Abstract
Allergic diseases are frequently exacerbated between midnight and early morning, suggesting a role for the biological clock. Mast cells (MC) and eosinophils are the main effector cells of allergic diseases and some MC-specific or eosinophil-specific markers, such as tryptase or eosinophil cationic protein, exhibit circadian variation. Here, we analysed whether the circadian clock is functional in mouse and human eosinophils and MC. Mouse jejunal MC and polymorphonuclear cells from peripheral blood (PMNC) were isolated around the circadian cycle. Human eosinophils were purified from peripheral blood of non-allergic and allergic subjects. Human MC were purified from intestinal tissue. We found a rhythmic expression of the clock genes mPer1, mPer2, mClock and mBmal1 and eosinophil-specific genes mEcp, mEpo and mMbp in murine PMNC. We also found circadian variations for hPer1, hPer2, hBmal1, hClock, hEdn and hEcp mRNA and eosinophil cationic protein (ECP) in human eosinophils of both healthy and allergic people. Clock genes mPer1, mPer2, mClock and mBmal1 and MC-specific genes mMcpt-5, mMcpt-7, mc-kit and mFcεRI α-chain and protein levels of mMCPT5 and mc-Kit showed robust oscillation in mouse jejunum. Human intestinal MC expressed hPer1, hPer2 and hBmal1 as well as hTryptase and hFcεRI α-chain, in a circadian manner. We found that pre-stored histamine and de novo synthesized cysteinyl leukotrienes, were released in a circadian manner by MC following IgE-mediated activation. In summary, the biological clock controls MC and eosinophils leading to circadian expression and release of their mediators and, hence it might be involved in the pathophysiology of allergy.
Keywords: allergy, biological clock, circadian rhythm, eosinophils, mast cells
Introduction
The central clock is located in the suprachiasmatic nuclei of the anterior hypothalamus, which receives light information from the outside world by way of the retino–hypothalamic tract. Circadian timing is organized in a hierarchy of dispersed oscillators, i.e. peripheral clocks reside in tissues, such as liver, kidney and intestine.1–3 The circadian clock affects nearly all aspects of physiology and behaviour and so plays a pivotal role in gastrointestinal tract motility, the sleep–wake cycle, metabolism, immune function and allergic diseases.1
The first clock gene identified in mammals, circadian locomotor output cycles Kaput, encodes the transcription factor CLOCK, which dimerizes with BMAL1, brain and muscle ARNT-like protein 1.4 The CLOCK:BMAL1 heterodimer binds to E-box (5′-CACGTG-3′) and E-box-like promoter sequences to mediate the transcription of a large number of genes including Periods (Per1, Per2, Per3) and Cryptochromes (Cry1, Cry2). The PER and CRY proteins are part of the negative feedback loop. After translation they oligomerize and translocate to the nucleus to turn off CLOCK:BMAL1-mediated transcription.5
Mast cells (MC) and eosinophils are the two main effector cells in allergic diseases, such as asthma, allergic rhinitis or food allergy.6,7 Eosinophils exert their inflammatory effects by releasing an array of mediators including cytokines, chemokines, lipid mediators, growth factors8 and basic granule-stored proteins, such as eosinophil peroxidase (EPO), eosinophil cationic protein (ECP), major basic protein (MBP) and eosinophil-derived neurotoxin/eosinophil protein X (EDN/EPX).9 Some eosinophil-specific molecules, such as ECP and EPX, show an oscillatory expression pattern in sputum, serum and urine.10 Mast cells are capable of producing and releasing histamine, chymase, tryptase and eicosanoids as well as pro-inflammatory cytokines and chemokines.11 Some MC-specific markers, such as tryptase12 and histamine,13 have been found to exhibit circadian variation.
Symptoms of allergic diseases are worst between midnight and early morning suggesting the involvement of the circadian clock in allergic disorders. Moreover, some medications used for the treatment of asthma, such as glucocorticoids or β2-adrenoceptor agonists, reset clock gene expression in vivo.14 The timing of allergic symptoms suggests that the circadian clock controls the physiology of MC and eosinophils. Here, we analysed whether the circadian clock is involved in the functionality of eosinophils and MC in mice and humans.
Materials and methods
Animals, treatments and tissues
Eleven-week-old C57BL/6 male mice were housed in a temperature- and humidity-controlled facility (23–24°, 60% humidity). Mice were entrained to a light–dark cycle of 12 hr light and 12 hr darkness (LD) for 2 weeks with food available ad libitum. Mice were anaesthetized with isoflurane and blood and small intestine were removed every 4 hr around the circadian cycle in total darkness under dim red light. Small intestine and blood were removed to analyse lamina propria cells and polymorphonuclear cells (PMNC), respectively. Mice were humanely killed at the end of the experiment. The joint ethics committee (IACUC) of the Hebrew University and Hadassah Medical Centre approved the study protocol.
Isolation of mouse PMNC
Mouse peripheral blood was collected into EDTA tubes and PMNC were isolated by a Ficoll density gradient solution (ACCUSPIN™ System-Histopaque®-1077, Sigma Chemical Co. St Louis, MO). Diluted blood samples in PBS (1 : 1) were layered onto the low-gradient Ficoll solution (3·3 : 1). Tubes were then centrifuged at 400 g for 30 min at 4°. After centrifugation the supernatant was discarded and the interface between plasma and Histopaque®-1077 containing the PMNC was collected. The PMNC were centrifuged at 100 g for 10 min at 4° and washed twice with sterile PBS and the final pellet was resuspended with 0·5 ml TriReagent (Sigma, Rehovot, Israel).
