Abstract
EZH2 is the catalytic subunit of PRC2, a central epigenetic repressor essential for development processes in vivo and for the differentiation of embryonic stem cells (ESCs) in vitro. The biochemical function of PRC2 in depositing repressive H3K27me3 marks is well understood, but how it is regulated and directed to specific genes before and during differentiation remains unknown. Here, we report that PRC2 binds at low levels to a majority of promoters in mouse ESCs, including many that are active and devoid of H3K27me3. Using in vivo RNA–protein crosslinking, we show that EZH2 directly binds to the 5′ of nascent RNAs transcribed from a subset of these promoters and that these binding events correlate with decreased H3K27me3. Our findings suggest a molecular mechanism by which PRC2 senses the transcriptional state of the cell and translates it into epigenetic information.
Introduction
Polycomb-group (PcG) protein complexes and, in particular, Polycomb repressive complex 2 (PRC2) are epigenetic regulators that sense the establishment of transcriptional repression during the early phases of development and maintain it after expression of the establishing factors wanes, often for very long periods of time and through several rounds of cell division. The cellular memories orchestrated by PRC2 are critical to the maintenance of cell identity, which is an essential requisite for the existence of multicellular life1-3.
The central biochemical function of PRC2 is that of depositing methyl groups on histone H3 lysine 27 (H3K27me3), which in turn leads to the formation of facultative heterochromatin and transcriptional repression4-7. Once established, this repressive structure is thought to propagate itself via a positive feedback loop based in part on the recognition of H3K27me3 by the PRC2 subunit EED8. However, the manner by which PRC2—and, more generally, the PcG axis—selects genes to be repressed remains the subject of intense interest, which is magnified by the recent implication of PRC2 and H3K27 in a number of malignancies9-12.
EZH2 is the most prominent catalytic subunit of PRC2 in pluripotent cells13. In addition to binding to a number of protein factors that are either part of the core PRC2 complex (SUZ12, EED, RBBP4, and RBBP6) or perform accessory functions in specific cell types2, EZH2 was also one of the first chromatin-associated proteins reported to bind to long noncoding RNAs (lncRNAs)14-16. However, genome-wide unbiased approaches revealed that RNA binding of EZH2 is promiscuous at best, with large fractions of the transcriptome reported to interact by both microarray- and sequencing-based techniques17,18. Although in some cases binding to these RNAs has been shown to influence chromatin recruitment, the fact that so many RNAs can bind is a clear indication that our current models to explain the function of PRC2–RNA interactions are lacking important components and/or that immunoprecipitations and in vitro binding approaches may not suffice to determine with the required specificity the nature of the true interactions in vivo. Moreover, the recruitment of PRC2 to a handful of loci by lncRNAs hardly represents a satisfying model to explain how this complex controls repressive chromatin states in an epigenetic manner.
To better understand the role of protein–RNA contacts in PRC2 function, we sequenced RNA that crosslinked to EZH2 in mouse ESCs and analyzed its genomic origin as compared to the distribution of PRC2 on chromatin. We show that PRC2 occupies a large fraction of active promoters in mouse embryonic cells, where it binds to nascent transcripts. RNA–EZH2 binding events correlate with a decrease of H3K27me3 at these promoters, suggesting a molecular mechanism by which PRC2 senses and maintains the appropriate epigenetic state at different genes.
Results
EZH2 directly contacts RNA in ESCs
Although it has been widely speculated that protein–RNA interactions are integral to PRC2 function, the experimental evidence in support of this observation so far originated either from candidate-based approaches14,16,19,20 or unbiased strategies based on native RNA immunoprecipitation (RIP)17,18, which suffer from an undesirably low signal-to-noise ratio21. In fact, the binding of EZH2 and of several other chromatin-associated proteins to ncRNAs is rather promiscuous in vitro (S.K., R.B., unpublished), suggesting that additional specificity determinants must exist in vivo, and strongly supporting the use of in vivo crosslinking strategies to capture functionally relevant RNA–protein interactions on chromatin.
