Fig. 1). The numbers of nuclei of each type vary in different species. For example, Paramecium tetraurelia have two micronuclei and one macronucleus, whereas T. thermophila has just one of each.
Figure 1.
The ciliate life cycle and fates of their nuclei. (A) Vegetative cells multiply by binary fission, duplicating micro- and macronuclei. Sexual reproduction results in the loss of the parental somatic macronucleus and differentiation of new nuclei. (B) Conjugation: (a) cells pair inducing micronuclear meiosis that ends with selection of one of the haploid products as the gametic nucleus and degeneration of those remaining (dashed line nuclei); (b) an additional replicative division of the selected nucleus produces genetically identical haploid nuclei, and one from each mate is then transferred to its partner; (c) karyogamy produces a diploid zygotic (gray shading) nucleus in each mate; (d) two replicative divisions of these zygotic nuclei in each mate produce undifferentiated micro- and macronuclei; (e) nuclear differentiation: two nuclei become new micronuclei, whereas two begin differentiating into new macronuclei, and the parental macronuclei are resorbed; (f) pair-separation and caryonidal division; (g) resumption of vegetative growth. Nuclei with dashed outline are targets for destruction.
Macro- and micronuclei divide by separate mechanisms during vegetative growth (Fig. 1A). Micronuclei divide via conventional closed mitosis. Macronuclei, in contrast, divide by a poorly understood amitotic mechanism that does not involve spindle formation or visible condensation of the centromere-less, somatic chromosomes. After DNA synthesis, the macronucleus simply splits into two roughly equal halves. There does not appear to be any mechanism to ensure equal segregation of macronuclear chromosomes to the two daughter cells. Instead, it is likely that the high ploidy level (∼800C in P. tetraurelia, ∼45C in T. thermophila) prevents lethal gene loss for a number of vegetative divisions. Most species have a finite vegetative life span, and clonal cell lines will eventually die if they do not engage in sexual reproduction before they become senescent.
2. CONJUGATION: DIFFERENTIATION OF THE GERMLINE AND SOMATIC GENOMES
Mature ciliate cells, which have reached an appropriate clonal age, will become sexually reactive on mild starvation and pair with cells of compatible mating types to initiate conjugation, a sexual process (Fig. 1B). If no compatible partner is available, some species will undergo a self-fertilization process called autogamy. In either case, nuclear reorganization ensues, starting with meiosis of micronuclei (i.e., containing the germline genome). The sequence of nuclear events is similar, with some variation, among all species and is depicted in Figure 1B for T. thermophila (Ray 1956; Sonneborn 1975; Martindale et al. 1982).
Postmeiotic development starts with the selection of a single haploid nucleus in each cell to pass on the genome. The selected nucleus undergoes an additional replicative division that produces two genetically identical gametic nuclei (Fig. 1B, step b). In the case of conjugation, the two mates exchange one of their two haploid nuclei, and subsequent karyogamy (i.e., the fusion of two haploid nuclei) therefore generates genetically identical zygotic nuclei in each conjugant (Fig. 1B, steps c,d). In ciliates that perform autogamy, two gametic nuclei within a single cell fuse to produce an entirely homozygous diploid genome. In either case, the resulting diploid zygotic nucleus (Fig. 1B, step d) divides twice more, and the four products differentiate, two into new micronuclei and two into new macronuclei (Fig. 1B, step e). On completion of nuclear development, cells return to vegetative growth with a special first caryonidal division (Fig. 1B, step f) to distribute new micro- and macronuclei to the two daughter cells of each conjugant, whereas the parental macronuclei are discarded (Davis et al. 1992).
3. MACRONUCLEI AND MICRONUCLEI: A MODEL FOR ACTIVE VERSUS SILENT CHROMATIN
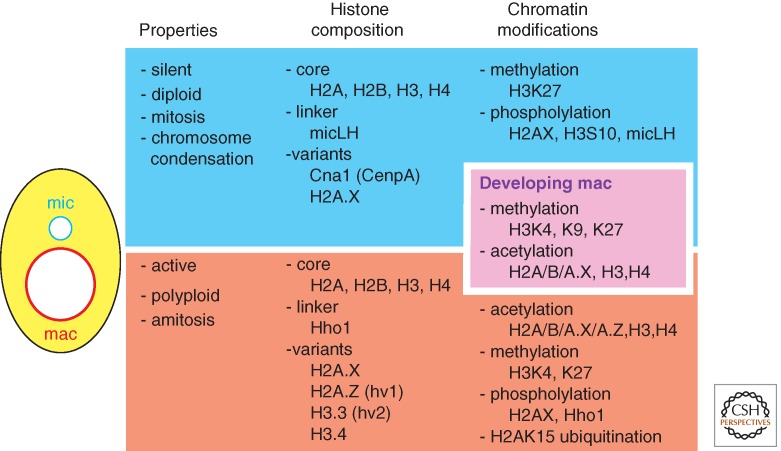
The basic concept of epigenetics is that individual copies of a DNA sequence can possess different activities and such differential states can be stably maintained. The nuclear dimorphism of ciliates is a natural example of homologous sequences that are maintained in a common cytoplasm, yet possess opposite activity states. The macronucleus serves as a model for the transcriptionally active state, the micronucleus for the repressed or silent state (Fig. 2). Early biochemical and immunohistochemical studies, primarily in Tetrahymena, compared properties of these different nuclei. Both histone variants and chromatin modifications were found to correlate with these different activity states and revealed the importance of chromatin structure in epigenetic regulation.
Figure 2.
Nuclear dimorphism of ciliates. The germline micronucleus (mic), the developing macronucleus (mac), and the somatic macronucleus contain different histone complements and modifications. Those known to occur specifically in each or in the developing somatic genome are listed.
3.1. Macro- and Micronuclei Contain Distinct Histone Variants
Isolation of histone proteins separately from the macronucleus and micronucleus of Tetrahymena identified some of the first histone variants. Histone variant (hv) 1 and hv2, which correspond to the common variants H2A.Z and H3.3, respectively (see Henikoff and Smith 2014), were detected exclusively within macronuclei, although the major histones are also present in both macro- and micronuclei (Fig. 2). Because of their macronucleus-specific localizations, these variants were suggested to be important for maintaining transcriptional activity (Allis et al. 1980; Hayashi et al. 1984). The hv2 (H3.3) variant and the more recently identified histone H3 variant H3.4 were shown to be constitutively expressed, a property critical for its DNA replication-independent deposition into chromatin, and indeed these histone H3 isoforms serve as replacement histones after transcription-associated nucleosome removal (Yu and Gorovsky 1997; Cui et al. 2006). Consistent with this, studies with multicellular eukaryotes showed the association of H3.3 deposition with transcription (Ahmad and Henikoff 2002). Like many eukaryotes, Tetrahymena has the CenpA homolog Cnp1p, the centromere-specific histone H3 variant. Cnp1p only localizes to the micronucleus and is essential for the micronuclear chromosome segregation. Consistent with the fact that the macronucleus divides by amitosis, the macronucleus is devoid of Cnp1p (Cervantes et al. 2006; Cui and Gorovsky 2006).
In addition to the different complement of core histone variants, the macro- and micronucleus contain different linker histones, the macronuclear linker histone H1 (Hho1p) and the micronuclear linker histone (micLH; Allis et al. 1984; Wu et al. 1986; Hayashi et al. 1987). Neither linker histone gene is essential for cell viability, but individual gene knockouts result in an increase in volume of the nucleus in which each acts. Thus both are critical for full chromatin compaction, possibly by stabilizing higher order chromatin structure (Shen et al. 1995). Loss of the Hho1p also leads to changes in expression of specific genes implicating this linker histone in maintaining proper transcriptional regulation (Shen and Gorovsky 1996).
3.2. Chromatin Modifications Correlate with Activity States and Biological Processes
3.2.1. Acetylation
The hyperacetylation of histones in the macronucleus and lack of this modification from the micronucleus provided early evidence correlating this posttranslational modification with gene activation (Vavra et al. 1982). The enzymes that performed chromatin acetylation remained unknown in any organism until the mid-1990s when C. David Allis and coworkers purified the first type A (nuclear) HAT from Tetrahymena (Brownell and Allis 1995; Brownell et al. 1996). These researchers started with highly purified macronuclei to separate this activity from type B cytoplasmic HAT activity, and followed their purification using an in-gel assay in which purified histones were polymerized into the polyacrylamide gel matrix used to fractionate protein extracts. After electrophoresis, proteins were renatured and incubated with radiolabeled acetyl-CoA to reveal a polypeptide with an apparent molecular mass of 55 kDa that could incorporate the acetyl moiety into the histone matrix.
The real breakthrough came after microsequencing the purified protein and cloning the gene. This Tetrahymena HAT was found to be homologous to a well-characterized transcriptional regulator of baker’s yeast, the Gcn5 protein. Before this discovery, transcriptional activators were primarily thought to act by recruiting RNA polymerase to promoters, but this work established that transcriptional activators also possess enzymatic activity, modifying chromatin, or other transcriptional regulators, thus changing the state of the template. Quickly thereafter, known regulators of many different eukaryotes were shown to act as HATs.
3.2.2. Methylation
Histone methylation patterns are specific for particular nuclei or developmental stage indicating the different modification states have different biological roles (Fig. 2). Histone H3 lysine 4 (H3K4) methylation is restricted to the macronuclei in growing Tetrahymena cells (Strahl et al. 1999), which was one of the first observations linking this modification with transcriptional activity. Furthermore, the trimethylation of H3K4 (H3K4me3) was found on the same tail as multiple acetylation marks, revealing coupling between different modifications separately associated with active chromatin (Taverna et al. 2007). Interestingly, the mono- and dimethylation of H3K4 (H3K4me1/2) were not strongly linked to acetylation on the same histone tails. This indicates that the number of methyl groups added to the same lysine residue results in differential regulation (see Cheng 2014).
H3K9me2/3, which is mainly associated with heterochromatin silencing in other eukaryotes, and H3K27me3, which is associated with polycomb repression (see Grossniklaus and Paro 2014), are abundant in developing macronuclei. In particular, they are enriched on germline-limited sequences that are subsequently eliminated from the somatic macronuclear genome (Taverna et al. 2002; Liu et al. 2007). These germline-limited sequences include most repetitive sequences that other eukaryotes package as heterochromatin. The developmentally regulated establishment of these heterochromatin modifications provides a useful model with which to elucidate their targeting to specific sequences (described in detail in Section 8). H3K27me3 is also found in the macro- and micronuclei of vegetative cells (Liu et al. 2007; Taverna et al. 2007), but whether and how it contributes to the inactive state of chromatin in vegetative cells has not been explored.
3.2.3. Phosphorylation
Purification of 32P-radiolabeled histones from macro- and micronuclei showed that linker histones and core histones H2A and H3 are highly phosphorylated in Tetrahymena (Allis and Gorovsky 1981). Multiple sites of Hho1 are phosphorylated and this modification was shown to participate in the regulation of specific gene transcription (Mizzen et al. 1999). Using mutational analysis, Dou and Gorovsky found that this requirement for phosphorylation could be mimicked by the addition of negatively charged amino acids into Hho1 (Dou et al. 1999). The charged residues, however, did not need to be present in the corresponding positions of the phosphorylated amino acid, but the complementary effect required a cluster of charged sites (Dou and Gorovsky 2000; Dou and Gorovsky 2002). These studies indicated that phosphorylation per se was not required, but that a critical charge density promotes proper transcription.
