Abstract
Matrix-assisted laser desorption/ionization (MALDI) mass spectrometric imaging (MSI) has been employed as a detection method for both capillary electrophoresis (CE)-MALDI and liquid chromatography (LC)-MALDI analysis. Based on our previous studies, here we report a new interface to couple LC to MSI by employing an automated matrix sprayer. The LC trace is directly collected on a ground stainless steel MALDI plate and dried. The matrix is sprayed on the MALDI plate using a programmable matrix sprayer. With the highly uniform matrix layers produced from the sprayer, MS image signal quality is significantly improved with enhanced signal-to-noise ratios for analyte peaks. With the programmable matrix application and imaging MS data acquisition, the new LC-MSI platform exhibits highly stable and reproducible performance. A total of 87 bovine serum albumin (BSA) tryptic peptides and 295 putative neuropeptides from blue crab pericardial organs have been observed with LC-MSI analysis, exhibiting better performance in terms of peptide coverage than regular LC-MALDI with discrete spot collection and our previously reported LC-MSI interface with matrix delivered by a capillary. In addition to relative quantitation with isotopic labeling as we previously demonstrated, we performed the first absolute quantitation using the new LC-MSI platform and obtained accurate quantitation results for neuropeptides, indicating great potential for quantitative analysis of complex samples.
Introduction
Coupling separations to matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) presents unique challenges in comparison to coupling separations to electrospray ionization (ESI)-MS analysis. New developments and applications of both LC-ESI and CE-ESI interfaces have seen tremendous growth in the past decade and are commercially available.1-3 In contrast, LC-MALDI and CE-MALDI interfaces are still limited to either direct or indirect offline fraction collection through home-built interfaces or commercially available spotters.4-12 The challenge lies in the inherent nature of MALDI: compared to LC-ESI and CE-ESI, MALDI couplings lack automation and reproducibility. These problems can be partially solved by employing a spotter so that LC or CE flow can be mixed with matrix through a connector and then spotted onto the MALDI plate automatically and more uniformly. However, in order to retain temporal resolution from separation dimension, hundreds of spots would need to be collected from a single run which requires prolonged experimental time and tremendous work for data analysis. To date, there is still a lack of LC-MALDI and CE-MALDI platforms that exhibit similar degree of separation efficiency and automation compared with LC-ESI-MS and CE-ESI-MS.
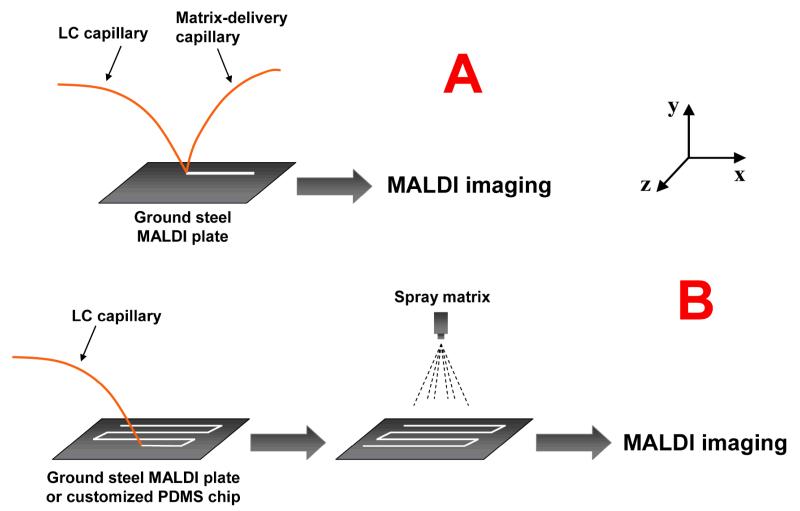
As a widely-practiced soft ionization method, MALDI features higher tolerance to impurities and traces of additives and exhibits complementary ionization efficiency to ESI.13 It is therefore worth developing new platforms for separation-MALDI couplings with increased automation, reproducibility and data analysis capacity. Recently, we published our work employing MALDI-mass spectrometric imaging (MSI) for LC-MALDI14 and CE-MALDI15,16 couplings. Although almost exclusively used for tissue imaging since its introduction,17-19 MALDI-MSI is inherently compatible with separations as a detection method. With the micrometer-scale step resolution from MSI, continuous LC or CE trace can be collected and imaged, so that the temporal resolution from separation dimension can be preserved. Meanwhile, instead of collecting hundreds of individual mass spectra, only one LC or CE trace image with all separated peak information will be generated by imaging software, and data analysis can be performed based on either mass-to-charge ratio or retention time (as reflected on the image) with great ease. We have reported both LC-MSI and CE-MSI interfaces for complex peptide analysis.14-16 As shown in Fig. 1A, two separate capillaries were employed to deliver CE flow and matrix flow, respectively. With ground stainless steel MALDI plate, no pre-applied matrix is necessary. A homogenous CE trace can be directly collected on the plate surface. The drawback of this interface is the use of two capillaries with different angles, which causes sample diffusion on the MALDI plate and prevents us from customizing a plate with prolonged trace patterns.