Because UV-crosslinking and immunoprecipitation approaches are extremely sensitive to contamination from abundant RNA-interacting proteins that may remain undetected by western blot, we generated ESC lines expressing, in an inducible manner, physiological levels of EZH2 fused to three different epitope tags (N3–EZH2, Supplementary Fig. 1a,b), allowing for tandem (and triple) affinity purifications, which, we reasoned, would minimize chances of contamination. As previously reported, N-terminal tagging of EZH2 did not perturb its incorporation into the PRC2 complex or its enzymatic activity (data not shown), and did not affect its distribution on chromatin (Supplementary Fig. 1c,d), consistent with our earlier studies13,22,23, as well as a recent electron microscopy reconstruction of the PRC2 holoenzyme24
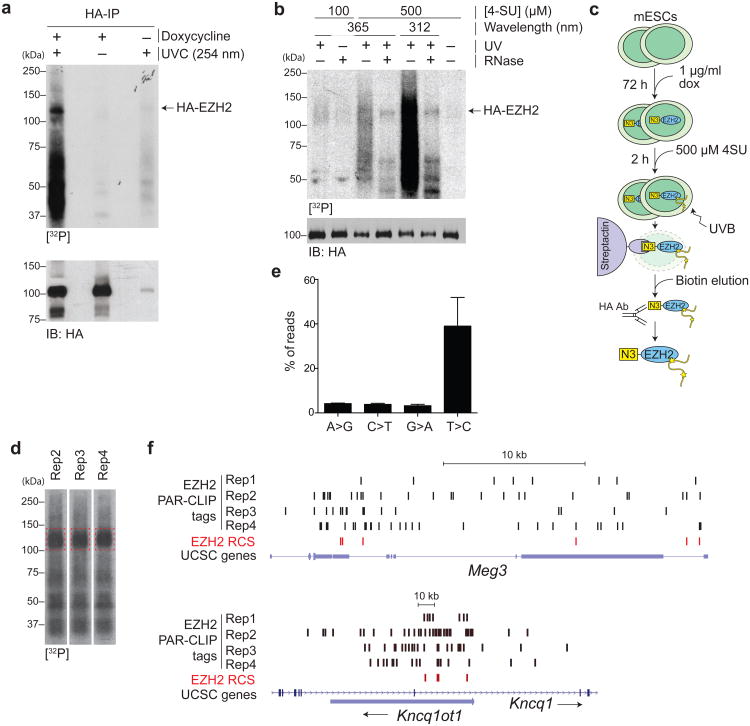
UVC irradiation followed by HA CLIP25 on N3–EZH2-expressing ESCs revealed the presence of a labeled protein migrating slightly above the predicted molecular weight for N3–EZH2. The radiolabeled band was only visible upon induction of N3–EZH2 expression and UVC irradiation (Fig. 1a), and therefore we concluded that it corresponded to RNA crosslinked to EZH2. Because the short-wavelength UVC irradiation appeared to cause extensive photodamage to EZH2 (Fig. 1a), in the following experiments we resorted to PAR-CLIP26, a modification of the CLIP technology that allows for crosslinking with UV light at longer wavelengths and lower energy. After HA IP in stringent detergent conditions that disrupted the core PRC2 complex (S.K., R.B., unpublished), autoradiography revealed that EZH2 was crosslinked to 32P-labeled material that was sensitive to RNase treatment (Fig. 1b). We also noted that higher 4-SU concentrations and irradiation with UVB (312 nm) rather than UVA resulted in more efficient crosslinking (Fig. 1b) and chose these conditions for the following experiments. Together, these data support the conclusion that EZH2 establishes direct physical contacts with RNA in ESCs that can be identified by CLIP or PAR-CLIP approaches.
Figure 1.