Enrichment of mouse lamina propria cells
Mouse jejunum was recovered and intestinal cells, located from the top of the villus to the bottom of the crypt, were removed using a previously described method.15 Briefly, the lumen of the intestine was washed with saline and turned inside out with a Pasteur pipette. The everted segment was slipped off the pipette and placed in saline at room temperature in a Petri dish and filled with the first solution (solution A), which contained 27 mm sodium citrate, 1·5 mm KCl, 96 mm NaCl, 8 mm KH2PO4, 5·6 mm Na2HPO4 at pH 7·3, and shaken at 75 r.p.m. at 37°. After 15 min, solution A was discarded and the jejunum was then filled with solution B containing PBS, 1·5 mm EDTA and 0·5 mm dithiothreitol. This solution selectively releases intestinal cells from their basement membrane; the cells at the top of the villus are the first to come off. After each removal, new solution B was added to the lumen for a different incubation duration and removed after additional cells were released from the lining of the intestine. Solution B, in the lumen, was changed eight times to give nine intestinal fractions. Fraction 9, which was isolated from the crypts at the base of the villi, contained enriched lamina propria MC16,17 and was centrifuged at 900 g for 5 min at 4° and the final pellet was resuspended with 0·5 ml TriReagent (Sigma) for RNA analyses or with lysis buffer for protein analyses.
Isolation of human peripheral eosinophils and sample collection
Eosinophils were isolated from peripheral blood of healthy non-allergic people and people with allergy against grass and flower pollen, respectively, by means of centrifugation on a density gradient of Pancoll (PAN-Biotech, Aidenbach, Germany) and using an eosinophil isolation kit (Miltenyi Biotech, Bergisch-Gladbach, Germany). All volunteers had regular sleep–wake patterns and none of them had travelled across time zones 3 months before or during the study. At the time of blood collection all allergic donors showed symptoms of allergy and took no medication 1 week before blood collection. Blood from all donors was withdrawn at 8·00 am. Permission to conduct the study was obtained from the local ethics committee. Purified eosinophils (purity of > 99·5%) were resuspended in RPMI-1640 supplemented with 10% heat-inactivated fetal calf serum (Gibco® Life-Technologies, Darmstadt, Germany), 100 μg/ml streptomycin and 100 U/ml penicillin (PAN-Biotech). Immediately afterwards, eosinophils were harvested every 3 hr over a period of 24 hr.
Isolation, culture and treatment of human intestinal mast cells
Human intestinal mast cells (hiMC) were isolated from macroscopically normal surgery tissue specimens derived from patients who underwent bowel resection, as described in detail elsewhere.18 Permission to conduct the study was obtained from the local ethics committee. Freshly isolated hiMC with a purity > 90% were analysed immediately after purification over a period of 24 hr. Human intestinal MC with a purity < 90% were cultured with 25 ng/ml stem cell factor in combination with 2 ng/ml interleukin-4 (IL-4; PeproTech, Rocky Hill, NJ) for at least 10 days to reach purity > 99% before long-term circadian rhythm measurements. For this purpose, hiMC were synchronized by addition of 40 μm dexamethasone (Sigma-Aldrich Chemie, Steinheim, Germany).19
Stimulation of hiMC through IgE receptor (FcεRI) cross-linking
Synchronized hiMC were treated with myeloma IgE for 90 min and stimulated every 4 hr with 1 μg/ml anti-human IgE for 10 or 90 min to analyse the release of histamine or cysteinyl leukotrienes (cysLT), respectively, in collected supernatants.
Release of histamine and cysLT
ELISA kits were used to measure the amount of histamine (IBL International GmbH, Hamburg, Germany) and cysLT (Enzo Life Sciences GmbH, Lörrach, Germany) in supernatants according to the manufacturer's instructions. The per cent release of histamine was calculated as follows: (histamine activity in the supernatant fraction/total histamine activity in the cellular and supernatant faction) × 100.
RNA extraction and quantitative real-time RT-PCR
Total RNA was extracted from human MC and eosinophils using an RNeasy mini kit (Qiagen, Hilden, Germany) and from mouse blood PMNC and enriched lamina propria MC using TriReagent (Sigma). Real-time RT-PCR was performed as described previously.3,20 For RT-PCR, total RNA was treated for 15 min at 37° with DNase I (Promega, Madison, WI). The reaction was stopped with 25 mm EDTA (Invitrogen™, Life Technologies, Darmstadt, Germany) for 10 min at 70°. After denaturation, cDNA was synthesized by adding 20 pmol random primers (Fermentas, St Leon-Rot, Germany), 5 mm dNTP (Genaxxon Bioscience, Biberach, Germany) and superscript™ III (Invitrogen™, Life Technologies) or MMuLV (Promega) reverse transcriptase. Finally, the reaction was inactivated by heating at 70° for 15 min. To quantify mRNA expression, real-time PCR were performed in optical tubes. Primers for all genes were tested alongside the normalizing gene Gapdh. Sequences of the specific human and mouse primers used are listed in Table 1. Primers were designed with Primer Express v.2 (Applied Biosystems, Foster City, CA) and validated by a standard curve and dissociation curve of the product. Reaction mixture without cDNA was used as negative control. All reactions were performed using the ICycler (Bio-Rad Labratories, Munich, Germany) or ABI Prism 7300 Sequence Detection System (Applied Biosystems). The fold change in target gene expression was calculated by the 2−ΔΔCt relative quantification method (Applied Biosystems).
Table 1.