Fig. 1. The interfaces coupling separations to mass spectrometric imaging.
A: LC/CE-MSI interface with matrix-delivery capillary. LC or CE flow is mixed with matrix flow and collected directly on the surface of the ground stainless steel MALDI plate. B: LC/CE-MSI interface with sprayed matrix for enhanced sensitivity and reproducibility.
In order to increase the homogeneity and automation during matrix application, in this work, we evaluated a new matrix application method for LC-MSI coupling that employs an automated matrix sprayer. As schematically shown in Fig. 1B, LC flow is collected directly on the ground stainless steel MALDI plate without mixing with matrix so that sharp peaks could be obtained due to the minimized sample diffusion. The dried LC trace is then covered by matrix with an automated sprayer so that highly homogenous traces can be formed with significantly enhanced reproducibility. The uniform matrix layers from matrix sprayer results in increased MS signal-to-noise ratios compared to regular LC-MALDI and our previous LC-MSI systems. Furthermore, enhanced reproducibility is achieved due to the automated and programmable matrix application procedure. We have applied this new platform to the analysis of both protein digests and neuropeptide extracts and demonstrated its advantages over standard LC-MALDI couplings. We also demonstrated its application to absolute quantitation of crustacean cardioactive peptide (CCAP) from a complex biological matrix as a result of the enhanced sensitivity and reproducibility. These results highlight the utility of the new LC-MSI interface with sprayed matrix as a novel and powerful tool for the analysis of a complex mixture of peptides.
Experimental
Chemical and materials
Acetic acid, ammonium hydroxide, hydrochloric acid, sodium hydroxide, acetone, acetonitrile, urea and ammonium bicarbonate were obtained from Fisher Scientific (Pittsburgh, PA). Lauryl methacrylate (LMA, 96%), ethylene dimethacrylate (EDMA, 98%), butandiol (99%), 1-propanol (99.5%), 3-(trimethoxysilyl) propyl ethacrylate (98%), 2,2′-Azobis(2-methylpropionitrile) (AIBN, 98%), trifluoroacetic acid (TFA), formic acid (FA), iodoacetamide (IAA) and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO). 2,5-dihydroxybenzonic acid (99%, DHB) were from Acros Organics (Geel, Belgium). D/L-dithiothreitol (DTT) and sequencing grade modified trypsin were from Promega (Madison, WI). Ethanol (200 proof) was purchased from Decon Laboratories (King of Prussia, PA). The CCAP (PFCNAFTGCa, m/z 956.38) standard peptide was purchased from the American Peptide Company (Sunnyvale, CA). A C18 Ziptip column from Millipore was used for sample cleaning, and all water used was doubly distilled on a Millipore filtration system (Bedford, MA).
Sample preparation
The tryptic peptide mixture was obtained by digestion of bovine serum albumin (BSA) with trypsin. The following solutions were used during digestion: 8 M urea, 1 M DTT as reducing reagent, 200 mM IAA as alkylating reagent. All of these solutions were dissolved in 25 mM ammonium bicarbonate. During digestion, 30 μg of BSA was reconstituted in 20 μL of 8 M urea, followed by adding 1 μL of DTT with gentle vortexing. After 1 h reducing at 37 °C, 20 μL of IAA was added to alkylate for 1 h at room temperature in dark. To consume leftover alkylating reagent, 4 μL of DTT was added to the tube followed by 120 μL of 25 mM ammonium bicarbonate solution to dilute urea. In-solution digestion was performed overnight at 37 °C after adding 1 μg of trypsin. After digestion was completed, 1 μL of formic acid was added to the sample with gentle vortexing to quench the reaction.
