Abstract
In a search for genes that regulate circadian rhythms in mammals, the progeny of mice treated with Nethyl-N-nitrosourea (ENU) were screened for circadian clock mutations. A semidominant mutation, Clock, that lengthens circadian period and abolishes persistence of rhythmicity was identified. Clock segregated as a single gene that mapped to the midportion of mouse chromosome 5, a region syntenic to human chromosome 4. The power of ENU mutagenesis combined with the ability to clone murine genes by map position provides a generally applicable approach to study complex behavior in mammals.
Progress has been made at the physiological and cellular levels in our understanding of circadian systems (1), yet the molecular mechanism of circadian clocks has not been fully elucidated (2). The isolation of “clock mutants” and the widespread requirement for protein synthesis in circadian clock systems imply that gene expression is an integral component of the oscillator (2). Recent molecular work with the Drosophila period (per) and Neurospora frequency (frq) genes suggests that a circadian cycle of per and frq transcription, respectively, may lie at the heart of the oscillator mechanism in these species (3). However, no information exists concerning the molecular elements of the clock system in mammals. In the absence of specific mechanistic information, genetics has been a powerful approach to uncover unknown elements. We report here the isolation of a mutation in the mouse that changes two central properties of circadian rhythms: the intrinsic period length and the persistence of rhythmicity. Taken together, our results define a gene, named Clock (for circadian locomotor out-put cycles kaput) that is essential for normal circadian behavior.
Because the majority of clock mutants isolated in other organisms have been semidominant (4), we screened heterozygotes directly in the mouse. With the mutagen ENU, average forward mutation frequencies of 0.0015 per locus per gamete (1 in 700) can be achieved in the mouse (5). Male mice of the inbred strain C57BL/6J (B6) were treated with a single injection of ENU and after recovery of fertility were mated with untreated B6 females (6). First generation (G1) offspring would be heterozygous for any induced mutations but otherwise possess an isogenic B6 background (Fig. 1A). Normal B6 mice exhibit a robust circadian rhythm of wheel-running activity; we used this behavioral assay to screen for circadian mutants (Fig. 1B). Activity rhythms were monitored during exposure to a light-dark cycle (LD) to assess synchronization or entrainment behavior and in constant darkness (DD) to determine the circadian period of the locomotor activity rhythm (7). Laboratory mice typically have circadian periods of less than 24 hours, with B6 mice having periods that average between 23.3 and 23.8 hours (8).
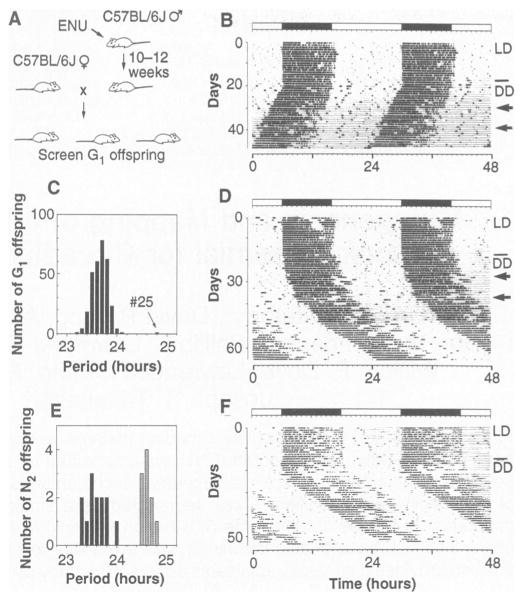
Fig. 1. ENU mutagenesis screen.
A behavioral assay was used to test for the presence of circadian rhythm mutations in firstgeneration offspring of ENU-treated males. (A) The mutagenesis procedure. Young male B6 mice were injected intraperitoneally with ENU at 150 mg/kg. On recovery of fertility, they were bred with B6 females to produce G1 offspring heterozygous for any induced mutations. Dominant or semidominant mutations could then be detected in G1 offspring. (B) Representative activity record of a wild-type G1 female from the screen. The record is double-plotted so that 48 hours are shown for each horizontal trace, and each day’s record is presented both to the right and beneath that of the preceding day. Wheel revolutions are recorded as pen deflections so that the dark regions represent times of activity. The animal was kept in LD14:10 for the first 20 days (illustrated by the bar above the record), then transferred to constant darkness (DD). Light pulses of 5 min were given on the days indicated by arrows. On transfer to DD, this animal exhibited a period of 23.7 hours. (C) Distribution of period among 304 G1 offspring tested for circadian phenotype. One animal, #25, exhibited a period that deviated from the mean by more than an hour. (D) Activity record of G1-25, the founder animal. This animal exhibited a period of 24.8 hours after 30 days in DD. (E) Distribution of period among 23 B6 N2 offspring from the founder animal. Thirteen offspring exhibited periods within the normal range, whereas 10 had periods longer than normal, comparable with that of the founder animal. (F) Representative activity record of a heterozygous male B6 N2 offspring. This animal had a period of 24.8 hours in DD.
We tested a total of 304 G1 offspring of ENU-treated males (9). The distribution of period lengths of the activity rhythms from the G1 mice was normal, with a mean of 23.7 hours and a standard deviation of 0.17 hours (Fig. 1C). One animal (G1-25) expressed a circadian period that gradually lengthened over the first 30 days of DD, stabilizing at 24.8 hours, more than six standard deviation units from the mean (Fig. 1D). The 303 other G1 mice all had wild-type period values ranging between 23.2 and 24.1 hours. Animal G1-25, a male, was mated with three normal B6 females, producing 23 backcross (N2) progeny among four litters.
To determine whether the long period was heritable, we measured the activity rhythms of these B6 N mice (10). Period lengths fell into two classes: 13 mice, both male and female, had periods in the normal range between 23.3 and 24.0 hours, whereas 10 mice, also of both sexes, had periods between 24.5 and 24.8 hours, corresponding to the founder animal’s period (Fig. 1, E and F). A 1:1 phenotypic ratio of normal to long periods would be expected among the progeny for a fully penetrant autosomal dominant period mutation; the observed ratio of 13:10 was not significantly different by a X2 test (see Table 1, cross 1), indicating the long-period phenotype was heritable. Ultimately, seven different types of crosses (Table 1) produced 362 progeny which were phenotypically tested (11). The pedigree included crosses with two other inbred strains, BALB/cJ (BALB) and C3H/HeJ (C3H) (12). All observed phenotypic ratios for Clock were consistent with those predicted for a single-locus semidominant autosomal mutation (see Table 1 for X2 analysis) (13).