Quantitative real-time PCR primer sequences
| Gene | Forward primer | Reverse primer |
|---|---|---|
| mPer1 | 5′-CCGAATACACACTTCGAAACCAG-3′ | 5′-TCCCGTTTGCAACGCAG-3′ |
| hPer1 | 5′-GGACCGACCCCTCATGCT-3′ | 5′-CCCGCCAACTGCAGAATCT-3′ |
| mPer2 | 5′-CGGGCTATGAAGCGCCTAG-3′ | 5′-GGTTGTTGTGAAGATCCTCTTCTCA-3′ |
| hPer2 | 5′-TAACTGTAGCTTCAGCGCGT-3′ | 5′-ACCTGTGTAAGCACACACAC-3′ |
| hCry1 | 5′-CAATGGTGAACCATGCTGAG-3′ | 5′-GCCTCCATTCCCATTAGGAT-3′ |
| mBmal1 | 5′-CAAGAATGCAAGGGAGGCC-3′ | 5′-TTGTCCCGACGCCTCTTTT-3′ |
| hBmal1 | 5′-ATTGAAACACCTCATTCTCAGGG-3′ | 5′-CTTACGACAAACAAAAATCCATCTG-3′ |
| mClock | 5′-CCTAGAAAATCTGGCAAAATGTCA-3′ | 5′-CCTTTTCCATATTGCATTAAGTGCT-3′ |
| hClock | 5′-ACAGCTGCTGACAAAAGCCAA-3′ | 5′-TGTGTTTATACGATTATCTGACCCAGAA-3′ |
| mc-kit | 5′-CTGCCATGACAGTTGCCGT-3′ | 5′-CTGTTAAATGGGCACTTGGTTTG-3′ |
| hc-kit | 5′-GCCCAATATAAAAGGCAAAT-3′ | 5′-AGTGCAAATGGTTACTTCCA-3′ |
| mMcpt-5 | 5′-TCCACGACATCATGCTACTGAAG-3′ | 5′-CTAGGGTTAGCTTGGCTTTCTCC-3′ |
| mMcpt-7 | 5′-ATGGTGTAAACCTGCCGCC-3′ | 5′-TGGCAACTTGCACCTCCTTC-3′ |
| hTryptase | 5′-TCTACACCCGTGTCACCTAC-3′ | 5′-TAGGAAGCAGTGGTGTTTTG-3′ |
| mFcεRI | 5′-TCACTGGAAGGTCTGCCCCAG-3′ | 5′-CAAGAGACATGAACAGCAGTGCT-3′ |
| hFcεRI | 5′-GTAATAAAAGCTCCGCGTG-3′ | 5′-TGAAGCCTTTCCTGGTTCTC-3′ |
| mEcp | 5′-ATTTCCAGGACAACCAGCCC-3′ | 5′-CCTGTCTGCTTTGTGGCTCC-3′ |
| hEcp | 5′-CAGTTCTCACAGGAGCCACA-3′ | 5′-AGCCCTCCACACCCATAAG-3′ |
| hEdn | 5′-CTACAGCGCGGAGACTGG-3′ | 5′-GAGGTTTGACATGGAGTGAGC-3′ |
| mEpo | 5′-CCCCTGGAAAGAGGCGTAAT-3′ | 5′-AGACATCCCGGACAAGAGGA-3′ |
| mMbp | 5′-CAGCCTTGCCCTGGTGG-3′ | 5′-GCCTCCTTGAGTACACAGGGTC-3′ |
| hCxcl8 | 5′-CTGAGAGTGATTGAGAGTGG-3′ | 5′-ACAACCCTCTGAACCCAGTT-3′ |
| mGapdh | 5′-CAAGAGGTGGACACAGTGGAGA-3′ | 5′-CGGCCACTATATTCTTCAAGGC-3′ |
| hGapdh | 5′-TGGTCTCCTCTGACTTCAAC-3′ | 5′-CCTGTTGCTGTAGCCAAATT-3′ |
Western blot analysis
Western blotting was performed as described.19,21 Blots of mouse jejunum cells were incubated with anti-mouse MCPT-5 polyclonal antibody (Proteintech, Chicago, IL) and with anti-rabbit c-KIT polyclonal antibody (Abcam, Cambridge, UK). Anti-mouse actin (MP Biomedicals, Solon, OH) was used to detect actin, the loading control. Blots of human eosinophils and hiMC were probed with mouse anti-human ECP monoclonal antibody (Pharmacia & Upjohn Diagnostics, Uppsala, Sweden) or mouse anti-human tryptase monoclonal antibody (Chemicon® International, Millipore Corporation, Billerica, MA), respectively. Membranes were incubated a second time with anti-β-actin (Cell Signaling Technology, New England Biolabs, Frankfurt am Main, Germany) or anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA) to normalize the obtained signals.
Statistical analyses
All results are expressed as means ± SEM. The acrophase and P-value of gene oscillation were calculated using the free Cosinor analysis software (Version 2·3) available at http://www.circadian.org/softwar.html. The significance level for all analyses was set at P < 0·05. Statistical analysis was performed with JMP software (version 8·1, SAS Institute Inc. Cary, NC).
Results
Clock and eosinophil-specific molecules oscillate in murine blood PMNC
First, we studied whether clock genes are expressed in a circadian manner in PMNC of mice maintained in LD conditions. Expression of clock genes was determined every 4 hr around the circadian cycle by analysing PMNC isolated from peripheral blood. The mRNA for mPer1, mPer2, mBmal1 and mClock showed robust oscillation (P < 0·05) (Fig. 1). We next examined the expression of the eosinophil-specific molecules mMbp, mEpo and mEcp. We found that these genes also exhibited significant circadian oscillation in mouse eosinophils (P < 0·05) (Fig. 1).
Figure 1.