Neuropeptides were extracted from the pericardial organs (POs) of the blue crabs (Callinectes sapidus). Blue crabs were purchased from a local grocery store and kept in an artificial seawater tank at 10-12 °C without food. The dissection procedure has been previously described.20, 21 Briefly, the crabs were anesthetized with ice for 30 min before dissection. The POs were dissected in physiological saline, which was comprised of 440 mM NaCl, 11mM KCl, 26 mM MgCl2, 13 mM CaCl2, 11 mM Trizma base, and 5 mM maleic acid in pH 7.45. The neuropeptides were extracted with cold acidified methanol, which consisted of methanol, water and acetic acid in the ratio of 90:9:1. Five POs were combined and homogenized in 50 μL acidified methanol in ice and centrifuged. The extraction was repeated three times and the supernatants were combined and dried. Both the BSA tryptic peptide mixture and extracted neuropeptides were reconstituted with 10 μL of 0.1% TFA, desalted by Ziptip C18 column and stored at −80 °C before use.
Absolute quantitation of neuropeptide PFCNAFTGCa
The CCAP peptide standard was dissolved in 0.1% TFA and diluted to a series of stocking solutions with the following concentrations: 1×10−4 M, 3×10−5 M, 1×10−5 M, 3×10−6 M, 1×10−6 M, 3×10−7 M and 1×10−7 M. An aliquot of 0.4 μL of each stock solutions was spotted on the MALDI plate, dried and sprayed with matrix using a sprayer, making the actual amount of CCAP to 40 pmol, 12 pmol, 4 pmol, 1.2 pmol, 400 fmol, 120 fmol and 40 fmol, respectively. Three spots for each concentration were measured using the software Quantinetix (ImaBiotech, Loos, France). Briefly, a region of interest (ROI) was selected for each concentration spot using Quantinetix, and the combined intensity was calculated to generate a standard curve against loading amount. After establishing the standard curve, the peak corresponding to neuropeptide PFCNAFTGCa was extracted from a LC-MSI run of blue crab PO, and the combined intensities were calculated and fit to the calibration curve for absolute quantitation.
LC separation
The LC separation was performed with a homemade monolithic reversed-phase LC (RPLC) column. A lauryl methacrylate-co-ethylene dimethacrylate (LMA-EDMA) monolithic column was made with a 40 μm i.d./190 μm o.d. fused-silica capillary (Polymicro Technologies, Phoenix, AZ) as reported perviously.14 Briefly, the capillary was rinsed with acetone, 0.2 M NaOH, 0.2 M HCl and water, respectively, and was silanized with 20% 3-(trimethoxysilyl) propyl methacrylate (in 95% ethanol) for 90 min. With 24% LMA, 16% EDMA, 40% 1-propanol and 20% 1,4-butandiol (w/w, with 1% AIBN as initiator), polymerization was carried out in a water bath at 70 °C for 24 h. After polymerization, the column was washed with acetonitrile and water. A Waters nanoACQUITY UPLC system was used for RPLC fractioning. The mobile phases contained solution A (0.1% formic acid in deionized water) and solution B (0.1% formic acid in acetonitrile). The LC separation started at 95% solution A, changed to 80% A at 10 min, and 30% A at 30 min. The flow rate was set to 0.5 μL/min with 1 μL sample loading amount.
LC-MSI coupling with matrix sprayer application
The LC flow was collected directly on a ground stainless steel MALDI plate which moved along the x-axis at a velocity of 4.5 mm/min. A lamp was used to dry the traces quickly so that approximately 0.5-0.6 mm wide, highly homogenous LC traces were deposited on the MALDI plate with minimum diffusion. Following this, the MALDI plate was transferred to a TM-Sprayer from HTX Technologies (Carrboro, NC) to spray matrix (50 mg/mL DHB in 50% methanol) on the dried traces. The parameters were carefully adjusted and set as following: matrix flow rate: 125 μL/min; nozzle velocity: 1200 mm/min; temperature: 85 degrees Celsius; gas pressure: 13 psi; line spacing: 3 mm; number of layers: 2; dry time: 1 min.