Table 1.
Inheritance of Clock. The segregation of phenotype among seven different crosses is shown. The maternal genotype is listed first for each of the crosses. In cases in which reciprocal crosses were performed, the two crosses are shown first separately, then combined. Clock phenotype was assayed by monitoring the circadian rhythm of locomotor activity of offspring kept in DD. The Clock/+ heterozygotes were distinguished by a longer and less stable free-running period than wild types (+/+); the Clock/Clock homozygotes were distinguished by a very long periodicity and subsequent loss of significant periodicity in the circadian range. These genotypic classifications have been confirmed by test cross (crosses 1, 4, and 5) or by the genotype of flanking SSLP markers (crosses 6 and 7) (13). Expected values (Exp) for each class were calculated on the basis of an autosomal semidominant mode of inheritance. Obs, observed.
| Cross | Genotype* of parents | Total progency | Offspring genotype
|
χ2 | P | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| +/+
|
Clock/+
|
Clock/Clock
|
||||||||
| Exp | Obs | Exp | Obs | Exp | Obs | |||||
| 1 | B6 +/+ × B6 Clock/+ | 23 | 11.5 | 13 | 11.5 | 10 | 0.391 | >0.5 | ||
| 2A | B6 Clock/+ × B6 +/+ | 4 | 2 | 2 | 2 | 2 | 0 | >0.99 | ||
| 2B | B6 +/+ × B6 Clock/+ | 42 | 21 | 21 | 21 | 21 | 0 | >0.99 | ||
| 2 | Combined 2A and 2B | 46 | 23 | 23 | 23 | 23 | 0 | >0.99 | ||
| 3 | B6 Clock/+ × B6 Clock/+ | 20 | 5 | 7 | 10 | 10 | 5 | 3 | 1.6 | >0.3 |
| 4 | C3H +/+ × B6 Clock/+ | 28 | 14 | 13 | 14 | 15 | 0.036 | >0.7 | ||
| 5 | BALB +/+ × B6 Clock/+ | 30 | 15 | 15 | 15 | 15 | 0 | >0.99 | ||
| 6 | (BALB × B6)F1 Clock/+ × (BALB × B6)F1 Clock/+ | 115 | 28.75 | 26 | 57.5 | 61 | 28.75 | 28 | 0.495 | >0.7 |
| 7A | (BALB × B6)F1 Clock/+ × B6 +/+ | 48 | 24 | 29 | 24 | 19 | 2.083 | >0.1 | ||
| 7B | B6 +/+ × (BALB × B6)F1 Clock/+ | 52 | 26 | 29 | 26 | 23 | 0.692 | >0.3 | ||
| 7 | Combined 7A and 7B | 100 | 50 | 58 | 50 | 42 | 2.56 | >0.1 | ||
B6, C57BL/6J; C3H, C3H/HeJ; BALB, BALB/cJ.
Intercrosses between Clock/+ heterozygotes produced animals of three phenotypic classes in the F2 generation, indicating that the mutation was semidominant. In the F2 progeny of both of these intercrosses (Table 1, crosses 3 and 6), the wild-type (Fig. 2, A and E) and Clock/+ (Fig. 2, B and F) phenotypes were observed. However, approximately one-quarter of the F2 offspring had extremely long periods of 26 to 29 hours on initial transfer to DD (Fig. 2, C, D, G, and H). The long, 26- to 29-hour period was followed by a complete loss of circadian rhythmicity after about 2 weeks in DD (Fig. 2C). Thus, steady-state period, as estimated by a X2 periodogram (14), fell into three classes: short (mean ~23.3 hours) for wild types; long (mean ~24.4 hours) for Clock/+; and very long (mean ~27.3 hours) for Clock/Clock. The quantitative analyses of circadian phenotype of progeny segregating Clock from the two interstrain (BALB × B6)F1 Clock/+ crosses (Table 1, crosses 6 and 7) are shown in Table 2. Highly significant differences in period length were found between the wildtype and Clock/+ offspring, as well as between the Clock/Clock animals and the other two classes in the (BALB × B6)F2 mice (Table 2).
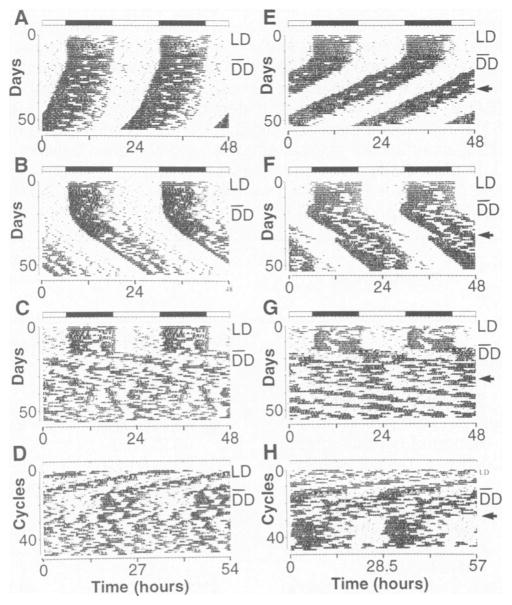
Fig. 2. Circadian activity records of F2 generation mice.
The activity LD records of six F2 offspring [B6 F DD A to D; (BALB × B6)F2, E to H] are shown. All animals were kept on LD12:12 for the first 7 to 10 days illustrated, then transferred to DD on the day indicated by a line in the right margin. (A) Activity record of a wild-type B6 F2 mouse. This animal had a period of 23.6 hours in DD. (B) Activity record of a heterozygous Clock/+ B6 F2 mouse. This animal had a period of 24.8 hours in DO. (C) Activity record of a homozygous Clock/Clock B6 F2 mouse. This individual had a period of 27.1 hours for the first 10 days in DD. There is a loss of circadian rhythmicity thereafter and only an ultradian rhythm is apparent. (D) The same activity record as illustrated in (C), plotted on a 27-hour time base. The approximately 27-hour periodicity during the first part of exposure to DD appears as vertically aligned activity bouts. (E) Activity record of a wild-type (BALB × B6)F2 mouse. This individual had a steady-state period of 23.1 hours in DD. A 6-hour light pulse was given on the day indicated by an arrow, resulting in a 1-hour phase advance in the activity rhythm. (F) Activity record of a heterozygous Clock/+ (BALB × B6)F2 mouse. This individual had a steady-state period of 24.7 hours in DD. A 6-hour light pulse was given on the day indicated by an arrow, resulting in a 3.3-hour phase advance in the activity rhythm. (G) Activity record of a homozygous Clock/Clock (BALB × B6)F2 mouse. This animal had a long period during the initial interval in DD, which damped out after about five cycles leaving an ultradian pattern. After a 6-hour light pulse indicated by the arrow, a period of 28.2 hours was observed, which then persisted for at least 15 cycles. (H) The same activity record as illustrated in (G) plotted on a 28.5-hour time base to show the transient restoration of a long periodicity after the light pulse by vertically aligning activity bouts.