A single position, serine 10, is phosphorylated in histone H3 (H3S10ph; Wei et al. 1998). This modification is cell cycle–dependent and is correlated with mitosis in many eukaryotes. In Tetrahymena, it is restricted to micronuclei during mitosis and meiosis. Replacing the normal histone H3 gene with a mutant form containing an alanine substitution at serine 10 (S10A) causes defects in micronuclear division resulting in lagging chromosomes and aneuploidy (Wei et al. 1999). However, the H3S10ph modification does not occur in the macronucleus, thus the S10A mutation does not affect macronuclear amitotic division. These results showed that H3 phosphorylation plays an important role in chromosome condensation and/or segregation during mitosis. The unique nuclear dimorphism of the ciliate again revealed key insight into the role of a chromatin modification.
4. HOMOLOGY-DEPENDENT GENE SILENCING IN CILIATES
Homology-dependent, RNA-mediated silencing mechanisms are widely used in eukaryotes for epigenetic regulation (see Martienssen and Moazed 2014). One adaptation of these mechanisms in ciliates leads to the ultimate means of gene silencing, DNA elimination (Section 6). The role of homologous RNAs in these genome rearrangements is covered in Section 8. More conventional silencing mechanisms were discovered in Paramecium when nonexpressed transgenes introduced into the vegetative macronucleus elicited a phenotype similar to a Mendelian mutant occurring in an endogenous gene homologous to the transgene. Feeding cells bacteria-expressing double-stranded RNA that is homologous to an endogenous gene results in its silencing, implicating RNAi-related mechanisms as mediators of homology-dependent silencing (Galvani and Sperling 2002).
4.1. Transgene-Induced Silencing
The Paramecium macronucleus is easy to transform by microinjection as any introduced DNA fragment will be maintained, replicating autonomously without the need for a specific origin. Transformation with high-copy, nonexpressed transgenes triggers posttranscriptional silencing of endogenous genes that possess sufficient sequence similarity (Ruiz et al. 1998; Galvani and Sperling 2001). Silencing is inefficient if the 3′UTR of the gene is present in the transgene, which suggests that improperly terminated transcripts are recognized as aberrant RNAs and directed to the RNAi machinery (Galvani and Sperling 2001). Silencing correlated with the accumulation of homologous short RNAs of approximately 23 nt in length (Garnier et al. 2004), indicating that an RNAi pathway is involved. The ∼23-nt short RNAs appear to target degradation of homologous mRNAs and thus can be considered small interfering (si)RNAs. Endogenous siRNAs of 23–24 nt have been identified in growing Tetrahymena cells produced from loci that may be pseudogenes (Lee and Collins 2006).
This endogenous silencing pathway in ciliates requires an RNA-dependent RNA polymerase (encoded by RDR3 in Paramecium and RDR1 in Tetrahymena) for efficient siRNA production. In Tetrahymena, Rdr1p is associated with the Dicer ribonuclease Dcr2p, indicating that siRNAs generation is coupled with making double-stranded RNA (dsRNA; Lee and Collins 2007). In Paramecium, RDR3 is required for the accumulation of endogenous siRNAs, some of which are derived from the regions between convergent genes. In Schizosaccharomyces pombe, regions of convergent transcription between adjacent loci are also regulated by RNA silencing mechanisms. In gene dense genomes as in yeast and ciliate macronuclei, it may be critical to abrogate possible readthrough transcription. These endogenous silencing pathways serve as a critical means to check the expression of aberrant RNAs.
4.2. Silencing Is Induced by dsRNA
dsRNA is likely to be the primary trigger for transgene-induced silencing observed in Paramecium. The silencing efficiency of transgenes correlates with the production of aberrant RNA molecules that correspond to both the sense and antisense strands of the injected sequence. dsRNA can also promote gene silencing when introduced by feeding Paramecium cells Escherichia coli transcribing both strands of a cloned gene (Galvani and Sperling 2002) using methodology developed for Caenorhabditis elegans (Timmons and Fire 1998; Timmons et al. 2001). Feeding of this dsRNA led to the accumulation of ∼23-nt siRNAs, the same size as those observed on transgene-induced silencing (Nowacki et al. 2005), indicating that both phenomena rely on a common RNAi pathway. Feeding heat-killed E. coli to spirotrich species that normally feed on algae also promotes gene silencing (Paschka et al. 2003), suggesting that this mechanism in conserved throughout the ciliate lineage.
Silencing induced by feeding dsRNA to Paramecium can be reversed by replacing E. coli with the normal food bacterium in the culture medium; similarly, direct microinjection of dsRNA into the cytoplasm induces only transient silencing of the homologous genes, presumably because the injected dsRNA is rapidly diluted out during vegetative growth (Galvani and Sperling 2002). Thus, despite the RNA-dependent RNA polymerase(s) activity in vegetative Paramecium cells, dsRNA molecules cannot be sufficiently or continually amplified to establish a heritable silencing state, unlike the apparent fate of dsRNA in C. elegans. Furthermore, heritable silencing may require H3K9 methylation-mediated transcriptional gene silencing, which is apparently absent from the vegetative macronucleus, at least in T. thermophila.
5. GENOME-WIDE REARRANGEMENTS OCCUR DURING MACRONUCLEAR DEVELOPMENT
Chromosomes of the macronucleus and micronucleus do not just differ in transcriptional activity and chromatin modifications as described in Section 3 (Fig. 2), but show several fundamental differences is size and ploidy: (1) The size and complexity of the macronuclear genome is reduced relative to that found in the micronucleus; (2) macronuclear chromosome number (per haploid genome) is greater; but (3) macronuclear chromosomes are significantly shorter than micronuclear chromosomes; and (4) the arrangement of some gene segments is scrambled in the micronucleus relative to the “correct” order found in the macronucleus that is needed for proper gene expression. The first three differences are common among ciliates species, whereas the fourth has, so far, only been documented in spirotrichous ciliates, including Stylonychia and Oxytricha.
The structural differences between these nuclei result from programmed genome rearrangement events that transform the germline micronuclear genome into the somatic genome during macronuclear development. The reduction in genome size of the macronucleus, relative to the micronucleus, results primarily from DNA elimination (Section 6.1), whereas the increase in chromosome number and shortening of chromosomes is the product of chromosome fragmentation (Section 6.2). The scrambled order of gene segments in the micronucleus is corrected in the macronucleus by DNA elimination events, which illustrates the extreme degree of genome reorganization that occurs in some ciliates (Section 6.3). These DNA rearrangement events are described below to provide the background for the discussion of the epigenetic mechanisms that operate during differentiation of these nuclei.
5.1. DNA Elimination
Programmed DNA elimination (Fig. 3) creates a gene-condensed somatic genome in the macronucleus, reducing its size and sequence complexity significantly relative to the genome found in the germline micronucleus. For example, the macronuclear genome of Tetrahymena is only ∼100 Mb, whereas its micronuclear genome is ∼150 Mb (Eisen et al. 2006; Coyne et al. 2008). The amount of DNA eliminated varies greatly between ciliate species. Upward of 20%–30% of the micronuclear DNA is removed in the oligohymenophorean ciliates Paramecium and Tetrahymena, whereas more than 95% of the germline genome is eliminated from macronuclei of spirotrichous ciliates, such as Euplotes, Stylonychia, and Oxytricha (reviewed in Jahn and Klobutcher 2002). The sequences eliminated are enriched in transposon-related repetitive sequences, but also include many single-copy, nongenic sequences. As a result, the macronuclear genomes become highly gene rich: 1 gene per ∼4 kb in Tetrahymena, ∼2 kb in Paramecium, and ∼2.5 kb in Oxytricha. This “streamlined” organization is consistent with genomes that are optimized for efficient gene expression.
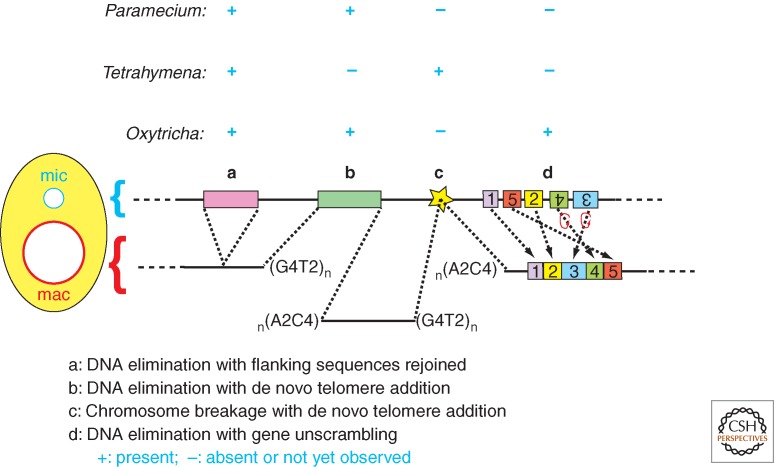
Figure 3.
Ciliate DNA rearrangements. The four classes (a–d) of DNA rearrangements that occur during development of a new macronucleus are illustrated as listed. The presence or absence of each class is indicated for the ciliate species Paramecium, Tetrahymena, and Oxytricha. Colored bars are internal eliminated sequences (IESs) removed by precise or imprecise deletions or alternatively by fragmentation coupled with telomere (G4T2)n addition. The conserved, 15-bp chromosome breakage sequence is denoted by the star. DNA unscrambling reorganizes the “exons” (numbered boxes) misordered in the micronucleus.
DNA elimination can be classified as three different types based on the precision and outcome of each event: imprecise deletions of internal DNA segments healed by the rejoining of flanking sequences (Fig. 3, class a); imprecise deletions of DNA segments followed by de novo telomere formation (Fig. 3, class b); and precise deletions followed by the rejoining of flanking sequences (Fig. 3, class a). One or more of these events have been observed in the commonly studied ciliates. The first is the predominant type observed in Tetrahymena, whereas all three are observed in Paramecium. The first and second types may result from alternative healing outcomes of imprecise excision events in this ciliate. Although precise deletion events are not common in Tetrahymena, they are the major rearrangement found in many ciliates.
The DNA segments removed from the developing macronuclear genome are commonly called internal eliminated sequences (IESs). Double-strand DNA breaks are generated at the boundaries of each IES and the flanking macronuclear-destined sequences (MDSs). These are then healed by ligation (Fig. 3, class a) or by de novo telomere formation (Fig. 3, class b). In Tetrahymena, the removal of ∼6000 IESs (per haploid genome) results in primarily imprecise deletion. In other words, the resulting macronuclear junctions formed by independent excision events can vary in position over tens of base pairs in different individuals and even for different copies of a chromosome in a same macronucleus. The IES/MDS boundaries often contain short (1–8 bp) direct repeats of variable sequence, one of which is retained in the macronuclear sequence. Although these repeats are not essential for excision, they may contribute to the precision of the joining process (Godiska et al. 1993).
Precise deletions of IESs occur at the same nucleotide positions in all copies of a macronuclear chromosome. This class of IESs is short (∼25–900 bp in Paramecium), single-copy, noncoding DNA segments. There are >45,000 such IESs per haploid genome of Paramecium; they are abundant within coding sequences, but also distributed throughout intergenic or intronic regions of germline chromosomes (reviewed in Jahn and Klobutcher 2002; Arnaiz et al. 2012). These precisely excised IESs, the so-called “TA” IESs are identified as having invariable 5′-TA-3′ repeat at each boundary, one copy of which remains within the macronuclear locus after excision (see Betermier 2004). The few nucleotide positions internal to the TA dinucleotides form a loosely conserved consensus (5′-TAGYNR-3′) in Paramecium (Klobutcher and Herrick 1995).
Both precise and imprecise deletions occur reproducibly: The same DNA segments are eliminated in different individuals, generation after generation; however, alternative choices at boundaries are observed for some IESs of both types. Besides the short, variable direct repeats, no common sequence has been identified in or around the eliminated DNA segments. So, how do Tetrahymena cells identify ∼6000 IESs? How do Paramecium cells precisely excise ∼60,000 IESs? These questions are addressed in Sections 8 and 9.