Mass spectrometry and imaging software
The MS images of collected LC traces were acquired with UltrafleXtreme MALDI-TOF/TOF from Bruker Daltonics (Bremen, Germany) equipped with 2000 Hz Smartbeam II laser. Positive reflectron mode was employed with the following parameters: ion source 1 voltage 25.00 kV, ion source 2 voltage 16.49 kV, reflector 1 voltage 20.90 kV, reflector 2 voltage 9.59 kV and lens voltage 8.65 kV. The step size was set to 100 μm ×100 μm (on the x/y axis) and 500 shots were accumulated in a random walk mode for each pixel. The imaging data was processed with the instrumental specific software, FlexImaging. The peak intensities from selected regions were obtained using Quantinetix from ImaBiotech (Loos, France) for absolute quantitation. For reproducibility test, a Waters nanoACQUITY UPLC system was used for separation and coupled to ESI-Q-TOF detection. The same monolithic RPLC column was used for separation with the same LC condition but using ESI as the ionization mode.
Results and discussion
LC-MSI interface with sprayed matrix
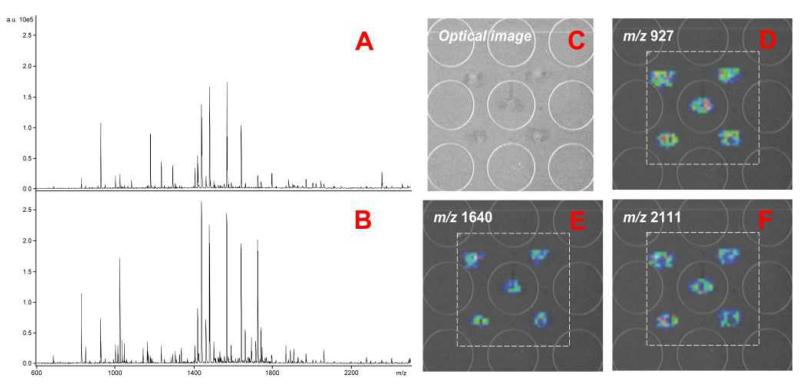
In this work, we have modified our previously reported interface15, 22 (Fig. 1A) by removing the matrix-delivery capillary. Instead, the flow from the LC column was collected directly on the MALDI plate and then sprayed with matrix. This new feature reflects our efforts made on increasing the sensitivity and reproducibility: (1) by collecting LC flow directly on the MALDI plate surface, the trace can be dried quickly without interference from matrix flow, thus minimizing diffusion. Highly uniform LC traces with approximately 0.5-0.6 mm width were observed that resulted in enhanced MS signal. (2) The automated matrix sprayer is programmable and generates highly reproducible spray and matrix coverage. As shown in Fig. 2A and 2B, sprayed matrix generates higher MS intensity for tryptic peptides compared to spotted matrix under similar condition. When integrated to MS imaging, intense image ion signals are also observed with very limited sample diffusion (Fig. 2C-2F). (3) With only one capillary sliding directly on the MALDI plate surface, the movement pattern of the MALDI plate is more flexible and programmable, so that customized trace pattern (e.g. spiral or serpentine pattern) can be designed to collect longer separations for improved peak capacity. Alternatively, a miniaturized chip can be designed to integrate LC-MALDI-MSI into a lab-on-a-chip format in the future.
Fig. 2. The MS spectra and images obtained from sample spots with either spotted or sprayed matrix.
A: Mass spectrum of BSA tryptic peptides with spotted matrix. B: Mass spectrum of BSA tryptic peptides with sprayed matrix. C: The optical image of 5 spots of BSA tryptic peptides with matrix sprayed. D-F: The images of 3 representative tryptic peptides for the 5 sample spots with matrix sprayed.