Table 2.
Phenotypic characteristics of Clock. Characteristics of the activity rhythm were subjected to quantitative analysis. Period estimates were performed with 10 consecutive days of data by a χ2 periodogram analysis (14). Steady-state periods were calculated with the 10-day interval during exposure to DD when an individual period value was stable. Change in period was calculated as the difference in period between the first and second 10-day intervals in DD. Amplitude is the logarithm of the PSD of the circadian (18 to 30 hour) peak from a Fourier analysis (18). Total activity counts are the average per 24 hours of 5 days during exposure to LD12:12 or 10 days during exposure to DD. Daytime activity is the percentage of the total activity that occurred during the 12-hour light period of 5 days during exposure to LD. Values shown are the mean ± standard error. The P values indicated are from a Student’s t test or a one-way analysis of variance (ANOVA) using a Tukey’s studentized range test. The Clock genotype was assigned by the circadian behavioral phenotype and by SSLP genotype. The genotypes of 100 of the [(BALB × B6)F1 × B6]N2 mice and 113 of the (BALB × B6)F2 mice were tested by the genotype of SSLP markers flanking the Clock locus. SSLP-nonrecombinant animals showed no recombination between flanking SSLP markers, whereas SSLP-recombinant animals showed recombination between the flanking markers. Thirteen [(BALB × B6)F1 × B6]N2 mice and 29 (BALB × B6)F2 mice were recombinant between the flanking SSLPs, and 2 (BALB × B6)F2 mice were not typed by SSLPs, so that the presumed Clock genotype was assigned by phenotype (13)
| Characteristic | Assigned genotype
|
||
|---|---|---|---|
| +/+ | Clock/+ | Clock/Clock | |
| [(BALB × B6)F1 × B6]N2 progeny | |||
| Total number of progeny | 58 | 42 | |
| SSLP-nonrecombinant | 49 | 38 | |
| SSLP-recombinant | 9 | 4 | |
| Steady-state period (hours) | 23.43 ± 0.026 | 24.63 ± 0.039* | |
| Range of periods (hours) | 23.2–23.9 | 24.2–25.2 | |
| Change in period (hours) | −0.102 ± 0.029 | 0.400 ± 0.083* | |
| (BALB × B6)F2 progeny | |||
| Total number of progeny | 26 | 61 | 28 |
| SSLP-nonrecombinant | 20 | 44 | 20 |
| SSLP-recombinant | 6 | 17 | 6 |
| Phenotype only | 0 | 0 | 2 |
| Steady-state period (hours) | 23.33 ± 0.077** | 24.42 ± 0.057** | 27.26 ± 0.224** |
| Range of periods (hours) | 22.6–23.9 | 22.4–25.3 | 25.0–29.9 |
| Change in period (hours) | (n = 18) | (n = 44) | |
| −0.017 ± 0.097 | 0.377 ± 0.052* | n.a.† | |
| Amplitude (log PSD) | (n = 18) | (n = 43) | (n = 20) |
| DD day 1–10 | 2.37 ± 0.151 | 2.57 ± 0.099 | 1.73 ± 0.136** |
| DD day 11–20 | 2.43 ± 0.188 | 2.66 ± 0.109 | 1.65 ± 0.110** |
| Total activity (counts/day) | (n = 17) | (n = 41) | (n = 19) |
| LD 12:12 | 12,290 ± 1,630 | 13,740 ± 1,070 | 13,660 ± 1,140 |
| DD day 1–10 | 15,470 ± 2,230 | 18,630 ± 1,440 | 18,000 ± 1,220 |
| DD day 11–20 | 16,790 ± 2,500 | 21,220 ± 1,550 | 19,320 ± 1,210 |
| Daytime activity (%) | 9.44 ± 2.51 | 8.42 ± 1.24 | 18.9 ± 2.55* |
P < 0.001
P < 0.05, all comparisons.
Not applicable.
A second phenotypic characteristic of Clock was the lability of the period expressed by Clock/+ mice in DD. Wild type mice exhibited either no change in period or a slight shortening of period in DD (Table 2) (15). In contrast, Clock/+ mice from either interstrain cross typically exhibited a lengthening of period (Table 2). In most Clock/+ mice, the period lengthened gradually over a number of weeks (for an example, see Fig. 2B), whereas in other cases the period changed abruptly (16). Finally, in a few Clock/+ mice there were intervals in DD during which either multiple rhythms were expressed or transient arrhythmicity occurred (17).
A third feature of Clock was seen in homozygous Clock/Clock mice: a failure to exhibit sustained circadian rhythmicity in DD (Fig. 3). Although a long period was initially expressed on transfer to DD for 5 to 15 cycles in most Clock/Clock mice, the amplitude and precision of the rhythm were low. Fourier analyses (18) were performed on all (BALB × B6)F2 progeny for the first and second 10-day intervals in DD. In wild type and Clock/+ mice, a peak in the circadian range was clear, with no decline in the amplitude of the power spectral density (PSD) between the two intervals (Table 2). In Clock/Clock mice, a circadian peak with reduced amplitude was present in most animals during the first 10-day interval (Table 2). The amplitude of the PSD in the circadian range then decreased during the second 10-day interval (Fig. 3, E to H, and Table 2). In 31 homozygous Clock mice, only one animal expressed a circadian rhythm that persisted longer than 20 days in DD. The loss of amplitude of the PSD in the circadian range was not due to a decrease in amount of activity. No differences were detected in the total amount of activity per 24 hours among the three phenotypic classes in the (BALB × B6)F2 progeny, either during exposure to LD or during the first or second 10-day intervals in DD (Table 2). In Clock/Clock homozygotes, a single 6-hour light pulse, given after the loss of circadian rhythmicity in DD, could restore the long periodicity temporarily (Fig. 2G and Fig. 3, A to D) (19). The mean logarithm of the PSD in the circadian range was 1.55 + 0.133 before and 2.08 ± 0.127 after a 6-hour light pulse (paired t test, P < 0.005, n = 14). Therefore, the loss of circadian rhythmicity in DD in Clock homozygotes can be temporarily reversed by a single light pulse.