5.2. Chromosome Fragmentation
Chromosome fragmentation occurs during macronuclear development resulting in macronuclei with more individual chromosomes than the micronuclei. Taking Tetrahymena as an example, each diploid micronucleus has five chromosome pairs whereas each macronucleus has nearly 200 different chromosomes (per haploid genome equivalent). Fragmentation is followed by the de novo addition of telomeric repeats to stabilize the shorten chromosomes. In many spirotrich ciliates, such as Euplotes, Stylonychia, and Oxytricha, chromosome fragmentations are so extensive that most macronuclear chromosomes are “gene-sized” nanochromosomes that possess a single open reading frame. In contrast, macronuclear chromosomes of Tetrahymena and Paramecium range in size from 20 kb to >1 Mb and contain many genes.
The recognition of chromosome fragmentation sites is not well understood. In Tetrahymena, a conserved 15-bp chromosome breakage sequence (CBS; Yao et al. 1987) is found at ∼200 loci within micronuclear chromosomes (Cassidy-Hanley et al. 2005; Hamilton et al. 2005) and is both necessary and sufficient to direct fragmentation and new telomere addition (Fig. 3, class c) (Fan and Yao 1996). Similarly, a weak consensus sequence is found near chromosome fragmentation sites of Euplotes (Klobutcher 1999). The presence of CBS and CBS-like sequences probably indicates that the sites of chromosome fragmentation are genetically programmed in these ciliates. In contrast, in Paramecium, all chromosome fragmentation may result from imprecise DNA elimination that is healed by telomere addition instead of joining of dsDNA breaks (Fig. 3, class b) (Le Mouël et al. 2003). This process is epigenetically regulated and is discussed in Section 7.1.
5.3. DNA Unscrambling
An amazing variation to precise DNA elimination has been observed in some spirotrich ciliates in which IES removal occurs simultaneously with DNA “unscrambling” (Fig. 3, class d) (Prescott 1999). In the micronuclear versions of scrambled genes, coding regions are not only interrupted by IESs, but are also disordered relative to the linear arrangement found in the reorganized macronuclear sequence. Two MDSs that will be joined to form an expressed gene can be located far apart, sometimes found in an inverted orientation relative to each other, and can even be found at different chromosomal loci. It has been estimated that as much as 20%–30% of genes in Stylonychia and Oxytricha are scrambled in the micronucleus.
The precision of reordering appears to be guided, at least in part by relatively long (11 bp on average) homologous repeats, shared by cognate MDS ends, which are unrelated in sequence to those at other MDS ends. These long repeats have been found at all scrambled sites so far studied and thus are likely critical for accurate unscrambling. Yet, these sequences are not unique to MDS/IES junctions, and this has led to the proposal that a template specifying the correct order may be involved (Prescott et al. 2003). Indeed, it has been shown that long RNA templates derived from the parental macronucleus direct DNA unscrambling in Oxytricha (Nowacki et al. 2008). The presumed mechanism of RNA template-directed DNA unscrambling process is explained in Section 11.
5.4. Transposases Are Required for DNA Elimination
Many DNA segments eliminated from the new macronucleus have similarity to transposable elements (e.g., Herrick et al. 1985; Baird et al. 1989; Wuitschick et al. 2002; Fillingham et al. 2004), and the boundaries of “TA” IESs in Paramecium and Euplotes resemble the termini of Tc1/mariner transposons found in virtually all eukaryotes (Klobutcher and Herrick 1997). This has spurred the hypothesis that DNA elimination is derived from transposon excision. Indeed, it has been shown that transposases have been recruited to mediate DNA elimination in Oxytricha, Paramecium, and Tetrahymena (Baudry et al. 2009; Nowacki et al. 2009; Cheng et al. 2010).
In both Paramecium and Tetrahymena, the DNA excision step of DNA elimination is initiated by a DNA double-strand break (DSB), producing a 4-base 5′ overhang (Saveliev and Cox 1995; Saveliev and Cox 1996; Gratias and Betermier 2003). In Paramecium, the overhang is centered on the 5′-TA-3′, which is commonly observed at both ends of IESs. This structure is similar to that of DSBs produced by the piggyBac transposase: 4-base 5′ overhangs carrying the duplicated 5′-TTAA-3′ (Mitra et al. 2008). Both Paramecium and Tetrahymena have genes encoding piggyBac transposase-related proteins, Pgm and Tpb2p, respectively. RNAi knockdown of these genes indicated that they are required for DNA elimination (Baudry et al. 2009; Cheng et al. 2010). Like the piggyBac transposase, recombinant Tpb2p shows endonucleolytic cleavage activity producing DNA DSBs possessing 4-base 5′ protruding ends (Cheng et al. 2010). Although Pgm and Tpb2p are structurally similar to piggyBac transposases, the genes encoding these proteins are retained in the macronucleus. In contrast, most of the transposon-related sequences are limited to the micronuclear genome. Therefore, the genes encoding these transposase-related proteins have been domesticated within their host genomes during ciliate evolution.
In Oxytricha, the telomere-bearing element (TBE) family transposases are necessary for DNA elimination (Nowacki et al. 2009). In contrast to the piggyBac transposase-related proteins in Paramecium and Tetrahymena, TBE transposases are encoded in TBE transposons in the micronuclear-limited sequences in Oxytricha. Therefore, although Paramecium and Tetrahymena perform DNA elimination using domesticated transposases, Oxytricha uses transposases in the (potentially) active transposons to achieve DNA elimination. The TBE transposases and the piggyBac transposases are evolutionarily not directly related. Therefore, different classes of ciliates may have independently acquired their abilities to use transposon-derived enzymes for DNA elimination. This hypothesis suggests that the mechanisms of DNA elimination of oligohymenophorean (including Paramecium and Tetrahymena) and spirotrich (including Oxytricha) ciliates may be evolutionarily unrelated.
6. DNA ELIMINATION IS GUIDED BY HOMOLOGY-DEPENDENT MECHANISMS
Sequence-specific information can be communicated between macronuclear genomes of different generations to influence development. This comes from extensive experimental evidence showing that patterns of DNA elimination from a new macronucleus are epigenetically regulated by the DNA sequence of the preexisting, already reorganized genome of the parental macronucleus. In this section, we introduce classic examples of this regulation, which is followed in Section 8 with an explanation of how these epigenetic phenomena can be mediated by RNAi-related mechanisms.
6.1. Epigenetic Inheritance of Alternative Rearrangements
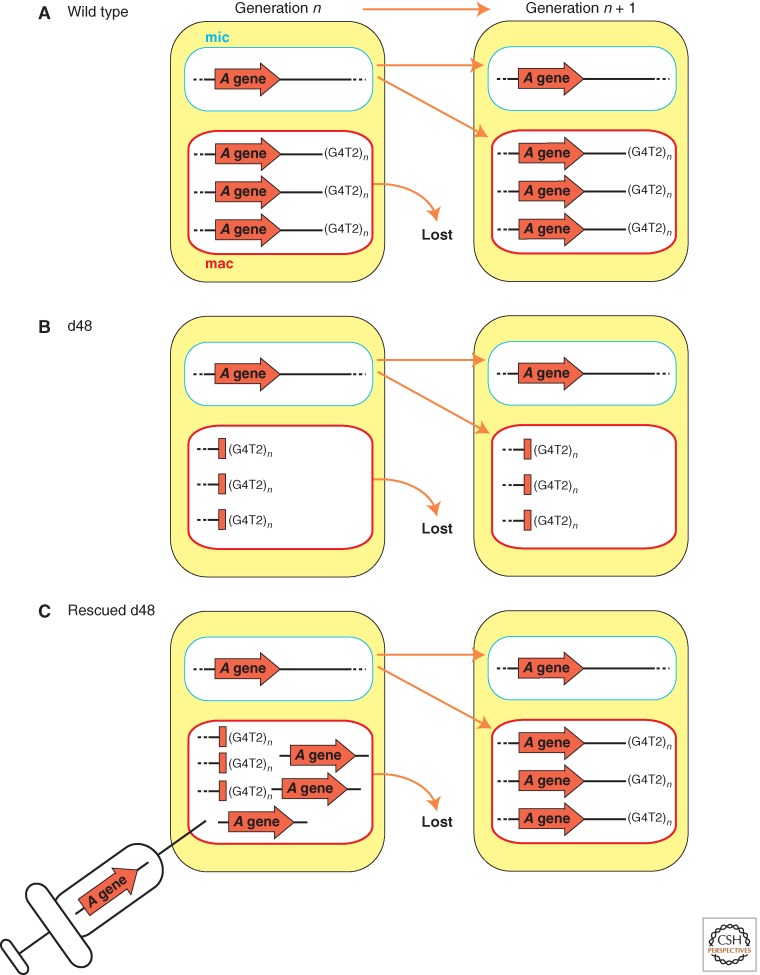
The first evidence that DNA rearrangements are subject to epigenetic regulation was uncovered when an aberrant DNA elimination pattern occurring in the previous sexual generation was shown to be inherited in new macronuclei of subsequent generations in Paramecium (Fig. 4). In wild-type macronuclei, the A surface antigen gene is located near a chromosome end, as chromosome fragmentation normally occurs downstream of this gene (Fig. 4A). A variant cell line, called d48, was found that failed to express the A gene because it lacked the gene itself, along with all downstream sequences in the macronucleus, as chromosome fragmentation placed the telomere at the 5′ end of the gene (Fig. 4B,left) (Forney and Blackburn 1988). This mutation was exclusive to the somatic macronucleus as the d48 strain micronucleus, when transplanted into a wild-type cell, gave rise to new macronuclei containing the full A gene after autogamy (Harumoto 1986; Kobayashi and Koizumi 1990). When the d48 strain was mated with a wild-type line, or allowed to undergo autogamy, the A gene of the macronucleus in the next generation was deleted (Fig. 4B,right) (Epstein and Forney 1984; Forney and Blackburn 1988). Reintroducing the A gene into the d48 macronucleus rescued the defect in A gene propagation during development (Fig. 4C). This restored, normal elimination pattern was maintained in the subsequent rounds of sexual reproduction (Koizumi and Kobayashi 1989; Jessop-Murray et al. 1991; You et al. 1991). These results indicated that the mere absence of the A gene from the parental macronucleus is sufficient to direct its future elimination from the new macronucleus. Similar experiments that focused on another telomere-proximal surface antigen gene, the B gene, showed that such epigenetic effects can be observed in other genomic regions (Scott et al. 1994).
Figure 4.
Epigenetic inheritance and experimental rescue of macronuclear A gene deletions. (A) Wild-type strain. (B) The d48 strain lacks the A gene in its macronucleus, but has a wild-type micronucleus. The A gene is reproducibly deleted during macronuclear development in each generation. (C) Transformation of the d48 strain macronucleus with A gene sequences restores amplification of the germline A gene in the developing macronucleus of sexual progeny.
The maternal macronuclear genome’s influence on DNA rearrangements showed marked sequence specificity. The A and B gene coding sequences are 74% identical overall. Nevertheless, injection of the A gene sequences into the macronucleus of a cell line that carried macronuclear deletions of both genes could only prevent A gene deletion from the new macronucleus, and similarly, injection with the B gene could only prevent its own deletion (Scott et al. 1994). In addition, the macronuclear deletion of A gene in the d48 strain could not be reverted by introduction of the G gene from another species of Paramecium, Paramecium primaurelia, that shares 78% identity with the A gene. On the other hand, the macronuclear deletion of the A gene could be rescued by transformation with a different allele of the A gene having 97% identity (Forney et al. 1996). Thus, the maternal rescue of macronuclear deletions is a homology-dependent process that requires a minimum level of sequence identity.