We compared the impact of different matrix application methods on LC-MSI coupling. In our previous design with matrix-delivery capillary, the matrix flow and LC flow are mixed at the tips of capillaries on the MALDI plate. This usually generated straight and continuous LC traces, but we frequently saw “marble-like” trace surface due to the irregular sizes of matrix crystals (Fig. S1A). When such type of LC trace was imaged, sample diffusion was observed with higher noise background (Fig. S1B). In this report, spraying matrix through a programmable sprayer generates very uniform matrix layer on top of the collected LC trace (Fig. S1C) and effectively reduces the level of noise. By extracting the peak, m/z 1292.9, as shown in Fig. S1B, limited sample diffusion with extremely low noise background was observed (Fig. S1D). We also employed the conventional “air brush” method to spray the matrix onto the MALDI plate manually; however, much larger and irregular crystal sizes were found after spraying making it difficult to mix with dried sample on the plate (Fig. S1E). These experiments indicate the benefits of employing an automated matrix sprayer for LC-MSI coupling.
In our previous report, we have demonstrated that the monolithic RPLC column can be readily coupled to LC-MSI with highly reproducible.14 Here, we further evaluated its reproducibility using sprayed matrix for analysis of BSA tryptic peptides. Three LC runs under the same conditions were performed in three consecutive days, followed by either ESI-QTOF detection or MALDI-MSI detection with sprayed matrix. The mass spectra were recorded and shown in Fig. S2. In both cases, almost identical spectra with comparable numbers of peptides were detected, indicating excellent stability and reproducibility of the LC-MSI system with sprayed matrix method.
Improved sensitivity and data analysis capability using LC-MSI with sprayed matrix
As we have demonstrated in Fig. 2 and Fig. S1, when the matrix is uniformly sprayed on the MALDI plate via a programmable sprayer, tiny and uniform matrix crystals can be formed and well mixed with the dried LC eluent on the MALDI plate. Increased sensitivity is obtained due to the well-mixed sample-matrix co-crystal, while the signal-to-noise ratio is also improved due to the uniformity of matrix. With BSA tryptic peptides, we have compared the number of peptides detected using standard LC-MALDI-TOF/TOF with collection of discrete spots, LC-MSI with matrix delivered with a capillary and LC-MSI with sprayed matrix, respectively. MS fingerprint search was performed using Mascot with the same parameters for each experiment (100 ppm error, 1 missed cleavage allowance and carbamidomethyl, ammonia loss and oxidation as variable modifications). As a result, 49 tryptic peptides (66% sequence coverage) were observed with LC-MALDI-TOF/TOF, 78 tryptic peptides (81% sequence coverage) were observed with LC-MSI with capillary matrix delivery and 87 tryptic peptides (86% sequence coverage) were observed with LC-MSI with sprayed matrix. We further applied LC-MSI with sprayed matrix to the analysis of neuropeptides extracted from crustacean pericardial organs. A total of 295 putative neuropeptides have been detected from a single LC-MSI run, including 85 previously identified neuropeptides, exhibiting the largest number of neuropeptides being identified compared to previous approaches. A detailed list of all putative neuropeptides is included as Table S1.
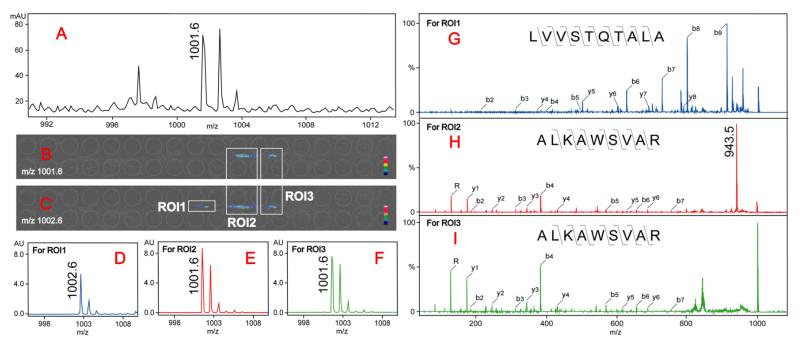
The enhanced matrix application significantly improved the ability to identify complex peak patterns from the mass spectrum. Combined with tandem MS for peptide de novo sequencing, we were able to distinguish and identify overlapping peptides as well as possible isomers with the same mass. As an example in Fig. 3A, a putative tryptic peptide peak m/z 1001.6 was observed in MS full scan, however, when trying to obtain its image after LC-MSI analysis, two distinguishable colored regions were observed with mass filter centered at m/z 1001.6 and a third colored region was observed at m/z 1002.6 in addition to the two isotopic peak images of m/z 1001.6 (Fig. 3B and 3C). The mass spectrum for each selected region of interest (ROI) can be extracted as shown in Fig. 3D-3F, indicating separated peaks with enhanced signal-to-noise ratios. Based on the distribution of ROIs, we can perform tandem MS acquisition and obtain MS/MS spectrum for each ROI. With the de novo sequencing results shown in Fig. 3G-3I, we conclude that peak m/z 1002.6 from ROI1 is a tryptic peptide LVVSTQTALA, while the colored regions from both ROI2 and ROI3 are tryptic peptide ALKAWSVAR. However, a fragment ion at m/z 943.5 is uniquely detected in its MS/MS spectrum from ROI2 (Fig. 3H) which results from the cleavage of C-N bond from Arginine side chain, whereas this fragment ion is missing in Fig. 3I. Considering the different retention times, we postulate that different secondary structures of peptide ALKAWSVAR (e.g., D/L isomers) may exist in liquid phase and are separated by RPLC.