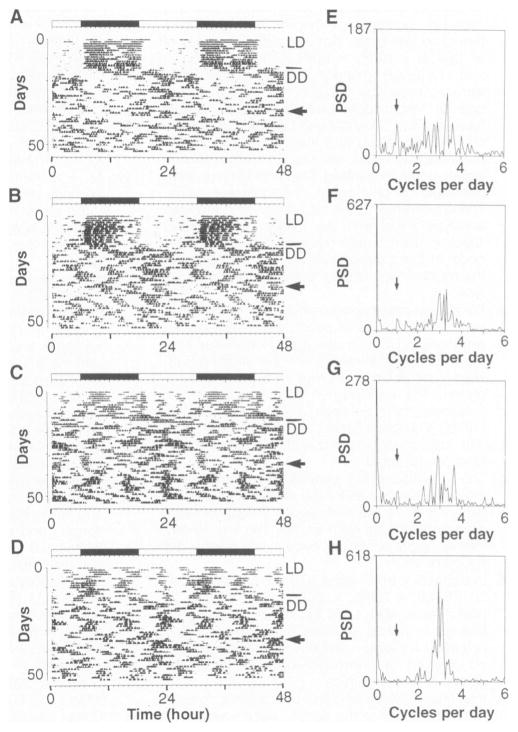
Fig. 3. Analysis of periodicity of homozygous Clock mutants.
The activity records of four homozygous Clock/Clock (BALB × B6)F2 individuals are shown (A to D). All animals shown were kept under LD12:12 for the first 12 days shown, then transferred to DD as indicated by the line in the right margin. A 6-hour light pulse was given on the day indicated by an arrow. Among the animals whose records are shown, no circadian rhythmicity persisted for more than about 10 cycles after either the initial transfer to DD or the light pulse. The corresponding Fourier analysis for each activity record is shown to the right (E to H). Fourier analyses encompass a 10-day interval corresponding to the 10 days preceding the light pulse (18). PSD, power spectral density. The frequency corresponding to one cycle per day, or a 24-hour period, is indicated by an arrow. Peaks corresponding to approximately 6- to 9-hour ultradian periods are seen for each animal.
Although circadian periodicities were abolished in the majority of Clock/Clock animals, the Fourier analyses indicated that a residual ultradian periodicity of about 6 to 9 hours remained (Fig. 3, E to H) (20). This ultradian periodicity resembled the residuum found in the locomotor records of rodents, including mice, bearing lesions of the master circadian pacemaker in the suprachiasmatic nucleus (SCN) (21). In analyses performed on other species of rodents (such as voles), ultradian locomotor rhythms do not appear to be generated by the circadian system (22). Therefore, the Clock mutation presents an additional situation in which circadian rhythmicity can be dissociated from ultradian periodicities in the locomotor behavior of rodents. In addition to SCN-lesioned mice (21), behavioral arrhythmicity has been reported in anophthalmic ZRDCT-An mice in association with a developmental loss of neurons in the SCN (23). We therefore examined the anatomy of the SCN of five to six (BALB × B6)F2 mice of each genotype but found no detectable differences among Clock genotypes (24). We concluded that the Clock phenotype cannot be attributed to any gross developmental or anatomical defects in the SCN.
Although the circadian rhythms of homozygous Clock/Clock mice were severely disrupted in DD, the locomotor behavior in LD was less affected. The phase angle of entrainment (25) was normal in Clock/+ mice. However, in Clock/Clock mice the onset of activity was more variable, and the distribution of activity in LD differed from that of wild-type or Clock/+ mice (Table 2 and Fig. 3, C and D). To verify that entrainment was occurring, we determined the phase of the circadian rhythm of the animals in DD after transfer from LD12: 12. In 22 out of 28 Clock/Clock mice in which a clear free-running rhythm was expressed, activity onset was in phase with the time of lights off, indicating that the phase of the rhythm was entrained (26). Given that the period is dramatically altered in Clock mutants, the modest differences found in LD are somewhat surprising. However, the limits of entrainment have been reported to be large in mice relative to other rodents (27), so the locomotor behavior measured in LD12:12 cycles may not be a sensitive indicator of possible changes in entrainment pattern.
Because Clock segregates as a single gene, we mapped its chromosomal location by linkage analysis. An intraspecific backcross [(BALB × B6)F1 × B6]N2 panel of 100 mice was used as a mapping cross (Fig. 4A). Clear segregation of Clock occurred, making it possible to assign the Clock genotype to all 100 animals (Fig. 4B). Simple sequence length polymorphisms (SSLPs) were used as DNA markers for genetic mapping of Clock (28). A preliminary linkage analysis was performed with a subset of the N2 animals (n = 20), with 47 polymorphic SSLPs distributed across the autosomes (29). Using the subset, we detected significant linkage with Clock (the logarithm of odds, or lod score > 3.0) (30) with only one marker, D5Mit24, indicating that Clock was linked to chromosome 5. Subsequent typing of the full N2 backcross panel confirmed that Clock was linked to D5Mit24, with a pairwise lod score of 19.1.