These studies of inheritance of Paramecium surface antigen genes suggest that the germline-derived genome to be rearranged in a newly forming macronucleus is compared to the existing parental macronuclear genome that was rearranged in the previous sexual reproduction. The result of this comparison is that any sequences that are absent from the parental macronucleus are targeted for elimination in the next generation. Two further predictions can be made from these conclusions: (1) Excision of an IES could be perturbed in the next generation if a IES copy is introduced into the parental macronucleus; and (2) any DNA sequences introduced only into the micronucleus may be targeted for elimination from the new macronucleus. Both are indeed the case (see Sections 7.2 and 7.3 below).
6.2. Homology-Dependent Inhibition of IES Elimination
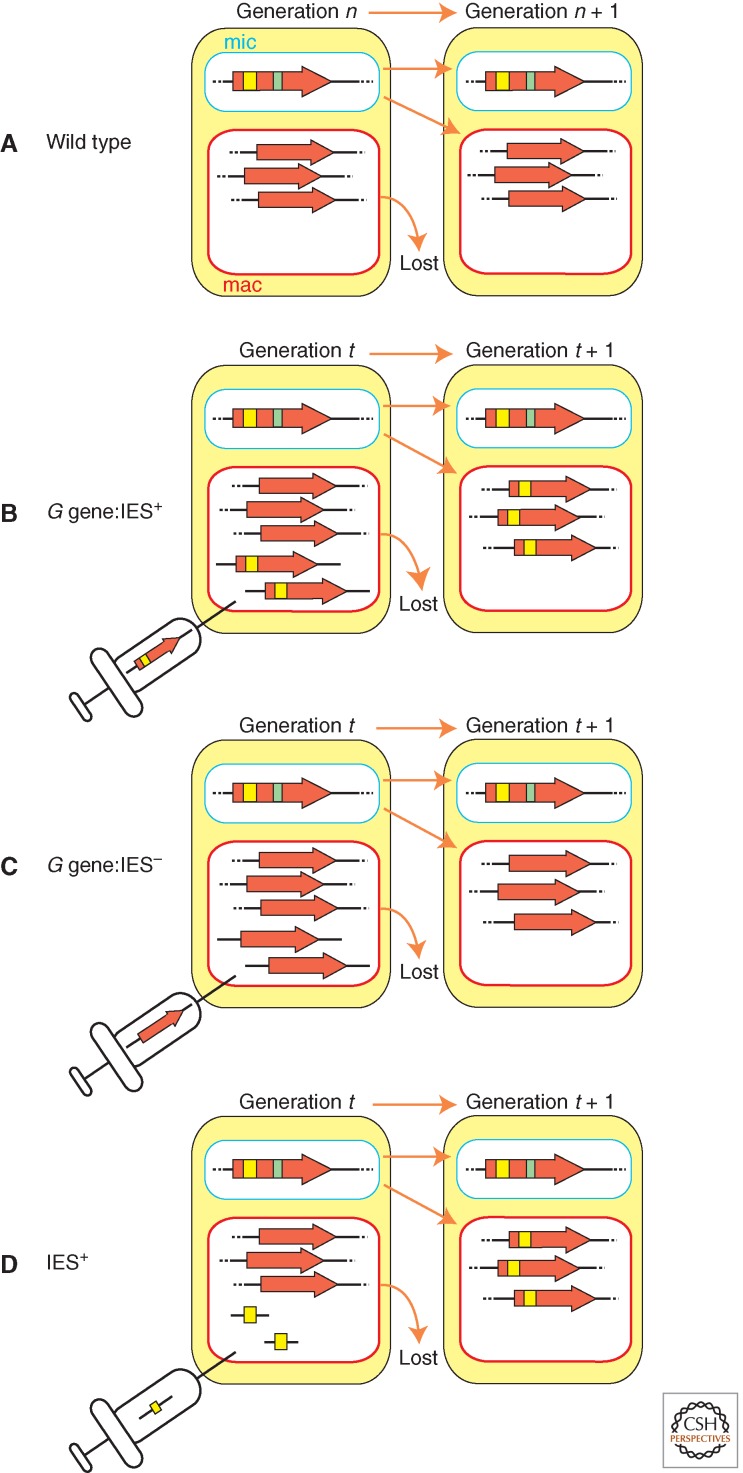
As illustrated in Figure 5, when wild-type Paramecium cells were transformed by microinjection of DNA containing a fragment of the G coding sequence that includes an IES, the transformed clones produced progeny lines that retained the IES in their newly formed macronuclear chromosomes (compare Fig. 5A and B). In contrast the progeny of cells transformed with the G coding sequence without the IES efficiently eliminated the IES from the new macronucleus (Fig. 5C) (Duharcourt et al. 1995). Transformation with DNA containing only the IES without any flanking sequences also caused IES retention showing that the IES copies in the parental macronucleus alone inhibit its excision from zygotic macronuclei (Fig. 5D) (Duharcourt et al. 1995). However, not all IESs are controlled by similar maternal effects in Paramecium. Excision of only five out of 13 IESs in the G or A surface genes were inhibited by introduction of the corresponding IESs into the parental macronucleus (Duharcourt et al. 1998). Therefore, IESs are of two types: those for which elimination is maternally regulated and those not affected by the genome content of the parental macronucleus.
Figure 5.
Homology-dependent inhibition of IES excision by the maternal macronucleus. (A) During development of wild type, IES (yellow and green bars) are excised efficiently. (B) Transformation of the maternal macronucleus with a high copy number of an IES (initial transformants = Generation t) can inhibit the elimination of the homologous IES during the subsequent (Generation t + 1, and future) rounds of new macronuclear differentiation. (C) Similar transformation with the IES– form of the gene does not change the wild-type rearrangement. (D) Transformation with IESs lacking flanking sequences results in inhibition of IES excision.
Experiments analogous to those above revealed that DNA within the Tetrahymena parental macronucleus can regulate the elimination of the homologous sequences from the developing macronuclear genome. The well-characterized M and R IESs were introduced into the macronuclei of wild-type strains. As per the results in Paramecium illustrated in Figure 5D, when these Tetrahymena cells were induced to conjugate, the progeny of the cells containing copies of the M or R IESs in the parental macronuclei failed to efficiently eliminate the chromosomal copies of the M or R IESs, respectively, during macronuclear development (Chalker and Yao 1996). The inhibition of DNA elimination was sequence-specific as significant inhibition of excision of nonhomologous IESs was not observed. As observed in Paramecium, sequences homologous to the IES itself were sufficient for this inhibition and the immediately flanking DNA had no effect. Importantly, this induced failure of DNA elimination was heritable as subsequent generations also retained genomic copies of the IES in their macronuclei. It is not clear whether Tetrahymena has any nonmaternally controlled IESs like Paramecium does.
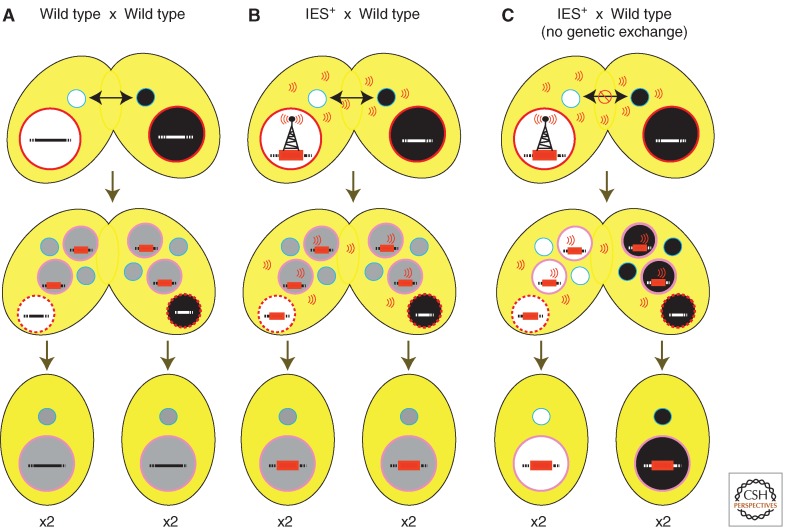
Because the macronuclear and micronuclear chromosomes do not directly interact, the “signal” that mediates the inhibitory effect of the introduced IESs in the parental macronucleus must be transmitted to the new macronucleus through cytoplasm. Tetrahymena conjugating pairs extensively share cytoplasm. Even when one strain possessing the IES in its parental macronucleus was crossed to a wild-type partner, elimination of the IES in all new macronuclei, including those that differentiated within the cytoplasm of the wild-type partner, was blocked (compare Fig. 6A and 6B) (Chalker et al. 2005). Exchange of germline genomes (see Fig. 1B) is not required to transmit the inhibitory signal from one mating partner to another as blocking reciprocal exchange of micronuclei between pairs did not block the transmission of the inhibitory signal (Fig. 6C) (Chalker et al. 2005). Therefore, the influence of the parental macronuclear genome is transmitted through factors able to freely move through the cytoplasm.
Figure 6.
Inhibition of IES excision by the parental macronucleus is mediated by factors transmitted through the cytoplasm. (A) During macronuclear differentiation following mating of wild-type cells, IESs (e.g., the red bar) are excised efficiently; however, (B) the presence of an IES (IES+) in the maternal macronucleus of one partner can signal its presence (depicted by radio tower emissions) to the developing macronuclei within both partners, inhibiting excision of the homologous IESs in all four resulting progeny. (C) The IES+ maternal macronucleus can inhibit IES excision in its wild-type mating partner even when genetic exchange is blocked.
6.3. “Spontaneous” Elimination of Foreign Sequences Introduced into the Micronuclear Genome
In Tetrahymena, when the neo gene is integrated into a micronuclear chromosome, it may be deleted from the genome of new macronuclei during successive rounds of conjugation (Yao et al. 2003; Liu et al. 2005; Howard-Till and Yao 2007). The neo gene is derived from the bacteria transposon Tn5, and thus is unlikely to contain by chance some specific signal to spontaneously induce its own elimination. Therefore, this result presents the possibility that any “foreign” sequence introduced into the micronuclear genome of Tetrahymena may be recognized as an IES.
Deletion of the neo gene is generally inefficient relative to the nearly 100% elimination observed for naturally occurring IESs. Furthermore, the efficiency of neo elimination from different micronuclear loci varied significantly (Liu et al. 2005; Howard-Till and Yao 2007). Not only did the genomic environment surrounding the neo gene influence elimination of this transgene, but multiple neo insertions into the micronuclear genome enhanced its DNA elimination (Liu et al. 2005; Howard-Till and Yao 2007). Thus, both the presence of a foreign sequence in the micronucleus as well as its repetitiveness in the genome affected DNA elimination. These position and copy number effects on transgene elimination will be discussed in Section 8 in the light of small RNA-directed regulation of DNA elimination mechanism. More generally, neo gene elimination suggests that IESs are recognized by more than sequence alone.
7. DNA ELIMINATION IS GUIDED BY SMALL RNA-DIRECTED TRANS-NUCLEAR COMPARISON OF WHOLE GENOMES
The homology-dependent effects described in Section 6 show that a cross talk occurs between the genomes of the parental macronucleus and the new macronucleus during nuclear differentiation, which can profoundly alter DNA elimination patterns and cellular phenotypes. The observations that only highly homologous sequences are affected suggest strongly that this cross talk is mediated by nucleic acids. As discussed in Sections 7.2 and 7.3, it is likely that the interplay between parental macronuclear transcripts and short RNAs derived from the meiotic micronucleus ultimately directs DNA elimination.