Fig. 3. Identification and characterization of overlapped tryptic peptides with LC-MSI.
A: MS full scan showing the m/z 1001.6 peak. B and C: the images of mass filters centered at m/z 1001.6 (B) and m/z 1002.6 (C). The colored regions of putative peptides separated in the images are marked with white boxes as regions of interest (ROIs). D-F: The extracted ion spectra from three ROIs showing the separated putative peptides. G-I: CID fragmentation spectra of selected parent ions from three ROIs showing different peptide sequence (ROI1) and secondary structures (ROI2 and ROI3) separated by LC-MSI.
Absolute quantitation with LC-MSI
Quantitation with MSI has been an area of intensive research because of its great potential in combining quantitation with spatial distribution.23-26 However, due to the inherent nature of MALDI imaging, it has been very difficult to accurately quantify single or multiple analytes from complex biological matrices. Unlike tissue imaging, when employing MSI as a detection method for LC separation, we could perform a routine isotopic labeling of the sample for relative quantitation which has been demonstrated in previous reports.14,15 With the newly developed LC-MSI using sprayed matrix application, highly uniform matrix layers can be applied to the entire MALDI plate that enables absolute quantitation by generating calibration curves from peptide standards with known concentrations. Because the LC trace and standard solution are both dissolved in 0.1% FA/acetonitrile and then dried on the MALDI plate, there is no need to calculate distinction coefficient between standards and real samples. The uniformly deposited matrix enables highly reproducible and comparable ionization among different areas of MALDI plate. By using a MSI quantitation software Quantinetix (ImaBiotech, Loos, France), we could obtain the peak intensities from selected regions of interest (ROIs) for both relative and absolute quantitation.
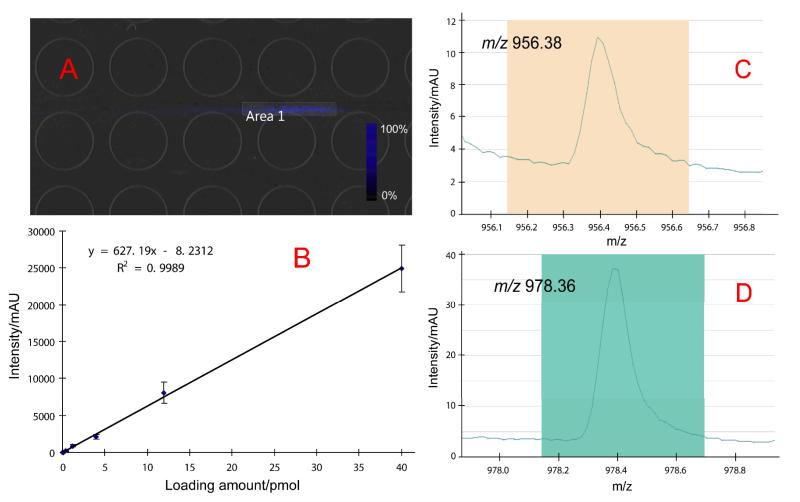
Based on the relative quantitation previously demonstrated,14,15 here we further explored integrating absolute quantitation to LC-MSI based analysis by taking advantage of the improved signal-to-noise ratios as well as quantitative imaging software. With the sprayed matrix, we have been able to obtain “cleaner” spectra with increased MS sensitivity. Together with Quantinetix software, it has become possible to combine absolute quantitation with LC-MSI system. An example of the absolute quantitation of CCAP peptide (PFCNAFTGCa) with m/z 956.38 and its sodium adduct at m/z 978.36 is demonstrated in Fig. 4. A series of standard peptide solutions in known concentrations was made and spotted on the MALDI plate. A region of interest (ROI) for each of the spot was selected with the targeted peak extracted to calculate the accumulated intensities, which were then plotted against loading amount to generate a standard curve. As shown in Fig. 4B, a good linear fitting has been observed with R2=0.9989 (n=3 for each point). We then performed a LC-MSI analysis of extracted neuropeptides from blue crab PO, and the peaks of CCAP and its sodium adduct were extracted from the image. In Quantinetix, we selected an area including