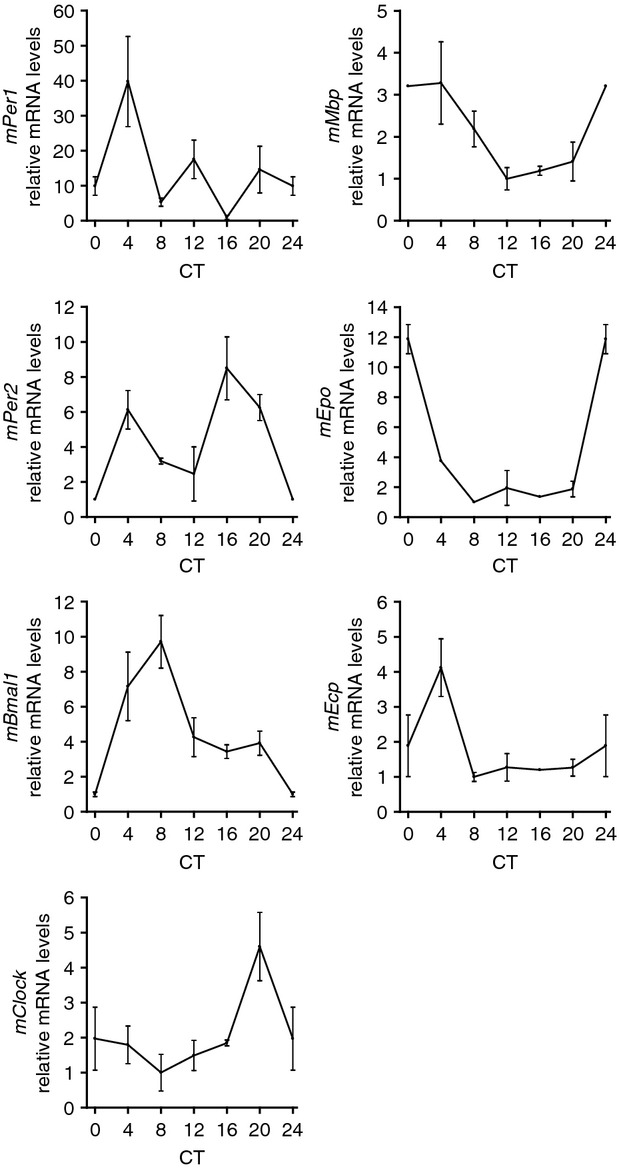
Circadian mRNA expression of clock genes (mPer1, mPer2, mBmal1 and mClock) and eosinophil-specific molecules (mMbp, mEpo and mEcp) in murine polymorphonuclear cells (PMNC). PMNC were isolated around the circadian cycle from mouse peripheral blood. Total RNA was extracted, reverse-transcribed and analysed by quantitative real-time PCR. Clock gene expression levels were normalized using mGapdh, n = 4 (measured each in triplicates), CT = circadian time, CT0 is the time when the light came on during the light–dark cycle. PMNC purity was > 95%.
Clock- and eosinophil-specific molecules oscillate in eosinophils of healthy and allergic subjects
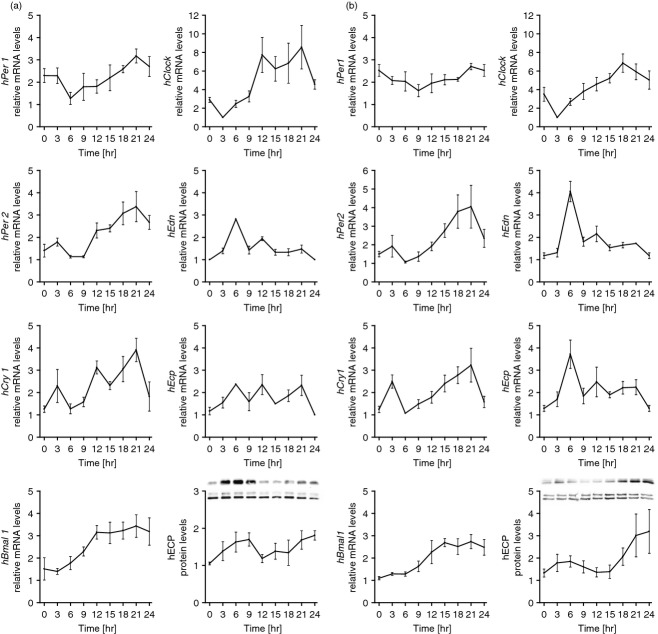
After having established clock functionality in mouse eosinophils, we studied whether clock genes are expressed in a circadian manner in human eosinophils. Eosinophils were isolated and purified from peripheral blood of non-allergic and allergic donors. Cells were harvested and analysed every 3 hr around the circadian cycle. All clock genes hPer1, hPer2, hCry1, hBmal1 and hClock were expressed and hPer1, hPer2, hBmal1 and hClock showed significant circadian rhythm (P < 0·05) in eosinophils isolated from non-allergic (Fig. 2a) and allergic (Fig. 2b) donors. Single donor analysis revealed significant circadian rhythm (P < 0·05) in 0/4 non-allergic donors for hPer1, and in 2/4 non-allergic donors for each of hPer2, hBmal1 and hClock. For allergic donors, single donor analysis revealed significant circadian rhythm (P < 0·05) of 1/5 (hPer1) and 2/5 (hPer2, hBmal1 and hClock). Furthermore, we analysed the expression of eosinophil-specific mediators and found circadian rhythm of hEdn and hEcp in eosinophils from allergic and non-allergic subjects. In parallel, protein expression of ECP also revealed a circadian oscillation (Fig. 2a,b). We also found oscillation of the clock genes hPer1 and hBmal1 in eosinophils when analysed over a period of 48 hr (see Supplementary material, Fig. S1). Hence, the circadian clock is expressed in human eosinophils and cell-specific mediators show oscillation. Importantly, we found no disruption in the circadian expression of clock genes or eosinophil-specific molecules during allergy.
Figure 2.
Circadian mRNA expression of clock genes (hPer1, hPer2, hCry1, hBmal1 and hClock) and eosinophil-specific molecules [hEdn and hEcp mRNA and eosinophil cationic protein (ECP)] in human blood eosinophils of non-allergic subjects (a) and allergic subjects (b). Purity of eosinophils was > 99·5%. Eosinophils were cultured and harvested every 3 hr around the circadian cycle. Total RNA was extracted, reverse-transcribed and analysed by quantitative real-time PCR. Clock gene expression levels were normalized using hGapdh, n = 4 (non-allergic donors) and n = 5 (allergic donors), each measured in triplicates. Protein levels of ECP were normalized using GAPDH. Representative Western blots for ECP are shown (top row) alongside the loading control (bottom row).
Clock genes and MC-specific molecules oscillate in mouse jejunum
We next set out to establish whether a circadian clock is functional in mouse MC. As mature MC are only found in tissues and we and others have previously shown a functional circadian clock in intestinal tissue,3,22 we isolated cells from intestinal tissue. Mice were maintained under LD conditions and the expression of clock genes was determined every 4 hr around the circadian cycle. mPer1, mPer2, mClock and mBmal1 mRNA showed robust oscillation in mouse lamina propria cells (P < 0·02) (Fig. 3). We next examined the expression of the MC-specific molecules mFcεRI α-chain, mMcpt-7, mMcpt-5 and mc-kit mRNA and mMCPT-5 and mc-Kit proteins (Fig. 3) and found that these genes and proteins exhibited significant circadian oscillation in MC (P < 0·02).