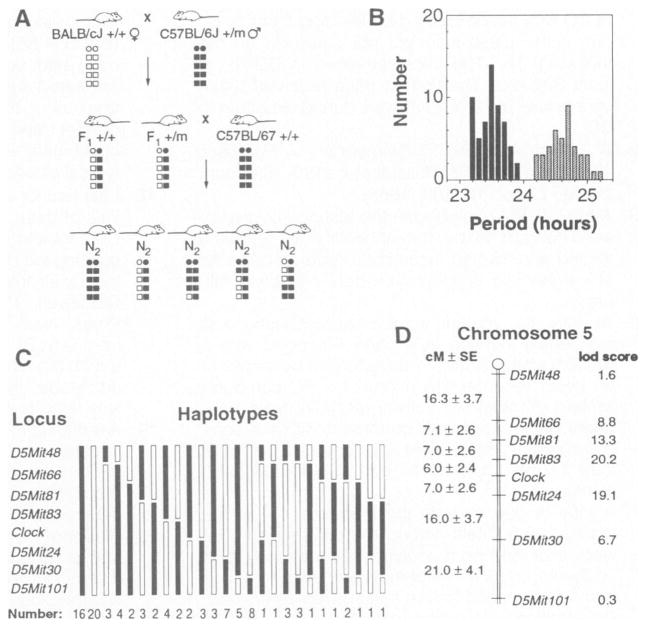
Fig. 4. Mapping of the Clock mutation.
A panel of 100 [(BALB × B6)F1 × B6]N2 mice was used to map Clock by genetic linkage and haplotype analysis. (A) The mating system used to produce the backcross panel. A schematic illustration of a pair of chromosomes is shown for each animal: circles represent centromeres, squares represent alleles at loci along the length of the chromosome, open symbols represent BALB alleles, and filled symbols represent B6 alleles. Clock heterozygous (+/m) B6 males were bred with wild-type (+/+) BALB females. The offspring were behaviorally tested and the Clock heterozygous F1 individuals were backcrossed to wild-type B6 to produce the N2 generation. One chromosome of each pair of autosomes in the N2 offspring is of B6 provenance (from the B6 parent), whereas the other (from the F1 parent) may be of either parental type or recombinant. Because the Clock mutation descends from a B6 animal, N2 offspring that are Clock/+ should carry B6 alleles at loci that flank the Clock locus, while +/+ offspring should carry BALB alleles at these loci. (B) Distribution of the period phenotype among the 100 animals. The steady-state period clearly distinguished animals between the two genotypes: 58 individuals were wild type and 42 individuals were heterozygous. See Table 1, cross 7, for analysis. (C) The assortment of haplotypes for chromosome 5 observed among the 100 N2 generation mice. Thirty-six animals had nonrecombinant haplotypes, 48 had singly recombinant haplotypes, and 16 exhibited doubly recombinant haplotypes. (D) Map of chromosome 5 showing the position of Clock relative to SSLP markers. Distances between loci are shown to the left in centimorgans ± standard error (cM ± SE), calculated with the program Map Manager (31). The lod scores to the right of each locus represent the logarithm of the odds for the pairwise test of linkage of the locus to Clock.
To position Clock on chromosome 5, we selected additional SSLP markers located both proximally and distally to D5Mit24. The position of Clock relative to these markers could then be inferred by haplotype analysis, placing Clock distal to D5Mit83 and proximal to D5Mit24 (Fig. 4C) (31). The genetic linkage map derived from this analysis shows that Clock lies on the midportion of chromosome 5, 6.0 + 2.4 centimorgans (cM) distal to D5Mit83 and 7.0 + 2.6 cM proximal to D5Mit24 (Fig. 4D). Multipoint analysis with the MAPMAKER/EXP 3.0 program shows that this map order has a logarithm-likelihood ratio of greater than 6 over the next best map order (32). The linkage of Clock to D5Mit24 and D5Mit83 was independently confirmed in a second mapping cross by genotyping 88 animals from the (BALB × B6)F2 progeny. Strong linkage to chromosome 5 was again found with 8 recombinants out of 226 meioses for D5Mit83 (lod score = 53.0) and 25 recombinants out of 226 meioses for D5Mit24 (lod score = 33.9). Thus, in two different mapping crosses Clock was positioned between D5Mit24 and D5Mit83 on the midportion of mouse chromosome 5. This entire region of mouse chromosome 5 is syntenic with human chromosome 4, suggesting that the human homolog of Clock, if it exists, is likely to map between 4pl6.3 and 4q22 (33).
In summary, Clock defines a mammalian gene that regulates two fundamental properties of the circadian clock system. Both the intrinsic circadian period and the persistence of circadian rhythmicity in DD are determined by the Clock genotype. Therefore, we propose that the Clock gene product is a candidate for a component of the circadian clock mechanism. Because a wild type allele of Clock is necessary for sustained circadian rhythmicity, Clock defines an essential gene for this behavior. No anatomical defects in the SCN have been observed in association with the Clock mutation, and homozygous Clock mutants appear fully viable on a hybrid genetic background (34). To the extent that we have characterized it, Clock appears to be a behavioral mutation limited to circadian rhythmicity.
The phenotypic characteristics of Clock are reminiscent of the long period and arrhythmic alleles of per in Drosophila and frq in Neurospora (4). The per0 and frq9 alleles are null mutations that abolish or disrupt circadian rhythms in their respective organisms, whereas the perLand frq7 alleles are point mutations that cause very long circadian periods of 28 to 30 hours. The perL and frq7 alleles are semidominant, and like Clock/+ mice, the periods of perL/+ flies are about 1 to 1.5 hours longer than those of wild-types (35). The robustness or strength of circadian rhythms in perL homozygous flies is also weaker, with lower amplitude and more variable rhythms as seen in homozygous Clock mice (36). Furthermore, the reduction in circadian amplitude is accompanied by the emergence of ultradian rhythms in both perL and Clock mutants (36). Thus, there are a number of phenotypic similarities between Clock and perL. It is tempting to speculate that Clock, like perL, could be a partial loss of function (a hypomorph) rather than a null allele (2, 4, 37). Genuine per homologs have not been successfully cloned in mammals; if they do exist, they may be poorly conserved at the nucleotide level (4).
The phenotype of Clock is different from that of the golden hamster tau mutation, the only other clock mutant in mammals (38). The tau mutation shortens the circadian period by about 2 hours in heterozygotes and by about 4 hours in homozygotes. Unlike Clock, however, tau does not lead to a loss of circadian rhythmicity under constant conditions. However, tau is remarkably similar to the perS allele in Drosophila. Both mutations are semidominant, shorten circadian period to the same extent, and increase the amplitude of the phase-response curve to light (4, 38). For this reason, it is logically possible that Clock and tau may define homologous genes in the mouse and the hamster, respectively. Unfortunately, because of the paucity of genetic information in the golden hamster, little progress has been made in understanding the molecular nature of tau.