the targeted ions (Fig. 4A) and extracted the accumulated intensity for calculation (Fig. 4C and 4D). We then calculated that 0.556 pmol of CCAP was present in the 1 μL LC loading amount, suggesting that each blue crab PO contains approximately 1.112 pmol (or 1.062 ng) of CCAP. Normalization against an isotopic labeled peak is usually necessary for tissue imaging to eliminate interference from biological matrices, however because the LC flow and standard solution are both mixtures of acidified water and acetonitrile, no normalization is made for LC-MSI. This result has demonstrated the ability to determine the absolute amount of trace-level analytes from complex biological matrix using LC-MSI platform.
Fig. 4. Absolute quantitation of crustacean cardio-active peptide (CCAP) PFCNAFTGCa.
A: The image of CCAP m/z 956.38 and its sodium adduct m/z 978.36 sharing the same distribution in MS full scan after LC-MSI separation. An area covering the MS signal was selected using Quantinetix. B: A standard curve was generated from the standard peptide solution which was spotted on the MALDI plate. The peak intensity obtained from panel B was fit to the curve for quantitation. C and D: extracted ions of m/z 956.38 and m/z 978.36 from the selected area. In Quantinetix, the intensity of the targeted peak from the selected area in panel A was shown. Peak intensity and standard deviation (shown as error bars in panel B) can be obtained from the software for quantitation.
Conclusions
We have developed a new interface to couple liquid chromatography to mass spectrometric imaging. By employing a sprayer to apply matrix, we have been able to generate highly uniform matrix layer and enhanced MS signal-to-noise ratios. Higher peptide coverage has been observed with both tryptic peptides and neuropeptide extracts. Furthermore, we demonstrated absolute quantitation using LC-MSI system for the first time. This work reflects our efforts to develop an interface for LC/CE-MALDI coupling that has comparable performance with ESI coupling in terms of sensitivity, reproducibility and automation. With the programmable and fully controllable matrix application together with image-based data analysis strategy, we have been able to apply this novel technique to the global analysis of complex peptide samples typically analyzed with ESI MS. This technique can be further improved by employing a high resolution MALDI-LTQ-Orbitrap MS instrument where multi-stage tandem MS (MSn) information in data dependent mode is enabled. With the chemical information extractable from each MS spectrum combined with database search, we expect to further expand the utility of the newly developed LC-MSI system to large-scale and high-throughput proteomics studies.
Supplementary Material
Acknowledgement
This work is supported by the National Science Foundation grant (CHE-0957784) and National Institutes of Health grants (1R01DK071801, 1R56DK071801). The acquisition of the TM sprayer was funded by an NIH shared instrument grant 1S10RR029531. Z. Zhang thanks Erin Gemperline in the Li Research Group for critical reading of the manuscript and making helpful suggestions. L. Li acknowledges an H.I. Romnes Faculty Research Fellowship.