Figure 3.
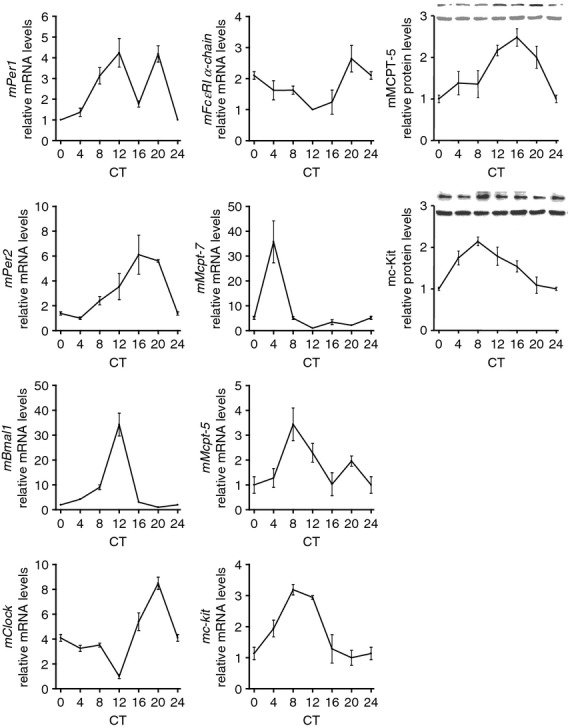
Circadian mRNA expression of clock genes (mPer1, mPer2, mBmal1 and mClock) and mast cell (MC) -specific molecules (mFcεRI α-chain, mMcpt-7, mMcpt-5 and mc-kit mRNA and mMCPT-5 and mc-Kit protein) in enriched mouse lamina propria mast cells. Mast cell purity was > 90%. Cells were collected around the circadian cycle. Total RNA was extracted, reverse-transcribed and analysed by quantitative real-time PCR. Clock gene expression levels were normalized using mGapdh, n = 4 (measured each in triplicates). Protein levels of mMCPT-5 and mc-Kit were normalized using β-actin, n = 4, CT = circadian time, CT0 is the time when the light came on during the light–dark cycle. Representative Western blots for mMCPT-5 and mc-Kit are shown (top row) alongside the loading control (bottom row).
Clock and MC-specific molecules exhibit circadian oscillation in hiMC
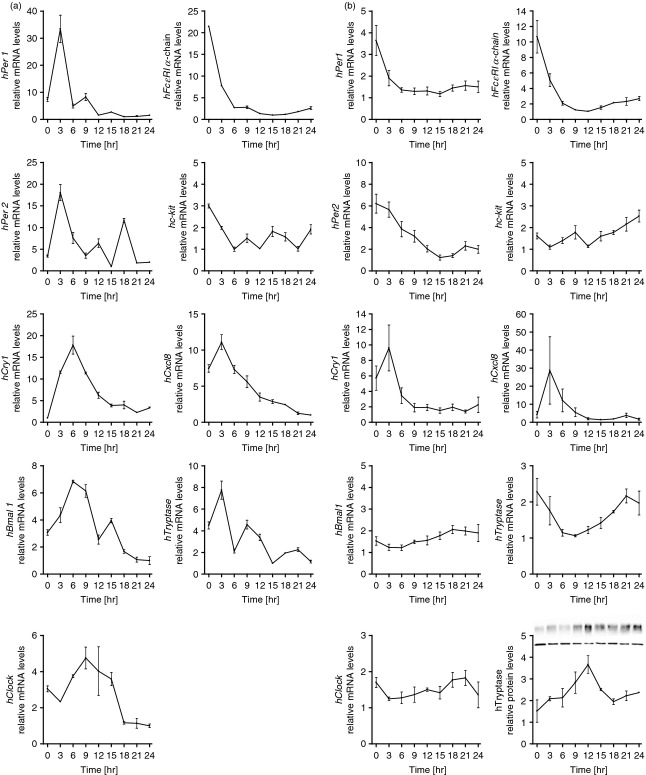
After confirming clock functionality in mouse jejunum and verifying the oscillation of MC-specific molecules, we set out to study the role of the biological clock in MC isolated from human intestinal tissue. Freshly isolated hiMC were cultured and harvested every 3 hr over a period of 24 hr. Analysis of clock genes revealed that all clock genes hPer1, hPer2, hCry1, hBmal1 and hClock were expressed and showed oscillation (Fig. 4a). Moreover, MC-specific molecules, i.e. hFcεRI α-chain, hc-kit and hTryptase, were found to exhibit oscillation (Fig. 4a). To analyse the hiMC with > 99% purity, long-term cultured hiMC were examined. Cultured hiMC were treated with dexamethasone to synchronize the cells and subsequently harvested every 3 hr over a period of 24 hr. Analysis of clock genes revealed that all clock genes, hPer1, hPer2, hCry1, hBmal1 and hClock, were expressed and that hPer1, hPer2 and hBmal1 showed significant circadian rhythm (P < 0·05) (Fig. 4b), but with reduced amplitude compared with freshly harvested cells (Fig. 4a). Furthermore, we examined the expression of MC-specific molecules, i.e. hFcεRI α-chain, hc-kit and hTryptase (Fig. 4b), and found that hFcεRI α-chain and hTryptase exhibited significant circadian rhythm (P < 0·05), also with reduced amplitude compared with freshly harvested cells (Fig. 4a). Correspondingly, tryptase protein expression also showed oscillation (Fig. 4b). Recently, we reported that MC are a potent source of chemokines, such as CXCL8.20 Analysis of hCxcl8 mRNA expression showed that in addition to MC proteases, hCxcl8 was also expressed in a circadian manner (Fig. 4a,b). We also found oscillation of the clock genes hPer1 and hBmal1 in hiMC when analysed over a period of 48 hr (see Supplementary material, Fig. S2), similar to what was observed in eosinophils (see Supplementary material, Fig. S1).
Figure 4.