Genetics may be one of the few ways in which we can identify genes involved in the clock mechanism in mammals. Because the two examples of cloned clock genes (Drosophila per and Neurospora frq) have no known homologs and are expressed at low abundance, it is unlikely that these genes could have been identified and their function studied by nongenetic means. Given the substantial genetic and physical mapping resources available for the mouse (33), it should be feasible to identify the molecular nature of Clock and other clock genes by the method of positional cloning (39). The power of ENU mutagenesis combined with the ability to clone genes by map position provides an approach to study complex behavior in mammals.
Acknowledgments
Supported by a grant from the MacArthur Foundation Mental Health Research Network I; the NSF Center for Biological Timing, and a State of Illinois Higher Education Cooperative Act award to J.S.T., F.W.T., and L.H.P.; NIH grants R01- DK40493 and P30-CA07175 to W.F.D.; and NIH postdoctoral training grant awards T32 NS071040 and T32 DC00015 to M.H.V. We thank our colleagues in the NSF Center for Biological Timing for their advice and support; J. Friedman for general advice on genetic mapping; E. Lander, W. Dietrich, A. Weaver, H. Katz, and J. Miller for their helpful advice on SSLPs; G. Suyeoka for assistance with SSLP typing; and A. Shedlovsky and 1. Riegel for critical reading of the manuscript.
Contributor Information
Martha Hotz Vitaterna, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA.
David P. King, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA
Anne-Marie Chang, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA.
Jon M. Kornhauser, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA
Phillip L. Lowrey, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA
J. David McDonald, McArdle Laboratory for Cancer Research and Laboratory of Genetics, University of Wisconsin, Madison, WI 53706, USA.
William F. Dove, McArdle Laboratory for Cancer Research and Laboratory of Genetics, University of Wisconsin, Madison, WI 53706, USA
Lawrence H. Pinto, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA
Fred W. Turek, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA
Joseph S. Takahashi, National Science Foundation Science and Technology Center for Biological Timing, Department of Neurobiology and Physiology, Northwestern University, Evanston, IL 60208, USA.
REFERENCES AND NOTES
- 1.Klein DC, Moore RY, Reppert SM, editors. Suprachiasmatic Nucleus: The Mind’s Clock. Oxford University Press; New York: 1991. [Google Scholar]; Takahashi JS. Curr Opin Neurobiol. 1991;1:556. doi: 10.1016/s0959-4388(05)80028-5. [DOI] [PubMed] [Google Scholar]; Takahashi JS, Murakami N, Nikaido SS, Pratt BL, Robertson LM. Recent Prog Horm Res. 1989;45:279. doi: 10.1016/b978-0-12-571145-6.50010-8. [DOI] [PubMed] [Google Scholar]; Turek FW. 1994;49:43. doi: 10.1016/b978-0-12-571149-4.50007-6. ibid. [DOI] [PubMed] [Google Scholar]; Michel S, Geusz ME, Zaritsky JJ, Block GD. Science. 1993;259:239. doi: 10.1126/science.8421785. [DOI] [PubMed] [Google Scholar]
- 2.Rosbash M, Hall JC. Neuron. 1989;3:387. doi: 10.1016/0896-6273(89)90199-2. [DOI] [PubMed] [Google Scholar]; Takahashi JS. Science. 1992;258:238. doi: 10.1126/science.1384127. [DOI] [PubMed] [Google Scholar]; Curr Opin Genet Dev. 1993;3:301. [Google Scholar]; Young MW, editor. Molecular Genetics of Biological. Rhythms Dekker; New York: 1993. [Google Scholar]
- 3.Hardin PE, Hall JC, Rosbash M. Nature. 1990;343:536. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]; Proc Nati Acad Sci USA. 1992;89:11711. [Google Scholar]; Edery I, Rutila JE, Rosbash M. Science. 1994;263:237. doi: 10.1126/science.8284676. [DOI] [PubMed] [Google Scholar]; Aronson BD, Johnson KA, Loros JJ, Dunlap JC. :1578. doi: 10.1126/science.8128244. ibid. [DOI] [PubMed] [Google Scholar]
- 4.Hall JC. Annu Rev Genet. 1990;24:659. doi: 10.1146/annurev.ge.24.120190.003303. [DOI] [PubMed] [Google Scholar]; Dunlap JC. Annu Rev Physiol. 1993;55:683. doi: 10.1146/annurev.ph.55.030193.003343. [DOI] [PubMed] [Google Scholar]
- 5.Russell WL, et al. Proc Nati Acad Sci USA. 1979;76:5818. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hitosumachi S, Carpenter DA, Russell WL. 1985;82:6619. doi: 10.1073/pnas.82.19.6619. ibid. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rinchik EM. Trends Genet. 1991;7:15. doi: 10.1016/0168-9525(91)90016-j. [DOI] [PubMed] [Google Scholar]
- 6.The mutagenesis procedure has been described [ Shedlovsky A, Gu6net JL, Johnson LL, Dove WF. Genet Res. 1986;47:135. doi: 10.1017/s0016672300022977.]. B6 mice were treated with ENU 150 mg per kilogram of bodyweight at 7 to 10 weeks of age and subsequently bred at McArdle Laboratories at the University of Wisconsin, Madison, WI. At 4 to 15 weeks of age, the G1 offspring were shipped to Northwestern University, Evanston, IL, and behaviorally tested after at least 3 weeks of acclimation to the new housing conditions. The expected mutation rate induced by ENU with this treatment procedure is about 1 in 1500 per locus per gamete. All animal care and experimental treatments were in accordance with institutional guidelines at both the University of Wisconsin, Madison, and Northwestern University
- 7.Mice were individually housed in cages equipped with running wheels within ventilated, light-tight chambers with timer-controlled lighting. Activity was monitored by an event recorder (Esterline- Angus; 216 mice) or by an on-line PC computer system (Stanford Software Systems, Chronobiology Kit; 148 mice). Mice were kept for at least 1 week on a light-dark cycle [14 hours light: 10 hours dark (LD14:10) for the first 41 mice; 12 hours light: 12 hours dark (LD12:12) for the remaining 323] to assess the phase of their locomotor behavior with respect to the LD cycle. Transfer to DD was accomplished by allowing lights to go out at the usual time but not come on on the following day. The mice remained in DD for at least 3 weeks. The first 41 mice received 5-min light pulses at 10-day intervals during exposure to DD.