7.1. Linking Short RNAs to DNA Elimination
An initial indication that DNA elimination uses homologous RNA molecules was the observation that Tetrahymena IESs were bidirectionally transcribed early during conjugation (Chalker and Yao 2001). This bidirectional nature of transcription also suggested the potential involvement of an RNAi-related mechanism in DNA elimination. Indeed, it was shown that an Argonaute family protein encoded by TWI1 is exclusively expressed during conjugation and is required for DNA rearrangement (Mochizuki et al. 2002). Argonaute proteins play key roles in RNAi-related processes by interacting with small RNAs (approximately 20–30 nt) that target Argonaute proteins to their complementary RNAs for gene silencing (Ghildiyal and Zamore 2009). Twi1p interacts with a species of small (∼29 nt) RNA that is exclusively expressed during conjugation in Tetrahymena (Mochizuki et al. 2002; Mochizuki and Gorovsky 2004). Disruption of the TWI1 gene destabilizes these small RNAs and abolishes DNA elimination in the developing macronucleus (Mochizuki et al. 2002), indicating that these small RNAs play a pivotal role. The ∼29-nt small RNAs are named scan (scn) RNAs because, as explained in Section 9.2, they “scan” the genomes for complementary sequences to direct DNA elimination.
Characterization of a gene, DCL1, encoding a Dicer-like ribonuclease further exposed the importance of these small RNAs in DNA elimination (Malone et al. 2005; Mochizuki and Gorovsky 2005). Dcl1p is expressed at high levels early in conjugation and localizes to premeiotic micronuclei, which indicated that generation of scnRNAs is temporally and spatially compartmentalized. Disruption of DCL1 caused loss of scnRNA production, accumulation of the micronuclear transcripts, and ultimately failure of DNA elimination. These observations strongly support that scnRNAs are produced from the bidirectional micronuclear transcripts by Dcl1p and participate in DNA elimination.
In Paramecium, two Argonaute proteins, Ptiwi01 and Ptiwi09, and two Dicer proteins, Dcl2 and Dcl3, were identified as the functional counterparts of Tetrahymena Twi1p and Dcl1p, respectively (Lepere et al. 2009; Bouhouche et al. 2011). Double knockdown of the genes encoding these Argonaute or Dicer proteins causes both the loss of ∼25-nt small RNAs and defects in DNA elimination. Thus, despite clear differences in IES size and structure between these ciliates, the ∼25-nt small RNAs are Paramecium scnRNAs, and both Paramecium and Tetrahymena use an RNAi-related mechanism for DNA elimination.
7.2. Trans-Nuclear Comparison of Whole Genomes by Small RNAs
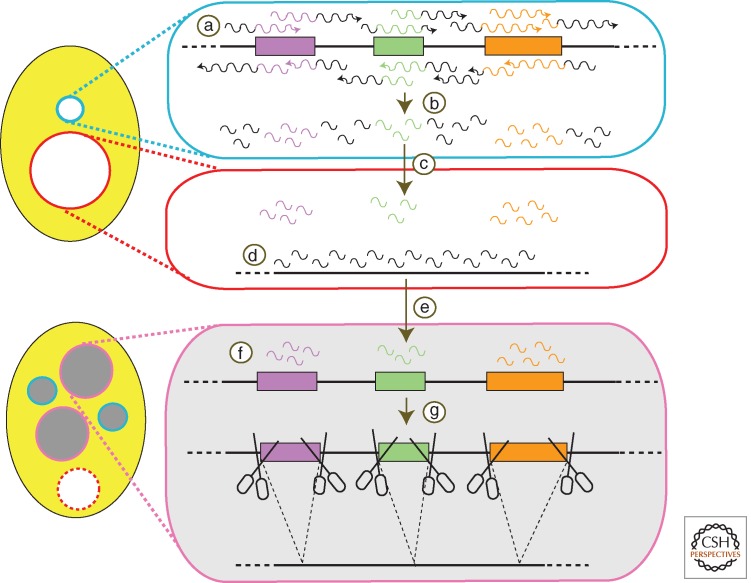
The presumed roles of scnRNAs in DNA elimination as well as how they may participate in mediating epigenetic phenomena has been described in several models (e.g., Mochizuki et al. 2002; Mochizuki and Gorovsky 2004; Aronica et al. 2008; Lepere et al. 2009). In the simplified model shown in Figure 7, the micronuclear genome is transcribed bidirectionally, randomly, and homogeneously (Fig. 7, step a) in early stages of conjugation. The transcripts produced form dsRNAs that are processed into scnRNAs (Fig. 7, step b). These scnRNAs are initially channeled to the parental macronucleus (Fig. 7, step c) in early–mid stages of conjugation. There they “scan” the existing rearranged genome for homology. The scnRNAs that pair with the parental macronuclear sequences are removed from the pool of active scnRNAs (Fig. 7, step d). This process is called “scanning” or scnRNA selection. The remaining scnRNAs, which are homologous to the micronuclear-limited sequences, are then transported to the developing macronucleus (Fig. 7, step e) where they target their complementary sequences for DNA elimination (Fig. 7, steps g and h) in late stages of conjugation.
Figure 7.
A template model for control of DNA deletion. Bidirectional transcription (a) of a large portion of the germline genome occurs early in development and leads to the production of scnRNAs (b). These are then transported into the maternal macronucleus (c) where any encounter homologous sequences (d), which triggers their removal from the active pool. The remaining micronucleus-specific RNAs are redirected to the developing macronucleus (e), where they mark homologous sequences (f), signaling their removal from the genome (g).
This model is supported by the following observations: (1) In Tetrahymena, although scnRNAs complementary to both MDSs and the micronuclear-specific sequences (mostly IESs) are produced in the early conjugation stage, only the latter scnRNA population becomes gradually enriched in the mid-conjugation stages (Mochizuki and Gorovsky 2004; Aronica et al. 2008); (2) in the mid-conjugation stages, after the bulk of scnRNA production, Argonaute proteins (Ptiwi01 and Ptiwi09 in Paramecium and Twi1p in Tetrahymena) localize initially in the parental macronucleus, then later move to the new macronucleus (Mochizuki et al. 2002; Bouhouche et al. 2011); and (3) the RNA-binding protein Nowa1/2, which is required for the elimination of maternally regulated IESs, is also translocated from the parental to new macronucleus in Paramecium (Nowacki et al. 2005).
The model explains most of the epigenetic effects described in Section 8, but does not account for the position and the copy number effects associated with transgene elimination (Section 7.3). If transcription, which produces the scnRNA precursors, occurs homogeneously from the meiotic micronuclear genome, a transgene should be efficiently eliminated regardless of its genomic location or copy number in that nucleus; but if this micronuclear transcription occurs nonuniformly, the genomic loci harboring transgenes will influence the likelihood of being eliminated. In Tetrahymena, scnRNAs are preferentially produced from IESs (Schoeberl et al. 2012); future efforts to understand what underlies biased scnRNA production will likely show why some transgenes are eliminated and others are not. In Paramecium, although Ptiwi01 and Ptiwi09 are required for the excision of both maternally controlled and nonmaternally controlled IESs, Dcl2 and Dcl3 are necessary only for the elimination of former class of IESs (Bouhouche et al. 2011). Therefore, scnRNAs may have some fundamental role in DNA elimination besides its epigenetic regulation, at least in Paramecium.
7.3. Involvement of the Parental Macronuclear Transcripts in DNA Elimination
To transmit sequence-specific epigenetic information from the parental to the new macronuclear genome, scnRNAs have to interact with these genomes by base pairing. In both fission yeast and plants, small RNAs target genomic site by interacting with cis-transcripts emerging from the loci. Two reports, one of Paramecium and one of Tetrahymena, suggest that scnRNAs interact with parental and developing genomes through long noncoding RNAs (ncRNAs) (Aronica et al. 2008; Lepere et al. 2008). In Tetrahymena, a study of the Twi1p-interacting, putative RNA helicase called Ema1p suggested that nascent noncoding transcripts mediate the interaction between chromatin and scnRNA–Twi1p complexes in the parental macronucleus and that this interaction is dependent on Ema1p (Aronica et al. 2008). Moreover, Ema1p-depleted strains showed defects in scnRNA selection and DNA elimination (Aronica et al. 2008). The importance of the parental macronuclear transcripts for proper IES elimination has been shown more directly in Paramecium (Lepere et al. 2008): down-regulation of noncoding transcripts from the parental macronucleus, by RNAi knockdown, blocked the scnRNA selection in the targeted regions and induced ectopic DNA elimination.
How scnRNA selection is achieved remains mostly speculation. One can hypothesize that the interaction between scnRNA and the parental macronuclear transcripts could remove the complementary scnRNAs from the pool by leading to their sequestration or degradation. Alternatively, scnRNAs that are not base paired with parental macronuclear transcripts could be selectively amplified. In Tetrahymena, northern blot hybridization studies of scnRNAs showed that those complementary to IESs were present at constant levels during early to late conjugation stages, whereas those complementary to MDSs were only detected in early stages (Aronica et al. 2008). These results suggest that the scnRNA selection process likely promotes scnRNA degradation, at least in Tetrahymena.
Although the interactions between scnRNAs and long noncoding RNAs within parental macronuclei result in scnRNA selection, similar interactions in the developing macronuclei likely induce DNA elimination. As we will explain in Section 10, these interactions effectively target heterochromatin formation on IESs, which appears to mark them for DNA elimination. The Twi1p-associated RNA helicase Ema1p is required for the interaction between Twi1p and nascent noncoding RNAs in the new macronucleus and the resulting heterochromatin formation (Aronica et al. 2008). It remains unclear how seemingly similar scnRNA–ncRNA interactions produce two dramatically different outcomes in different nuclei. The temporal (early vs. late) and spatial (parental vs. new macronucleus) separation of these different interactions may simply trigger alternative outcomes based on the downstream effectors recruited to the loci by the scnRNA–Argonaute complex.
8. scnRNA-INDUCED HETEROCHROMATIN FORMATION PRECEDES DNA EXCISION
In most eukaryotes, di- and trimethylated histone H3 lysine 9 (H3K9me2/3) is widely associated with transcriptionally repressed DNA that is partitioned in the nucleus as heterochromatin (see Section 10 of Allis et al. 2014). In Tetrahymena, H3K9me2/3 is not found in the transcriptionally silent micronucleus as one might presume, but is exclusively found in developing macronuclei, established immediately preceding DNA elimination, and particularly enriched on the histones associated with IESs (Taverna et al. 2002; Liu et al. 2007).
Different histone modifications are established on chromatin by specific “writers” and interact with different protein “readers” to enforce a regulatory state (see Cheng 2014; Patel 2014). In Tetrahymena, the writer for H3K9me2/3 appears to be the Polycomb group protein, enhancer-of-zeste-like (Ezl1p), a histone methyltransferase (Taverna et al. 2002; Liu et al. 2007). Ezl1p has been shown to be required for the accumulation of trimethylated H3K27 (H3K27me3) as well as H3K9me2/3 in the new macronucleus (Liu et al. 2007). The known readers for these modifications are the chromodomain-containing proteins Pdd1p and Pdd3p. Pdd1p binds to both H3K9me2/3 and H3K27me3, whereas Pdd3p only binds to H3K9me2/3 (Liu et al. 2007).