(a) Circadian expression of clock genes (hPer1, hPer2, hCry1, hBmal1 and hClock mRNA) and mast cell (MC) -specific molecules (hFcεRI α-chain, hc-kit, hCxcl8, hTryptase mRNA) in freshly isolated human intestinal MC (hiMC) harvested every 3 hr around the circadian cycle (one representative out of three experiments measured in triplicates is shown). (b) Circadian expression of clock genes (hPer1, hPer2, hCry1, hBmal1 and hClock mRNA) and mast cell-specific molecules (hFcεRI α-chain, hc-kit, hCxcl8, hTryptase mRNA and tryptase protein) in long-term cultured hiMC. Purity of freshly isolated hiMC was > 90%, purity of long-term cultured hiMC was > 99%. The hiMC were synchronized with 40 μm dexamethasone for 2 hr and subsequently harvested every 3 hr. Total RNA was extracted, reverse-transcribed and analysed by quantitative real-time PCR. Clock gene expression levels were normalized using hGapdh, n = 4 (measured each in triplicates). Protein levels of tryptase were normalized using β-actin. Representative Western blot for tryptase is shown (top row) alongside the loading control (bottom row).
Histamine and cysLT exhibit a daily release by hiMC
To connect the circadian clock to hiMC functionality, hiMC synchronized with dexamethasone were activated by IgE receptor cross-linking every 4 hr around the circadian cycle. Both pre-stored histamine (Fig. 5a) and de novo synthesized cysLT (Fig. 5b) were released in response to IgE-mediated stimulation in a circadian manner by hiMC. Interestingly, the highest release of both histamine and cysLT was detected at time-point 20 hr.
Figure 5.
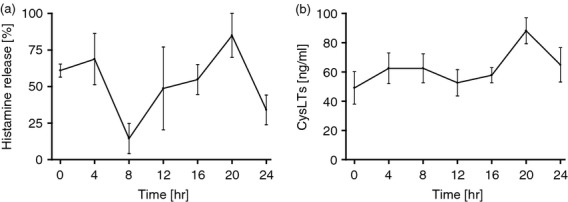
Circadian release of pre-stored histamine (a) and de novo synthesized cysteinyl leukotrienes (cysLT) (b) by human intestinal mast cells (hiMC) in response to FcεRI cross-linking. The hiMCs were synchronized with 40 μm dexamethasone for 2 hr and stimulated by FcεRI cross-linking every 4 hr around the circadian cycle. The release of histamine is shown in per cent of the total cell content for every time-point, n = 2, release of cysLT, n = 4, hiMC purity was > 99%.
Discussion
We found that essential elements of the biological clock in eosinophils and MC exhibit circadian expression in mice and humans. These results are supported by the findings that allergic attacks, such as asthma and allergic rhinitis, are frequently exacerbated between midnight and early morning, suggesting a role of the biological clock. Circadian clock genes have been shown to be expressed in a number of immune cells, such as peripheral blood mononuclear cells,23,24 macrophages25 as well as mouse and human CD4+ T cells.26 In peripheral blood mononuclear cells, oscillation has been found for hBmal1, hCry1, hPer2 and in particular for hPer1.23,24 Circadian rhythm for hClock could not be detected in human oral mucosa and skin,27 but was found in whole blood cells.28 However, we detected that hPer1, hPer2, hBmal1 and hClock had a robust rhythmic expression in human eosinophils isolated from peripheral blood. Hence, rhythmic expression of clock genes may vary among different cells and tissues. In addition, circadian rhythms of the clock genes mPer1, mPer2, mClock and mBmal1 were detected in murine PMNC.
It is noteworthy that although the procedure of sampling was different for murine PMNC and human eosinophils, i.e. drawing blood every 4 hr from mice versus drawing blood at one time-point from volunteers followed by culturing and then harvesting every 3 hr, robust oscillations were achieved. These results demonstrate the existence of an endogenous pacemaker that keeps running even after removal of the tissue, e.g. blood or intestine, a characteristic of a circadian clock.
Circadian variations have been reported for eosinophil-specific mediators, such as serum ECP and serum and urine EPX with nocturnal and early morning peaks.10 Furthermore, ECP was significantly increased in nasal secretions of allergic children in the morning29 as well as EDN/EPX in sputum30 and in urine.31 These findings provide an explanation for the rhythmic mRNA expression of eosinophil-specific proteins likely to be regulated by the clock. Indeed, our results show circadian variations for hEcp and hEdn mRNA as well as for ECP in human eosinophils from peripheral blood and mMbp, mEpo and mEcp in murine blood PMNC, confirming the functionality of the biological clock in eosinophils.
Surprisingly both eosinophils from subjects suffering from allergy and eosinophils from healthy donors showed a functional circadian clock. Although disruption of circadian expression caused by sleep disturbances has been shown to exacerbate allergic reactions,32 the state of circadian rhythms during allergy has never been tested to our knowledge. According to our results we assume that the allergic reaction does not feed back to the circadian clock. However, a more rigorous around-the-clock sampling from humans should be performed. This is unlike metabolic factors that are controlled by the circadian clock but their disruption leads to disruption of circadian rhythms.33 It is noteworthy that clock genes can respond to many different stimuli and the rhythm of immune genes observed in vitro could exhibit major differences in terms of phase and amplitude when analysed in vivo.