- 8.Schwartz WJ, Zimmerman P. J Neurosci. 1990;10:3685. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]; Possidente B, Stephan F. Behav Genet. 1988;18:109. doi: 10.1007/BF01067080. [DOI] [PubMed] [Google Scholar]
- 9.A total of 364 mice began the testing: 42 died or were judged to be too unhealthy to complete testing and 18 had incomplete records because of mechanical or electronic data collection failures.
- 10.As with the mutant screen, each mouse was individually housed in a cage equipped with a running wheel. Activity was recorded by means of an event recorder (15 mice) or a PC computer system (8 mice). All activity recording of subsequent generations was performed with the computer system. Mice were kept on LD12:12 for at least 1 week then transferred to DD for at least 3 weeks.
- 11.A total of 396 mice in the pedigree began the testing: 22 animals among the different crosses were excluded from analysis because they died or developed health problems and were removed from wheel cages before testing was completed. An additional 12 were excluded from analysis because of inadequate data collected due to mechanical difficulties. Expected ratios were calculated on the basis of a single semidominant autosomal mutation.
- 12.BALB mice have a significantly shorter average period than B6 (8), whereas C3H mice have periods similar to B6 [ Ebihara S, Tsuji K, Kondo K. Physiol Behav. 1978;20:795. doi: 10.1016/0031-9384(78)90308-6.Possidente B, Hegmann JP. 1982;28:199. doi: 10.1016/0031-9384(82)90126-3. ibid.]
- 13.In B6 N2 (Table 1, cross 1), (C3H × B6)F, (Table 1, cross 4), and (BALB × B6)F1 (Table 1, cross 5) mice, the genotype of Clock]+ mice was confirmed by test cross. The probable genotypes of all [(BALB × B6)F1 × B6]N2 (Table 1, cross 7) and 113 (BALB × B6)F2 (Table 1, cross 6) mice were assessed by the genotype of simple sequence length polymorphism (SSLP) markers flanking Clock (D5Mit24 and D5Mit83; see results on linkage analysis and mapping of Clock). For the remaining cases, animals were either recombinant between flanking SSLP markers or were classified by phenotypic characteristics alone. The only difficult cases for phenotyping involved wild-type mice with the longest periods and Clock+ heterozygous mice with the shortest periods. In these cases the lability of the period of the rhythms was usually indicative of the genotype.
- 14.For steady-state period estimates, a 10-day interval in which circadian period was stable during exposure to DD was analyzed. Period was estimated during this interval with the Stanford Software Systems Chronobiology Kit with a x2 periodogram. The periodogram is described by G. Sokolove and W. Bushnell [J. Theor. Biol. 72,131 (1978)]. Periods at 6-min increments ranging from 4 to 36 hours were tested, and no autocorrelation function was used.
- 15.The x2 periodogram analysis was performed in the same manner as for the steady-state period, except that the first and second 10-day intervals were analyzed for each animal. The first 10-day interval began on the first cycle in which lights did not come on.
- 16.In 5 (BALB × B6)F1 mice, 9 (BALB × B6)F2 mice, 10 [(BALB × B6)F1 × B6]N2 mice, 8 (C3H × B6)F1 mice, and 1 B6 N3 mouse a stepwise change in period of more than 0.5 hour was observed in Clockl+ progeny. Spontaneous period changes of this magnitude were never observed in wild-type mice.
- 17.Intervals in which at least two distinct periods were expressed in the activity pattern were observed in Clockl+ progeny of three crosses; five (BALB × B6)F1 mice, seven (BALB × B6)F2 mice, and seven [(BALB × B6)F1 × B6]N2 mice had intervals of several days in which multiple periodicities were apparent. In addition, two Clock]+ (BALB × B6)F2 and two ClockI+ (BALB × B6)N2 mice had transient intervals during which they appeared arrhythmic. Individual Clock]+ progeny from all of the crosses were observed to have greater day-to-day variability in the time of activity onset (that is, less precise rhythms) than was typical of wild-type mice.
- 18.Fast Fourier analysis was based on 10-day intervals of data, with the Stanford Software Systems Chronobiology Kit. The analysis involved first calculating the Hartley transform and then converting the result into the Fourier transformation [R. N. Bracewell, The Hartley Transform (Oxford Univ. Press, New York, 1986)]. Frequencies ranging from 0 to 24 cycles per day were tested, with a 6-min bin size. For statistical analysis of PSD amplitude, the logarithm of the PSD amplitude was used to maintain homogeneity of variance.
- 19.A 6-hour light pulse restored circadian rhythmicity (as assessed by an increase in amplitude of the PSD in the circadian range) in 12 of 14 Clock/Clock (BALB × B6)F2 mice that received this treatment.
- 20.Ultradian rhythms have been defined as those of higher frequency than circadian or 24-hour rhythms [J. Aschoff, in Handbook of Behavioral Neurobiology 4: Biological Rhythms, J. Aschoff, Ed. (Plenum, New York, 1981), pp. 3–10]. Periods ranging from 1 to 12 hours were considered ultradian for our analysis. In 22 of 28 Clock/Clock (BALB × B6)F2 mice, the PSD amplitude in the ultradian range was higher than that of the circadian range for the second 10-day interval in DD. The peaks in the PSD distribution corresponded to ultradian periods ranging from 6 to 9 hours.
- 21.Schwartz WJ, Zimmerman P. Physiol Behav. 1991;49:1283. doi: 10.1016/0031-9384(91)90364-t. [DOI] [PubMed] [Google Scholar]; Rusak B. J Comp Physiol. 1977;118:145. [Google Scholar]; Ibuko N, Inouye SIT, Kawamura H. Brain Res. 1977;122:33. doi: 10.1016/0006-8993(77)90660-6. [DOI] [PubMed] [Google Scholar]; Van Den Pol AN, Powley T. 1979;160:307. doi: 10.1016/0006-8993(79)90427-x. ibid. [DOI] [PubMed] [Google Scholar]; Gerkema MP, Groos GA, Daan S. J Biol Rhythms. 1990;5:81. doi: 10.1177/074873049000500201. [DOI] [PubMed] [Google Scholar]
- 22.Gerkema MP, Daan S, Wilbrunk M, Hop MW, Van der Leest F. J Biol Rhythms. 1993;8:151. doi: 10.1177/074873049300800205. [DOI] [PubMed] [Google Scholar]; Lehmann U. Int J Chronobiol. 1976;4:223. [PubMed] [Google Scholar]
- 23.Silver J. J Comp Neurol. 1977;176:589. doi: 10.1002/cne.901760409. [DOI] [PubMed] [Google Scholar]; Scheuch GC, Johnson W, Conner RL, Silver J. J Comp Physiol A. 1982;148:333. [Google Scholar]
- 24.M. H. Vitaterna et al., in preparation. Serial coronal sections were taken throughout the suprachiasmatic region and Nissl stained. The volume of the paired SCN was estimated, and no differences in volume were detected among the Clock genotypes.