DNA elimination requires the function of Ezl1p and Pdd1p, in addition to the RNAi-related proteins described in Section 7.1 (Coyne et al. 1996; Liu et al. 2007). This shows that heterochromatin formation is a prerequisite of DNA elimination. Furthermore, Tetrahymena strains expressing a histone H3 mutant in which lysine 9 is replaced by glutamine (H3K9Q) are defective for DNA elimination (Liu et al. 2004). Although the requirement for H3K27me3 could not be assessed directly by K27Q, K27R, and K27A mutations because severe defects during meiosis and developmental arrest occurred before the appearance of new macronuclei, replacement of H3S28 to glutamic acid (H3S28E), which disrupted the H3K27me3-Pdd1p interaction, inhibited DNA elimination (Liu et al. 2007). Furthermore, this mutant abolished the accumulation of H3K9me3 (Liu et al. 2007), whereas conversely H3K27me3 accumulated normally in the H3K9Q mutant (Liu et al. 2004) suggesting that H3K27 methylation may be upstream of H3K9 methylation. This leaves open the possibility that an unknown methyltransferase, not Ezl1p, directs H3K9me. Tethering Pdd1p to an ectopic chromatin site using a LexA–LexA operator system is sufficient to trigger DNA elimination at that site (Taverna et al. 2002), indicating that localization of Pdd1p to chromatin is sufficient to recruit all of the downstream factors necessary for DNA elimination. Therefore, the structure of heterochromatin, but not the RNAi-related mechanism or H3K9/K27me, is the direct trigger for DNA elimination. The transposase, Tpb2p, discovered in Tetrahymena is localized to heterochromatic foci (Cheng et al. 2010) in which DNA elimination is believed to take place, suggesting that it may directly bind to some component of heterochromatin. Therefore, Tpb2p may directly recognize heterochromatin structure to catalyze DNA excision.
The RNAi-related pathway acts upstream of the establishment of heterochromatin modifications, as Dcl1p and Twi1p are required for the accumulation and/or targeting of H3K9me2/3 and H3K27me3, whereas the writer Ezl1p is not required for scnRNAs accumulation (Liu et al. 2004; Liu et al. 2007). It may be that Ezl1p is recruited to chromatin in which Tw1ip–scnRNA complexes are interacting with nascent long ncRNA in the developing new macronucleus. Because the scnRNAs that are complementary to the micronucleus-limited sequences are selected in the parental macronucleus by the scnRNA selection (Section 8.2), the scnRNA–ncRNA interaction is expected to specifically recruit Ezl1p to induce the histone modifications only on IESs. The molecular mechanism underlying this recruitment is unknown because no direct interaction between Twi1p and Ezl1p has been detected. Nonetheless, we know that scnRNAs are the specificity factors targeting heterochromatin modifications to the correct genomic loci. The importance of heterochromatin in DNA elimination of other ciliates has yet to be established. It is interesting to note that many IESs in Paramecium are shorter than the length of DNA wrapped around a nucleosome, and thus a heterochromatin-independent mechanism is likely involved in the elimination of at least such short IESs.
9. DNA UNSCRAMBLING IS TEMPLATED BY MATERNAL RNAs
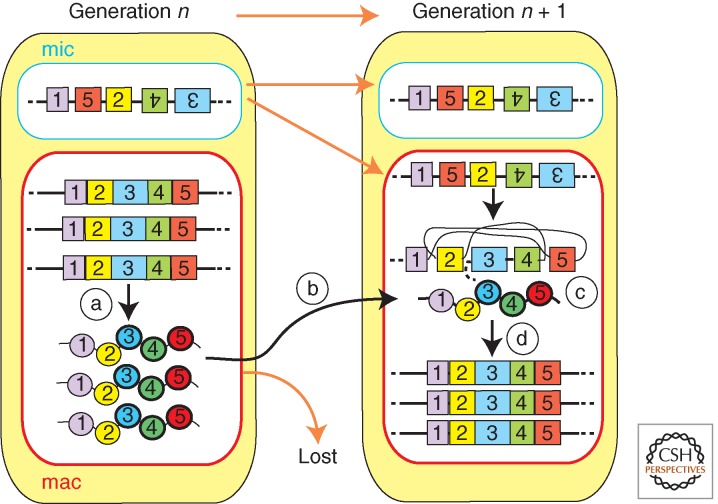
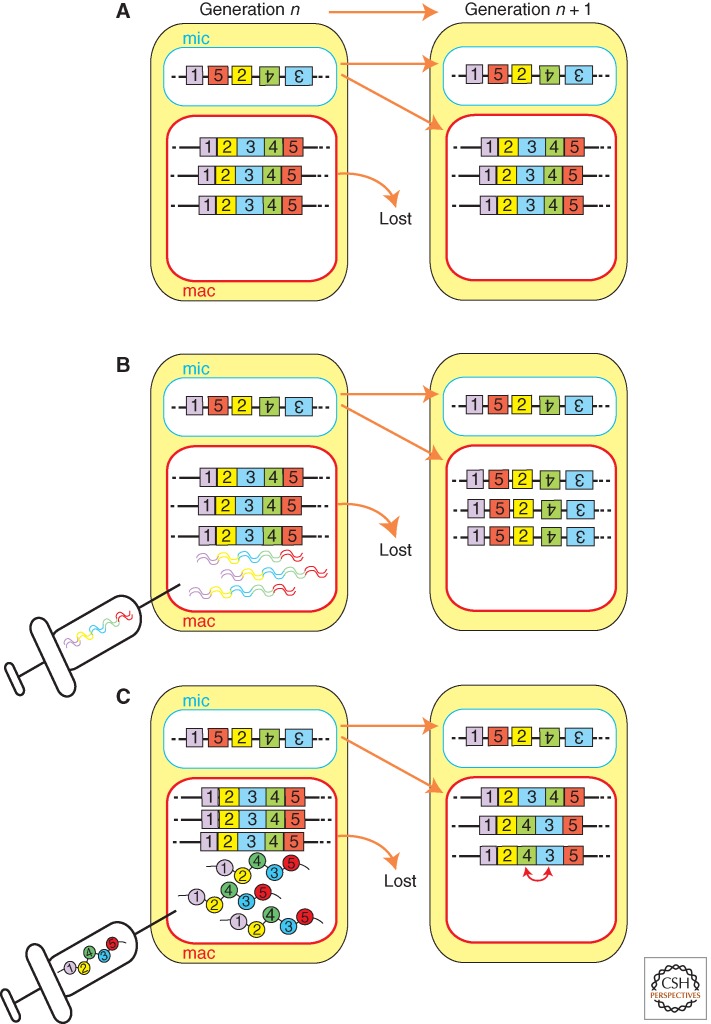
RNAs have roles in the DNA rearrangements of both oligohymenophorean and spirotrich ciliates; however, current evidence suggests that the mechanism(s) of RNA-directed DNA rearrangement of these two classes of ciliates may be fundamentally different. Although small, micronuclear RNAs direct DNA elimination in Paramecium and Tetrahymena; DNA elimination and DNA unscrambling (Figure 2D) of Oxytricha is regulated by long, macronuclear RNAs. Long RNA-guided DNA unscrambling was first proposed as a theoretical model (Fig. 8) (Prescott et al. 2003) and has since been shown experimentally (Fig. 9) (Nowacki et al. 2008). Transcription of the parental macronuclear genome occurs during early conjugation in Oxytricha (Nowacki et al. 2008). This transcription produces macronuclear long RNAs which likely span from telomere to telomere of “gene-sized” macronuclear nanochromosomes. RNAi knockdown of specific macronuclear long RNAs from a macronuclear nanochromosome, on injection of complementary dsRNAs, inhibited both DNA elimination and DNA unscrambling of the targeted loci in the new macronuclei of progeny cells (Fig. 9B). More strikingly, the injection of mutated artificial single-stranded RNAs was sufficient to reprogram the unscrambling order (Fig. 9C). Therefore, single-stranded long macronuclear RNAs can act as templates in DNA rearrangement in Oxytricha.
Figure 8.
The template model of DNA unscrambling. The exons (numbered boxes) of scrambled genes from the micronucleus (mic) are faithfully reordered during nuclear differentiation. Transcripts produced from the maternal macronucleus (a) are transported to the developing macronucleus (b) where any encounter with a homologous germline-derived scrambled gene (c) and template correct reordering of the DNA in the somatic genome of the progeny (generation n + 1) (d).
Figure 9.
Maternal RNAs template unscrambling. (A) Scrambled genes in the micronucleus (mic) must have their exons (numbered boxes) reordered during nuclear differentiation. (B) Injection of dsRNA (RNAi) homologous to transcripts produced from the maternal macronucleus perturbs both unscrambling and DNA rearrangement in the macronuclei of the progeny generation (n + 1). (C) Injection of scrambled RNA templates results in the parental macronucleus directing misordering of the homologous nanochromosome of the progeny.
Recently, it was reported that 27-nt small RNAs are accumulated during conjugation of Oxytricha (Fang et al. 2012; Zahler et al. 2012) and interact with the Argonaute protein Otiwi1 (Fang et al. 2012). Interestingly, these Otiwi1-bound small RNAs are produced from the parental macronucleus, not from the micronucleus as scnRNAs in Tetrahymena and Paramecium. Furthermore, injection of small RNAs complementary to an IES caused retention of the IES indicating that the Otiwi1-bound small RNAs in Oxytricha have an opposite role compared with scnRNAs in Tetrahymena and Paramecium: a protective role against DNA elimination (Fang et al. 2012). Although the division of labor between long macronuclear RNAs and Otiwi1-bound small RNAs in DNA rearrangement is currently unclear, Oxytricha clearly uses a fundamentally different strategy than Tetrahymena and Paramecium although these ciliates perform fundamentally similar DNA rearrangement processes.
The long macronuclear RNAs in Oxytricha have an additional surprising function in epigenetic genome regulation: These RNAs regulate not only the genome rearrangements, but also the chromosome copy numbers in the macronuclear development (Nowacki et al. 2010). In the macronucleus, chromosomes of the diploid zygotic nucleus are amplified by endoreplication. Although the copy number is the same for nearly all individual macronuclear chromosomes of Paramecium (∼800C) and Tetrahymena (∼45C), the copy numbers of different macronuclear chromosomes in Oxytricha can vary greatly, ranging from hundreds to tens of thousands. Because Oxytricha has more than 20,000 different macronuclear chromosomes per haploid genome (most containing a single gene), understanding the regulation of copy number has presented a challenge. It has been shown that the copy numbers of the new macronuclear chromosomes are influenced by the copy numbers of the corresponding chromosomes in the parental macronucleus. This copy number information is transmitted by the long macronuclear RNAs: More copies of a parental macronuclear chromosome produce more long macronuclear RNAs that correlate with the final copy number of that chromosome in the new macronucleus (Nowacki et al. 2010).
The mechanistic action of long macronuclear RNAs in directing DNA unscrambling and copy number regulation remains to be discovered. Their interactions with the developing somatic genome may facilitate the recruitment of TBE transposases to mediate DNA elimination/unscrambling. In turn, increasing the efficiency of DNA rearrangement may lead to increases in final chromosome copy number. Regardless of the actual molecular mechanism, the discovery that “RNA caches” participate in genome rearrangements undoubtedly broadens our horizons about the potential for RNAs to change the activity state of chromatin and even alter the physical structure of chromosomes.
10. BIOLOGICAL ROLES OF PROGRAMMED GENOME REARRANGEMENTS
DNA elimination occurs in all ciliates studied to date. Therefore, it must not be as wasteful a process as it may have seemed on its initial discovery, but likely offers important fitness advantages. Because many eliminated sequences are related to transposons, it has been suggested that DNA elimination is a defense mechanism against transposons (e.g., Yao et al. 2003; Fillingham et al. 2004). In this role, the epigenetic regulation of DNA elimination may act as a “genomic immune system” as discussed in Section 10.1. In addition, DNA elimination and DNA unscrambling may accelerate the emergence of new genes during the course of evolution, and even allow increase in the coding potential from specific loci by alternative “DNA splicing” (Fass et al. 2011). Moreover, the epigenetic regulation of DNA elimination/unscrambling may allow inheritance of acquired characteristics of the soma to future sexual generations.