Besides eosinophils, we tested the functionality of the circadian clock in MC. We demonstrated that in murine jejunal cells the clock mechanism is expressed and that MC-specific molecules, such as proteases, show a circadian rhythm. These findings are consistent with the existence of a clock mechanism in peripheral tissues. Moreover, our results support previous observations in which other immune system components, such as defensins and Toll-like receptors were found to oscillate in intestinal tissue.2,3,19 In addition, circadian clock genes have been shown to be rhythmically expressed in mouse bone marrow-derived MC synchronized by serum shock in vitro.34
We show for the first time around-the-clock expression in cells isolated from human tissue. Although some studies have shown altered expression of some clock genes, circadian expression in human tissues has never been shown because of the difficulty in getting around-the-clock tissue samples. Our report is the first to show around-the-clock oscillation in MC isolated from intestinal tissue. It should be noted that clock genes including hPer1, hPer2, hCry1 and hBmal1 as well as hClock are expressed in an oscillatory pattern in freshly isolated mature hiMC as well as in long-term synchronized cultured hiMC. Furthermore, MC-specific genes such as hFcεRI α-chain and hTryptase show circadian variation. Moreover, we found that in addition to MC-specific molecules, hCxcl8 is also expressed in a circadian manner. The long-term circadian analysis revealed that after at least 10 days in culture, clock genes and MC-specific molecules showed decreased amplitude. This phenomenon is a characteristic of the circadian mechanism leading to rhythms that drift away when there is no synchronizing cue.35
A trend towards a circadian pattern in mRNA levels of IL-13 and IL-6 has been reported for IgE-mediated activation of bone marrow-derived MC, whereas IL-13 mRNA and IL-6 mRNA did not display any oscillation pattern following ionomycin stimulation, suggesting that circadian regulation of MC activation may be a result of the circadian variation in FcεRI expression.34 We found that FcεRI α-chain is indeed expressed in a circadian manner in mature human and mouse MC. However, the basal expression of hCxcl8 also showed circadian oscillation, whereas the highest expression of hCxcl8 was 3 hr after the hFcεRI α-chain expression peak. Hence, oscillation of cytokine production may not only depend on FcεRI expression but also on its circadian expression.
Notably, we found that the MC mediators histamine and cysLT, responsible for allergic symptoms, are released in a circadian manner by hiMC following IgE-mediated activation. Interestingly, the highest release of both histamine and cysLT was detected at the same time-point. These results suggest that the circadian clock is connected to the functionality of MC. We conclude that the circadian clock is involved in the functional activity of hiMC, leading to circadian production and release of their mediators, and consequently in allergic diseases. Hence, the influence of the biological clock on the responsiveness of MC to IgE-mediated activation may explain how the biological clock contributes to circadian allergic symptoms.
Recently, a time-of-day-dependent variation in cutaneous anaphylactic reaction in mice was reported to be regulated by Per2 supporting an involvement of the circadian clock in MC-mediated reactions.36 Moreover, Dugas-Breit et al. suggest a possible diurnal pattern of serum tryptase levels in both allergic patients and healthy controls with lower levels in the afternoon and higher levels in the morning.12 We demonstrated circadian expression of tryptase in hiMC on mRNA as well as at the protein level. Moreover, mouse MC proteases, such as mMcpt-5 and mMcpt-7, were rhythmically expressed in mouse jejunum. Hence, our findings suggest that the diurnal pattern of tryptase concentration and the diurnal expression of other MC-specific proteins are dependent on clock functionality.
In this respect it is noteworthy that an endotoxin-driven inflammatory response in mice has been found recently to be regulated in a time-of-day-dependent manner. Specifically, a subset of cytokines, such as the pro-inflammatory IL-6, is regulated in a circadian manner.37 Macrophages were identified as key to the rhythmic response to systemic lipopolysaccharide administration. Mast cells share some characteristics with macrophages, such as the production of pro-inflammatory cytokines/chemokines and, as shown here, the regulation by the circadian clock. It is tempting to speculate that MC are not only involved in a rhythmic response during allergic disease but also in the course of other inflammatory responses, such as lipopolysaccharide-mediated inflammation.
In conclusion, the biological clock is functional in MC and eosinophils leading to circadian expression of their mediators and, therefore, may play a pivotal role in the pathophysiology of allergy.
Acknowledgments
The authors thank Yvonne Soltow for excellent technical assistance. This work was supported by the Deutsche Forschungs-gemeinschaft (LO 581/7-1 to Axel Lorentz and Oren Froy).
Glossary
- BMAL1
brain and muscle ARNT-like protein
- CLOCK
circadian locomotor output cycles Kaput
- CRY
Cryptochromes
- cysLT
cysteinyl leukotrienes
- ECP
eosinophil cationic protein
- EDN/EPX
eosinophil-derived neurotoxin/eosinophil protein X
- EPO
eosinophil peroxidase
- IL-4
interleukin-4
- LD
light–dark cycle of 12 h light and 12 h darkness
- MBP
major basic protein
- MC
mast cells
- hiMC
human intestinal MC
- PER
Periods
- PMNC
polymorphonuclear cells
Disclosures
All authors declare that they have no conflicts of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Circadian mRNA expression of hPer1 and hBmal1 in human eosinophils over 48 hr.