- 25.Phase angle of entrainment was measured as the time of activity onset relative to the time of lightsoff, determined from the initial free-run in DD [J. T. Enright, in (20), pp. 11–20]. Activity onset in wild-type and ClockI+ mice closely coincided with the time of lights-off.
- 26.The phase angle of entrainment of 22 of 28 (BALB × B6)F2 Clock/Clock homozygotes was determined from the initial free-running activity rhythm on release in DD. A line was eye-fit through times of activity onset. The point of intersection with the last day of LD was taken as the time of activity onset. The average time of activity onset preceded lights-off by 0.39 + 0.643 hour (mean + SE). This entrainment is comparable with the Drosophila perS and perL mutants, which can entrain to 24-hour LD cycles despite endogenous periods that differ from 24 hours by about 5 hours [ Hamblin-Coyle MJ, Wheeler DA, Rutila JE, Rosbash M, Hall J. J Insect Behav. 1992;5:417.], but differs from homozygous tau mutant hamsters, which have endogenous periods of about 20 hours and are unable to entrain to a 24-hour LD cycle 38
- 27.Aschoff J. Cold Spring Harbor Symp Quant Biol. 1960;XXV:11. doi: 10.1101/sqb.1960.025.01.004. [DOI] [PubMed] [Google Scholar]; Pittendrigh CS, Daan S. J Comp Physiol. 1976;106:291. [Google Scholar]; Davis FC, Menaker M. 1981;143:527. doi: 10.1007/BF00609919. ibid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich W, et al. Genetics. 1992;131:423. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dietrich W, et al. In: Genetic Maps. O’Brien SJ, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. [Google Scholar]
- 29.Animals were killed and their internal organs were dissected and frozen at −800C. Genomic DNA was prepared from livers with methods adapted from Ausubel FM, et al., editors. Current Protocols in Molecular Biology. Wiley; New York: 1993. For SSLP genotyping, methods are as described (28). A total of 47 SSLPs distributed among the autosomes were typed. Numbers in parentheses indicate the number of recombinants with Clock over the total number typed. D1Nds2 (8/20), DlMit3 (9/19), DlMitl7 (8/20), D2Mit43 (13/20), D2Mitl (8/20), D2Mit51 (8/20), D3Mit6 (7/20), D3Mitl7 (9/20), D4Mit31 (13/20), D4Mit53 (7/20), D5Mit24 (2/20), D5Mit30 (8/20), D5Mitl1 (3/20), D6Mit39 (10/20), D6Mitl (11/19), D7Mit31 (9/20), D7Mit25 (8/20), D8Mit13 (11/20), D8Mit4 (11/20), D8Mit25 (12/20), D9Mitl 1(10/19), D9Mit2 (12/20), D9Mitl8 (12/20), DlOMit31 (9/17), DlOMitl4 (10/19), D1lMit5 (10/17), D1lMitl2 (8/18), D1lMit2 (12/20), D12Mit5 (7/20), D12Nds2 (9/20), D13Mit30 (10/20), D13Mitl5 (14/20), D14Mit7 (8/20), D14Mit9 (7/18), D14Ndsl (11/20), D15Mit28 (8/20), D15Mit35 (13/20), D16Mit4 (8/18), D16Mit27 (11/20), D16Mit9 (8/16), D17Mit6 (11/20), D17Mit46 (11/20), D17Mit39 (11/20), D18Mit18 (16/20), D18Mit4 (12/20), D19Mitl (9/20), D19Mit1l (8/20).
- 30.Ott J. Analysis of Human Genetic Linkage. Johns Hopkins Univ. Press; Baltimore, MD: 1991. revised edition. [Google Scholar]
- 31.Haplotype analysis was performed with the program Map Manager, 2.4 [ Manley KF. Mamm Genome. 1993;4:303. doi: 10.1007/BF00357089.]. The phenotype of all recombinant animals was verified a second time, and no ambiguous cases were found.
- 32.Multipoint analysis was performed with the program MAPMAKER/EXP 3.0 [ Lander ES, et al. Genomics. 1987;1:174. doi: 10.1016/0888-7543(87)90010-3.].
- 33.Copeland NG, et al. Science. 1993;262:57. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]; Kozak CA, Stephenson DA. Mamm Genome. 1993;4:S72. doi: 10.1007/BF00360831. [DOI] [PubMed] [Google Scholar]
- 34.In the (BALB × B6)F2 generation, there was no evidence of segregation distortion. In the B6 F2 generation, fewer homozygous Clock/Clock mice were observed than would be expected (three rather than five). Although this was a small sampie, one cannot exclude the possibility that the homozygotes are not fully viable, at least with a B6 genetic background. An additional consideration in evaluating the survival of Clock mutants was that this generation was only three generations removed from the mutagenesis {the (BALB × B6)F2 and [(BALB × B6)F1 × B6]N2 were four generations removed} and as a result was still carrying numerous mutations in addition to Clock.
- 35.Smith RF, Konopka RJ. Mol Gen Genet. 1982;185:30. doi: 10.1007/BF00270625. [DOI] [PubMed] [Google Scholar]; Gailey DA, Villella A, Tully T. J Comp Physiol. 1991;169:685. doi: 10.1007/BF00194897. [DOI] [PubMed] [Google Scholar]
- 36.Dowse HB, Ringo JM. J Biol Rhythms. 1987;2:65. doi: 10.1177/074873048700200106. [DOI] [PubMed] [Google Scholar]
- 37.Baylies MK, Bargiello TA, Jackson FR, Young MW. Nature. 1987;326:390. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- 38.Ralph MR, Menaker M. Science. 1988;241:1225. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- 39.Collins FS. Nature Genet. 1992;1:3. doi: 10.1038/ng0492-3. [DOI] [PubMed] [Google Scholar]