10.1. DNA Elimination as a Genome Defense Mechanism
The studies of RNAi and related mechanisms in many different eukaryotes support the hypothesis that this pathway evolved as a cellular defense mechanism controlling the proliferation of viruses and transposons by degrading mRNAs, inhibiting translation, and/or targeting the formation of heterochromatin on these genomic invaders (Matzke and Birchler 2005). Transposable elements of ciliates are largely eliminated from the somatic macronucleus during development, which will effectively limit their impact. The finding in Tetrahymena that histone H3K9 and H3K27 methylation is small RNA-dependent and marks genomic regions destined for the programmed DNA elimination revealed that the role of RNAi in ciliate nuclear differentiation is fundamentally similar to its role in establishing heterochromatin in other eukaryotes; ciliates just go one step further and eliminate heterochromatin from their somatic genome.
The specific sequences that are targeted by small RNAs for heterochromatin formation followed by DNA elimination are selected by a global comparison of maternal germline and somatic genomes. This mechanism would efficiently protect against the deleterious effects of transposition in the germline: Any new transposon integrating into the germline would be recognized as alien by comparison with the somatic genome during sexual reproduction, leading to its removal from the transcribed somatic genome of progeny, thereby limiting its future spread.
It has been suggested that RNAi is a primitive immune system against parasitic elements such as viruses and transposons (Waterhouse et al. 2001). The elimination of DNA, mediated by an RNAi-related mechanism, is conceptually more similar to the cellular immune system in vertebrates, both of which learn the distinction between self and nonself. A huge repertoire of lymphocytes expressing different antibodies is initially generated, but in early development all lymphocytes that recognize available antigens (likely self-antigens) are eliminated from the future pool (explained in Busslinger and Tarakhovsky 2014). Once past this stage, the recognition of a cognate antigen (then likely to be alien) leads to the clonal expansion of the corresponding lymphocytes. By analogy, the DNA elimination mechanism can be considered to be a genomic immune system.
10.2. DNA Elimination and DNA Unscrambling as “DNA Splicing” Mechanism
DNA elimination occurs extensively from the coding regions. In this form, one can envisage DNA elimination removing “DNA introns” (= IESs) from the genome to assemble “DNA exons” (= MDSs), via “DNA splicing” events. Thus, like RNA splicing, IESs in the coding regions are removed with nucleotide level precisions. Because DNA unscrambling in Oxytricha is always accompanied with DNA elimination, this process is DNA splicing coupled with translocations and inversions of DNA exons. The biological consequences of DNA elimination and DNA unscrambling have obvious overlap with RNA splicing. RNA splicing offers two important advantages to cells. First, in evolutionary timescales, the presence of introns in DNA would allow genetic recombination to combine the exons of different genes and facilitate the emergence of new and useful proteins. Second, for individual organisms, the exon–intron organization of genes allows the same gene to produce multiple different proteins via alternative splicing. Thus, analogous to RNA splicing, DNA elimination/unscrambling has the potential to accelerate the emergence of useful genes over the course of evolution and allow individual loci to express multiple protein isoforms after alternative DNA elimination/rejoining.
The ability of DNA unscrambling, in which even translocations and inversions are possible, would greatly facilitate both evolutionary processes and alternative gene expression. Although alternative DNA elimination/unscrambling that produces different protein products has not been detected in ciliates, at least one case is known for which an IES in an exon results in alternative polyadenylation and splicing during development (Fass et al. 2011). Future large-scale genome sequence analyses will elucidate the extent of such outcomes.
The epigenetic regulation of DNA elimination and DNA unscrambling described in this article may also allow fixation of traits without alterations to the germline genome. As discussed above, DNA elimination/unscrambling can produce heritable variants of macronuclear chromosomes from a single micronuclear genome. Because the macronucleus is polyploid and its chromosomes are amitotically segregated during vegetative growth, variations in the macronuclear chromosomes can be selectively assorted based on positive or negative influences. So if a specific variant, or a combination of variants, is advantageous to the growth and/or survival of cells, it would dominate over other variants in the macronucleus during vegetative growth. Once a particular variant is fixed in the macronucleus, the pattern of DNA rearrangement in the next sexual generation can be biased toward this variant form by the epigenetic specification of DNA rearrangement patterns. This system could also work if an accidental rearrangement on macronuclear chromosomes during vegetative growth is beneficial to cells. In this way, acquired characters of somatic macronucleus can be inherited to the next sexual generations, and thus ciliates appear to naturally use a form of Lamarckian inheritance. Because the micronucleus retains the original genome, cells have a chance to return to an original character when the variant loses its advantage for survival owing to some environmental change.
11. EPIGENETIC REGULATION OF MATING-TYPE DETERMINATION
We finish this article by introducing yet another beautiful example of epigenetic inheritance in ciliates: the determination of mating type in Paramecium that was extensively studied by T.M. Sonneborn. Because the molecular mechanism behind this epigenetic inheritance has not been fully explained, we will let you imagine how it works.
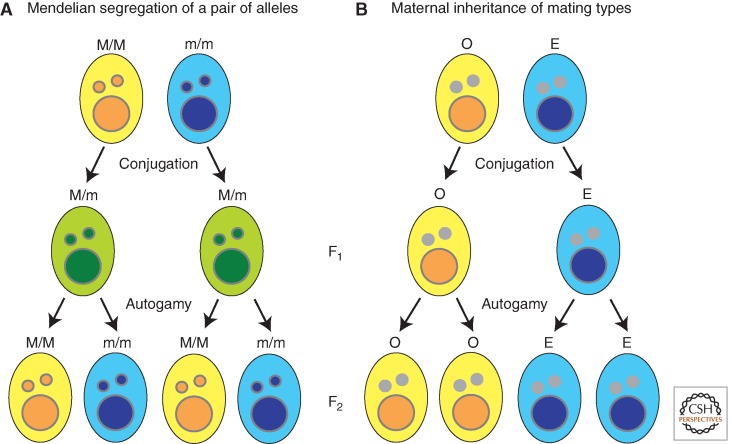
The two complementary mating types of P. tetraurelia, which are called O and E, show a cytoplasmic pattern of inheritance (Fig. 10). The O and E traits are terminally differentiated phenotypes that are determined during development of the somatic macronucleus from the totipotent germline. After conjugation, a vegetative clone descended from the O parent is almost always mating-type O, whereas one arising from the E parent is almost always of type E (Fig. 10B), although both exconjugants develop from identical zygotic genomes (Fig. 10A). However, if a significant exchange of cytoplasm occurs between mating cells, all progeny typically develop as type E (Sonneborn 1977). Thus, a cytoplasmic factor must exist that directs development of the E type. One Mendelian mutation affecting mating-type determination has been shown to perturb IES excision at other loci (Meyer and Keller 1996). This supports the hypothesis that alternative genome rearrangements may determine mating type. If this is the case, then epigenetic regulation of the rearrangement may underlie the cytoplasmic inheritance of this trait. The E-determining cytoplasmic factor would have the capacity to direct an alternative rearrangement of the mating-type gene, resulting in a macronuclear form of the gene that specifies type E.
Figure 10.
Epigenetic inheritance of mating-types in Paramecium. (A) Expected Mendelian segregation of genotypes and phenotypes. M and m alleles express yellow and blue phenotype, respectively. M/m F1 heterozygote conjugation progeny shows green (intermediate) phenotype. F2 homozygote progeny by autogamy shows either yellow or blue phenotype. (B) Mating type (O or E) is irreversibly determined during the development of the somatic macronucleus (large circle) from the totipotent germline micronucleus (small circle); however, the parental macronucleus directs differentiation of each exconjugant toward maintaining the existing mating type.
12. CONCLUSION
As future research illuminates the players (RNAs and proteins) mediating the many non-Mendelian phenomena that have been described in ciliates, more connections to known epigenetic mechanisms will become apparent. Clearly, further studies of DNA rearrangements will contribute new insights into RNA-based inheritance. The discovery of RNAi and other epigenetic mechanisms have largely come from efforts of researchers working in many model organisms aiming to explain examples of unexpected phenotypes or inheritance patterns. Many homology-dependent effects, including meiotic silencing in fungi and paramutation in plants (see Allshire and Ekwall 2014; Aramayo and Selker 2014; Pikaard and Mittelsten Scheid 2014), remain incompletely understood. It will be the combined insight provided by future experiments in all eukaryotes, including ciliates, which will expose the full scope of epigenetic processes.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ahmad K, Henikoff S 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 [DOI] [PubMed] [Google Scholar]
- Allis CD, Gorovsky MA 1981. Histone phosphorylation in macro- and micronuclei of Tetrahymena thermophila. Biochemistry 20: 3828–3833 [DOI] [PubMed] [Google Scholar]
- Allis CD, Glover CV, Bowen JK, Gorovsky MA 1980. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell 20: 609–617 [DOI] [PubMed] [Google Scholar]
- Allis CD, Allen RL, Wiggins JC, Chicoine LG, Richman R 1984. Proteolytic processing of h1-like histones in chromatin: A physiologically and developmentally regulated event in Tetrahymena micronuclei. J Cell Biol 99: 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739 [DOI] [Google Scholar]
- *.Allshire R, Ekwall K 2014. Epigenetics in Saccharomyces pombe. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Aramayo R, Selker EU 2014. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Mathy N, Baudry C, Malinsky S, Aury JM, Wilkes CD, Garnier O, Labadie K, Lauderdale BE, Le Mouel A, et al. 2012. The Paramecium germline genome provides a niche for intragenic parasitic DNA: Evolutionary dynamics of internal eliminated sequences. PLoS Genet 8: e1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K 2008. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev 22: 2228–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SE, Fino GM, Tausta SL, Klobutcher LA 1989. Micronuclear genome organization in Euplotes crassus: A transposonlike element is removed during macronuclear development. Mol Cell Biol 9: 3793–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Betermier M 2009. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev 23: 2478–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betermier M 2004. Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res Microbiol 155: 399–408 [DOI] [PubMed] [Google Scholar]
- Bouhouche K, Gout JF, Kapusta A, Betermier M, Meyer E 2011. Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res 39: 4249–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Allis CD 1995. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci 92: 6364–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD 1996. Tetrahymena histone acetyltransferase: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851 [DOI] [PubMed] [Google Scholar]
- *.Busslinger M, Tarakhovsky A 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bisharyan Y, Fridman V, Gerber J, Lin C, Orias E, Orias JD, Ryder H, Vong L, Hamilton EP 2005. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. II. Physical and genetic mapping. Genetics 170: 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Xi X, Vermaak D, Yao MC, Malik HS 2006. The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus. Mol Biol Cell 17: 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC 1996. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol Cell Biol 16: 3658–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev 15: 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Fuller P, Yao MC 2005. Communication between parental and developing genomes during tetrahymena nuclear differentiation is likely mediated by homologous RNAs. Genetics 169: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Vogt A, Mochizuki K, Yao MC 2010. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell 21: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cheng X 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao MC 1996. Genome downsizing during ciliate development: Nuclear division of labor through chromosome restructuring. Annu Rev Genet 30: 557–578 [DOI] [PubMed] [Google Scholar]
- Coyne RS, Thiagarajan M, Jones KM, Wortman JR, Tallon LJ, Haas BJ, Cassidy-Hanley DM, Wiley EA, Smith JJ, Collins K, et al. 2008. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics 9: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Gorovsky MA 2006. Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila. Mol Cell Biol 26: 4499–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Liu Y, Gorovsky MA 2006. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol Cell Biol 26: 7719–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MC, Ward JG, Herrick G, Allis CD 1992. Programmed nuclear death: Apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Dev Biol 154: 419–432 [DOI] [PubMed] [Google Scholar]
- Dou Y, Gorovsky MA 2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol Cell 6: 225–231 [DOI] [PubMed] [Google Scholar]