Figure S2. Circadian mRNA expression of hPer1 and hBmal1 in human intestinal mast cells over 48 hr.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Froy O, Chapnik N, Miskin R. Mouse intestinal cryptdins exhibit circadian oscillation. FASEB J. 2005;19:1920–2. doi: 10.1096/fj.05-4216fje. [DOI] [PubMed] [Google Scholar]
- 3.Froy O, Chapnik N. Circadian oscillation of innate immunity components in mouse small intestine. Mol Immunol. 2007;44:1954–60. doi: 10.1016/j.molimm.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Vitaterna MH, King DP, Chang AM, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–25. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–76. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 6.Minai-Fleminger Y, Levi-Schaffer F. Mast cells and eosinophils: the two key effector cells in allergic inflammation. Inflamm Res. 2009;58:631–8. doi: 10.1007/s00011-009-0042-6. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff S, Crowe SE. Gastrointestinal food allergy: new insights into pathophysiology and clinical perspectives. Gastroenterology. 2005;128:1089–113. doi: 10.1053/j.gastro.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Hogan SP, Rosenberg HF, Moqbel R, et al. Eosinophils: biological properties and role in health and disease. Clin Exp Allergy. 2008;38:709–50. doi: 10.1111/j.1365-2222.2008.02958.x. [DOI] [PubMed] [Google Scholar]
- 9.Kariyawasam HH, Robinson DS. The eosinophil: the cell and its weapons, the cytokines, its locations. Semin Respir Crit Care Med. 2006;27:117–27. doi: 10.1055/s-2006-939514. [DOI] [PubMed] [Google Scholar]
- 10.Wolthers OD, Heuck C. Circadian variations in serum eosinophil cationic protein, and serum and urine eosinophil protein X. Pediatr Allergy Immunol. 2003;14:130–3. doi: 10.1034/j.1399-3038.2003.02038.x. [DOI] [PubMed] [Google Scholar]
- 11.Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol. 2007;7:93–104. doi: 10.1038/nri2018. [DOI] [PubMed] [Google Scholar]
- 12.Dugas-Breit S, Przybilla B, Schopf P, Rueff F. Possible circadian variation of serum mast cell tryptase concentration. Allergy. 2005;60:689–92. doi: 10.1111/j.1398-9995.2005.00771.x. [DOI] [PubMed] [Google Scholar]
- 13.Friedman BS, Steinberg SC, Meggs WJ, Kaliner MA, Frieri M, Metcalfe DD. Analysis of plasma histamine levels in patients with mast cell disorders. Am J Med. 1989;87:649–54. doi: 10.1016/s0002-9343(89)80398-5. [DOI] [PubMed] [Google Scholar]
- 14.Burioka N, Miyata M, Endo M, et al. Alteration of the circadian rhythm in peak expiratory flow of nocturnal asthma following nighttime transdermal β2-adrenoceptor agonist tulobuterol chronotherapy. Chronobiol Int. 2005;22:383–90. doi: 10.1081/cbi-200053587. [DOI] [PubMed] [Google Scholar]
- 15.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis I. An indicator of cellular differentiation. J Biol Chem. 1973;248:2536–41. [PubMed] [Google Scholar]
- 16.Hammel I, Arizono N, Galli SJ. Mast cells in rat dermis and jejunal lamina propria show a five-fold difference in unit granule volume. Cell Tissue Res. 1991;265:329–34. doi: 10.1007/BF00398080. [DOI] [PubMed] [Google Scholar]
- 17.Forbes EE, Groschwitz K, Abonia JP, et al. IL-9- and mast cell-mediated intestinal permeability predisposes to oral antigen hypersensitivity. J Exp Med. 2008;205:897–913. doi: 10.1084/jem.20071046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sellge G, Bischoff SC. Isolation, culture, and characterization of intestinal mast cells. Methods Mol Biol. 2006;315:123–38. doi: 10.1385/1-59259-967-2:123. [DOI] [PubMed] [Google Scholar]
- 19.Sherman H, Froy O. Expression of human β-defensin 1 is regulated via c-Myc and the biological clock. Mol Immunol. 2008;45:3163–7. doi: 10.1016/j.molimm.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Feuser K, Thon KP, Bischoff SC, Lorentz A. Human intestinal mast cells are a potent source of multiple chemokines. Cytokine. 2012;58:178–85. doi: 10.1016/j.cyto.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Lorentz A, Wilke M, Sellge G, et al. IL-4-induced priming of human intestinal mast cells for enhanced survival and Th2 cytokine generation is reversible and associated with increased activity of ERK1/2 and c-Fos. J Immunol. 2005;174:6751–6. doi: 10.4049/jimmunol.174.11.6751. [DOI] [PubMed] [Google Scholar]
- 22.Hoogerwerf WA, Hellmich HL, Cornelissen G, et al. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133:1250–60. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–5. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 24.Kusanagi H, Hida A, Satoh K, et al. Expression profiles of 10 circadian clock genes in human peripheral blood mononuclear cells. Neurosci Res. 2008;61:136–42. doi: 10.1016/j.neures.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Keller M, Mazuch J, Abraham U, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106:21407–12. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollinger T, Leutz A, Leliavski A, et al. Circadian clocks in mouse and human CD4+ T cells. PLoS ONE. 2011;6:e29801. doi: 10.1371/journal.pone.0029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjarnason GA, Jordan RC, Wood PA, et al. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001;158:1793–801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takimoto M, Hamada A, Tomoda A, et al. Daily expression of clock genes in whole blood cells in healthy subjects and a patient with circadian rhythm sleep disorder. Am J Physiol Regul Integr Comp Physiol. 2005;289:R1273–9. doi: 10.1152/ajpregu.00126.2005. [DOI] [PubMed] [Google Scholar]
- 29.Aoyagi M, Watanabe H, Sekine K, et al. Circadian variation in nasal reactivity in children with allergic rhinitis: correlation with the activity of eosinophils and basophilic cells. Int Arch Allergy Immunol. 1999;120(Suppl. 1):95–9. doi: 10.1159/000053604. [DOI] [PubMed] [Google Scholar]
- 30.Panzer SE, Dodge AM, Kelly EA, Jarjour NN. Circadian variation of sputum inflammatory cells in mild asthma. J Allergy Clin Immunol. 2003;111:308–12. doi: 10.1067/mai.2003.65. [DOI] [PubMed] [Google Scholar]
- 31.Storm van's GK, Mattes J, Gruntjens T, et al. Circadian variation of urinary eosinophil protein X in asthmatic and healthy children. Clin Exp Allergy. 1999;29:1497–501. doi: 10.1046/j.1365-2222.1999.00731.x. [DOI] [PubMed] [Google Scholar]
- 32.Baiardini I, Braido F, Cauglia S, Canonica GW. Sleep disturbances in allergic diseases. Allergy. 2006;61:1259–67. doi: 10.1111/j.1398-9995.2006.01221.x. [DOI] [PubMed] [Google Scholar]
- 33.Froy O. Metabolism and circadian rhythms – implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Reece SP, Van Scott MR, Brown JM. A circadian clock in murine bone marrow-derived mast cells modulates IgE-dependent activation in vitro. Brain Behav Immun. 2011;25:127–34. doi: 10.1016/j.bbi.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Fujioka A, Takashima N, Shigeyoshi Y. Circadian rhythm generation in a glioma cell line. Biochem Biophys Res Commun. 2006;346:169–74. doi: 10.1016/j.bbrc.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura Y, Harama D, Shimokawa N, et al. Circadian clock gene Period2 regulates a time-of-day-dependent variation in cutaneous anaphylactic reaction. J Allergy Clin Immunol. 2011;127:1038–45. doi: 10.1016/j.jaci.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Gibbs JE, Blaikley J, Beesley S, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:582–7. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.