References
- 1.Lee MS, Kerns EH. Mass Spectrometry Reviews. 1999;18:187–279. doi: 10.1002/(SICI)1098-2787(1999)18:3/4<187::AID-MAS2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 2.Petrovic M, Hernando MD, Diaz-Cruz MS, Barcelo D. Journal of Chromatography A. 2005;1067:1–14. doi: 10.1016/j.chroma.2004.10.110. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar WM, Blackburn RK, Krise JM, Moseley MA. Journal of the American Society for Mass Spectrometry. 2003;14:971–979. doi: 10.1016/S1044-0305(03)00209-5. [DOI] [PubMed] [Google Scholar]
- 5.Ericson C, Phung QT, Horn DM, Peters EC, Fitchett JR, Ficarro SB, Salomon AR, Brill LM, Brock A. Analytical Chemistry. 2003;75:2309–2315. doi: 10.1021/ac026409j. [DOI] [PubMed] [Google Scholar]
- 6.Funke S, Azimi D, Wolters D, Grus FH, Pfeiffer N. J. Proteomics. 2012;75:3177–3190. doi: 10.1016/j.jprot.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Hattan SJ, Vestal ML. Analytical Chemistry. 2008;80:9115–9123. doi: 10.1021/ac8017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mateos J, Lourido L, Fernandez-Puente P, Calamia V, Fernandez-Lopez C, Oreiro N, Ruiz-Romero C, Blanco FJ. J. Proteomics. 2012;75:2869–2878. doi: 10.1016/j.jprot.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 9.Pes O, Preisler J. Journal of Chromatography A. 2012;1217:3966–3977. doi: 10.1016/j.chroma.2010.02.058. [DOI] [PubMed] [Google Scholar]
- 10.Preisler J, Hu P, Rejtar T, Karger BL. Analytical Chemistry. 2000;72:4785–4795. doi: 10.1021/ac0005870. [DOI] [PubMed] [Google Scholar]
- 11.Rejtar T, Hu P, Juhasz P, Campbell JM, Vestal ML, Preisler J, Karger BL. Journal of Proteome Research. 2002;1:171–179. doi: 10.1021/pr015519o. [DOI] [PubMed] [Google Scholar]
- 12.Soderberg CAG, Lambert W, Kjellstrom S, Wiegandt A, Wulff RP, Mansson C, Rutsdottir G, Emanuelsson C. Plos One. 2012;7:e38927. doi: 10.1371/journal.pone.0038927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domon B, Aebersold R. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Shan J, Li L. Journal of Chromatography A. 2013;1293:44–50. doi: 10.1016/j.chroma.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Ye H, Wang J, Hui L, Li L. Analytical Chemistry. 2012;84:7684–7691. doi: 10.1021/ac300628s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang ZC, Jia CX, Li LJ. Journal of Separation Science. 2012;35:1779–1784. doi: 10.1002/jssc.201200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amstalden van Hove ER, Smith DF, Heeren RMA. Journal of chromatography. A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 18.Greer T, Sturm R, Li LJ. J. Proteomics. 2011;74:2617–2631. doi: 10.1016/j.jprot.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heeren RMA, Smith DF, Stauber J, Kukrer-Kaletas B, MacAleese L. Journal of the American Society for Mass Spectrometry. 2009;20:1006–1014. doi: 10.1016/j.jasms.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Wang JH, Zhang YZ, Xiang F, Zhang ZC, Li LJ. Journal of Chromatography A. 2010;1217:4463–4470. doi: 10.1016/j.chroma.2010.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen RB, Hui LM, Cape SS, Wang JH, Li LJ. ACS Chem. Neurosci. 2010;1:204–214. doi: 10.1021/cn900028s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang JH, Ye H, Zhang ZC, Xiang F, Girdaukas G, Li LJ. Analytical Chemistry. 2011;83:3462–3469. doi: 10.1021/ac200708f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stauber J. Bioanalysis. 2012;4:2095–2098. doi: 10.4155/bio.12.187. [DOI] [PubMed] [Google Scholar]
- 24.Kallback P, Shariatgorji M, Nilsson A, Andren PE. J. Proteomics. 2012;75:4941–4951. doi: 10.1016/j.jprot.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 25.Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos JM, Bouzom F, Stauber J. J. Proteomics. 2012;75:4952–4961. doi: 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 26.Zhong M, Lee CY, Croushore CA, Sweedler JV. Lab Chip. 2012;12:2037–2045. doi: 10.1039/c2lc21085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.