- Dou Y, Gorovsky MA 2002. Regulation of transcription by H1 phosphorylation in Tetrahymena is position independent and requires clustered sites. Proc Natl Acad Sci 99: 6142–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Mizzen CA, Abrams M, Allis CD, Gorovsky MA 1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol Cell 4: 641–647 [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E 1995. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev 9: 2065–2077 [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Keller AM, Meyer E 1998. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol 18: 7075–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LM, Forney JD 1984. Mendelian and non-Mendelian mutations affecting surface antigen expression in Paramecium tetraurelia. Mol Cell Biol 4: 1583–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Yao M-C 1996. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol Cell Biol 16: 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF 2012. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 151: 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass JN, Joshi NA, Couvillion MT, Bowen J, Gorovsky MA, Hamilton EP, Orias E, Hong K, Coyne RS, Eisen JA, et al. 2011. Genome-scale analysis of programmed DNA elimination sites in Tetrahymena thermophila. G3 (Bethesda) 1: 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Felsenfeld G 2014. A brief history of epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham JS, Thing TA, Vythilingum N, Keuroghlian A, Bruno D, Golding GB, Pearlman RE 2004. A non-long terminal repeat retrotransposon family is restricted to the germ line micronucleus of the ciliated protozoan Tetrahymena thermophila. Eukaryot Cell 3: 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forney JD, Blackburn EH 1988. Developmentally controlled telomere addition in wild-type and mutant Paramecia. Mol Cell Biol 8: 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forney JD, Yantiri F, Mikami K 1996. Developmentally controlled rearrangement of surface protein genes in Paramecium tetraurelia. J Eukaryot Microbiol 43: 462–467 [Google Scholar]
- Galvani A, Sperling L 2001. Transgene-mediated post-transcriptional gene silencing is inhibited by 3' non-coding sequences in Paramecium. Nucleic Acids Res 29: 4387–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani A, Sperling L 2002. RNA interference by feeding in Paramecium. Trends Genet 18: 11–12 [DOI] [PubMed] [Google Scholar]
- Garnier O, Serrano V, Duharcourt S, Meyer E 2004. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol 24: 7370–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiska R, James C, Yao MC 1993. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev 7: 2357–2365 [DOI] [PubMed] [Google Scholar]
- Gratias A, Betermier M 2003. Processing of double-strand breaks is involved in the precise excision of paramecium internal eliminated sequences. Mol Cell Biol 23: 7152–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R 2014. Transcriptional silencing by Polycomb group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton E, Bruns P, Lin C, Merriam V, Orias E, Vong L, Cassidy-Hanley D 2005. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. I. Cloning and identification of functional sites. Genetics 170: 1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harumoto T 1986. Induced change in a non-Mendelian determinant by transplantation of macronucleoplasm in Paramecium tetraurelia. Mol Cell Biol 6: 3498–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood J 1985. The erratic career of cytoplasmic inheritance. Trends Genet 1: 298–300 [Google Scholar]
- Hayashi T, Hayashi H, Fusauchi Y, Iwai K 1984. Tetrahymena histone H3. Purification and two variant sequences. J Biochem (Tokyo) 95: 1741–1749 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Hayashi H, Iwai K 1987. Tetrahymena histone H1. Isolation and amino acid sequence lacking the central hydrophobic domain conserved in other H1 histones. J Biochem (Tokyo) 102: 369–376 [DOI] [PubMed] [Google Scholar]
- *.Henikoff S, Smith M 2014. Histone variants and epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick G, Cartinhour S, Dawson D, Ang D, Sheets R, Lee A, Williams K 1985. Mobile elements bounded by C4A4 telomeric repeats in Oxytricha fallax. Cell 43: 759–768 [DOI] [PubMed] [Google Scholar]
- Howard-Till RA, Yao MC 2007. Tudor nuclease genes and programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot Cell 6: 1795–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn CL, Klobutcher LA 2002. Genome remodeling in ciliated protozoa. Annu Rev Microbiol 56: 489–520 [DOI] [PubMed] [Google Scholar]
- Jessop-Murray H, Martin LD, Gilley D, Preer JR, Polisky B 1991. Permanent rescue of a non-Mendelian mutation of Paramecium by microinjection of specific DNA sequences. Genetics 129: 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA 1999. Characterization of in vivo developmental chromosome fragmentation intermediates in E. crassus. Mol Cell 4: 695–704 [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G 1995. Consensus inverted terminal repeat sequence of Paramecium IESs: Resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucl Acids Res 23: 2006–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G 1997. Developmental genome reorganization in ciliated protozoa: The transposon link. Prog Nucleic Acid Res Mol Biol 56: 1–62 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Koizumi S 1990. Characterization of non-Mendelian and Mendelian mutant strains by micronuclear transplantation in Paramecium tetraurelia. J Protozool 37: 489–492 [Google Scholar]
- Koizumi S, Kobayashi S 1989. Microinjection of plasmid DNA encoding the A surface antigen of Paramecium tetraurelia restores the ability to regenerate a wild-type macronucleus. Mol Cell Biol 9: 4398–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2006. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev 20: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol 14: 604–610 [DOI] [PubMed] [Google Scholar]
- Le Mouël A, Butler A, Caron F, Meyer E 2003. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in Paramecia. Eukaryotic Cell 2: 1076–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepere G, Betermier M, Meyer E, Duharcourt S 2008. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev 22: 1501–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepere G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E 2009. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res 37: 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mochizuki K, Gorovsky MA 2004. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci 101: 1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Song X, Gorovsky MA, Karrer KM 2005. Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryot Cell 4: 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD 2007. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21: 1530–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL 2005. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol 25: 9151–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Martienssen R, Moazed D 2014. RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale DW, Allis CD, Bruns P 1982. Conjugation in Tetrahymena thermophila: A temporal analysis of cytological stages. Exp Cell Res 140: 227–236 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA 2005. RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Meyer E, Keller AM 1996. A Mendelian mutation affecting mating-type determination also affects developmental genomic rearrangements in Paramecium tetraurelia. Genetics 143: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Fain-Thornton J, Craig NL 2008. piggyBac can bypass DNA synthesis during cut and paste transposition. Embo J 27: 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen CA, Dou Y, Liu Y, Cook RG, Gorovsky MA, Allis CD 1999. Identification and mutation of phosphorylation sites in a linker histone. Phosphorylation of macronuclear H1 is not essential for viability in Tetrahymena. J Biol Chem 274: 14533–14536 [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2004. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev 14: 181–187 [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA 2005. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev 19: 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110: 689–699 [DOI] [PubMed] [Google Scholar]
- Nowacki M, Zagorski-Ostoja W, Meyer E 2005. Nowa1p and Nowa2p: Novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia. Curr Biol 15: 1616–1628 [DOI] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF 2008. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF 2009. A functional role for transposases in a large eukaryotic genome. Science 324: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF 2010. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci 107: 22140–22144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka AG, Jönsson F, Maier V, Möllenbeck M, Paeschke K, Postberg J, Rupprecht S, Lipps HJ 2003. The use of RNAi to analyze gene function in spirotichous ciliates. Eur J Protistol 39: 449–454 [Google Scholar]
- *.Patel D 2014. A structural perspective on readout of epigenetic H and DM marks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Germot A, Moreira D 2000. The new phylogeny of eukaryotes. Curr Opin Genet Dev 10: 596–601 [DOI] [PubMed] [Google Scholar]
- *.Pikaard C, Mittelsten Scheid O 2014. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM 1999. The evolutionary scrambling and developmental unscrambling of germline genes in hypotrichous ciliates. Nucleic Acids Res 27: 1243–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Ehrenfeucht A, Rozenberg G 2003. Template-guided recombination for IES elimination and unscrambling of genes in stichotrichous ciliates. J Theor Biol 222: 323–330 [DOI] [PubMed] [Google Scholar]
- Ray C 1956. Meiosis and nuclear behavior in Tetrahymena pyriformis*. J Eukaryot Microbiol 3: 88–96 [Google Scholar]
- Ruiz F, Vayssie L, Klotz C, Sperling L, Madeddu L 1998. Homology-dependent gene silencing in Paramecium. Mol Biol Cell 9: 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev SV, Cox MM 1995. Transient DNA breaks associated with programmed genomic deletion events in conjugating cells of Tetrahymena thermophila. Genes Dev 9: 248–255 [DOI] [PubMed] [Google Scholar]
- Saveliev SV, Cox MM 1996. Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J 15: 2858–2869 [PMC free article] [PubMed] [Google Scholar]
- Schoeberl UE, Kurth HM, Noto T, Mochizuki K 2012. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev 26: 1729–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JM, Mikami K, Leeck CL, Forney JD 1994. Non-Mendelian inheritance of macronuclear mutations is gene specific in Paramecium tetraurelia. Mol Cell Biol 14: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Gorovsky MA 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86: 475–483 [DOI] [PubMed] [Google Scholar]
- Shen X, Yu L, Weir JW, Gorovsky MA 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell 82: 47–56 [DOI] [PubMed] [Google Scholar]
- Sonneborn TM 1937. Sex, sex inheritance and sex determination in Paramecium aurelia. Proc Natl Acad Sci 23: 378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM 1975. Paramecium aurelia. In Handbook of genetics (ed. King R), pp. 469–594 Plenum, New York [Google Scholar]
- Sonneborn TM 1977. Genetics of cellular differentiation: stable nuclear differentiation in eucaryotic unicells. Annu Rev Genet 11: 349–367 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci 96: 14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110: 701–711 [DOI] [PubMed] [Google Scholar]
- Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, Diaz RL, Shabanowitz J, Chait BT, Hunt DF, Allis CD 2007. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci 104: 2086–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112 [DOI] [PubMed] [Google Scholar]
- Vavra KJ, Colavito-Shepanski M, Gorovsky MA 1982. Histone acetylation and the deoxyribonuclease I sensitivity of the Tetrahymena ribosomal gene. Biochemistry 21: 1772–1781 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T 2001. Gene silencing as an adaptive defence against viruses. Nature 411: 834–842 [DOI] [PubMed] [Google Scholar]
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci 95: 7480–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97: 99–109 [DOI] [PubMed] [Google Scholar]
- Wu M, Allis CD, Richman R, Cook RG, Gorovsky MA 1986. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc Natl Acad Sci 83: 8674–8678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuitschick JD, Gershan JA, Lochowicz AJ, Li S, Karrer KM 2002. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res 30: 2524–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, Zheng K, Yao CH 1987. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell 48: 779–788 [DOI] [PubMed] [Google Scholar]
- Yao MC, Fuller P, Xi X 2003. Programmed DNA deletion as an RNA-guided system of genome defense. Science 300: 1581–1584 [DOI] [PubMed] [Google Scholar]
- You Y, Aufderheide K, Morand J, Rodkey K, Forney J 1991. Macronuclear transformation with specific DNA fragments controls the content of the new macronuclear genome in Paramecium tetraurelia. Mol Cell Biol 11: 1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Gorovsky MA 1997. Constitutive expression, not a particular primary sequence, is the important feature of the H3 replacement variant hv2 in Tetrahymena thermophila. Mol Cell Biol 17: 6303–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Neeb ZT, Lin A, Katzman S 2012. Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS One 7: e42371. [DOI] [PMC free article] [PubMed] [Google Scholar]