EZH2 binds to RNA in mESCs. (a) CLIP blots for HA-tagged EZH2 in control cells and cells induced with doxycycline and before or after irradiation with UVC. The autoradiography is shown at the top and the approximate position of HA-EZH2 is indicated. The corresponding HA immunoblot is shown at the bottom. (b) PAR-CLIP autoradiography (top) and western blot (bottom). Different 4-SU concentrations, UV wavelength, and RNAse treatments were tested. (c) Scheme of the purification strategy used for PAR-CLIP-seq experiments. (d) Autoradiography of 3 biological replicates (rep1–3) utilized for PAR-CLIP-seq library construction. The dashed red boxes indicate the position of the excised bands. (e) Histogram plot for the mutation frequencies in PAR-CLIP-seq reads. The bars represent the average % of unique mapped CLIP tags containing the indicated mutation from the 4 biological replicates + s.d. (f) Genome browser view of EZH2 CLIP tags mapping to the Meg3 lncRNA (top) or Kcnq1ot1 antisense ncRNA (bottom). The 4 biological replicates are plotted separately. RCSs called by PARalyzer are shown as red bars. UCSC gene models are displayed. Rep1–4, biological replicate.
To recover RNA amounts sufficient to construct libraries for deep sequencing while minimizing contaminations during the IP, we scaled up our PAR-CLIP strategy and performed tandem affinity purification with StrepTactin-coupled resin followed by HA IP (Fig. 1c), in 4 biological replicates. Deep sequencing of the CLIP tags eluted from the EZH2 bands (Fig. 1d) yielded ∼60 million reads, of which ∼30% could be aligned to the genome (Supplementary Table 1). Of these, 40% displayed the T>C transition diagnostic of 4-SU mediated crosslinking (Fig. 1e). Initial validations of the alignments confirmed that the replicates were highly concordant (Supplementary Fig. 2a,b) and that known EZH2-interacting lncRNAs, such as Kcnq1ot1 and Meg316,18 (S.K., R.B., unpublished), accumulated considerable numbers of CLIP tags (Fig. 1f). We pooled the 4 replicates into a single dataset to maximize sequencing depth and proceeded to analyze the genome-wide patterns of RNA–EZH2 binding interactions.
EZH2 preferentially binds to the 5′ of nascent RNAs
We identified 13,764 high-confidence RNA contact sites (RCSs) for EZH2 genome-wide using a software package that takes advantage of the T>C transitions produced by PAR-CLIP to identify bona fide interaction sites27. Although several RCSs localized to lncRNAs as expected17,28, the majority of them was in the body of protein-coding genes (Fig. 2a), suggesting the existence of unknown interactions between EZH2 and a vast number of RNAs. Visual inspection of the genes accumulating the largest number of RCSs revealed a striking bias for CLIP tags that mapped to the 5′ region of the mRNA with no apparent distinction between exons and introns (Fig. 2b, Supplementary Fig. 2c), suggesting that EZH2 (and therefore PRC2) made direct physical contact with nascent RNAs. This was consistent with a study in HCT116 cells, which reported several intronic CLIP tags associated with EZH2 despite shallower sequencing compared to our PAR-CLIP29.
Figure 2.
Genome-wide analysis of EZH2 CLIP in mouse ESCs. (a) Distribution of RCSs identified with PARalyzer relative to genomic features and their distribution. The number of features in each set is indicated at the top. (b) EZH2 CLIP tags mapping to two representative protein-coding genes. An EZH2 ChIP-seq track is shown, with the scale indicated to the right in reads per 10 million mapped (RP10M). Red bars show RCSs called by PARalyzer. Rep1–4, biological replicate. (c) Density profile of RCSs over the 250 unique RefSeq transcripts with the most RCSs, each divided in 100 bins. TSS, transcription start site; TTS, transcription termination site. The 10 kb upstream and downstream are included, each divided in 50 200 nts bins. The distribution of RNA-seq reads (gray dashed line) is shown for comparison. (d) Same as c but for CLIP tags (after duplicate removal) and comparing tags containing the T>C transition (solid black line) with tags not containing the mutation (dashed gray line). (e) Distribution of CLIP tags and RNA-seq reads on the first 50 bp of first exons (E1), first introns (i1), last introns (in) and last exons (En) from RefSeq transcripts with at least two exons. The bars represent the mean percentage of total reads mapping to these features from 2 (RNA-seq) or 4 (CLIP-seq) biological replicates + numerical range. (f) PAR-CLIP as in Fig. 1d after chasing the 4-SU with uridine for the indicated time. Autoradiography (top) and HA blot (bottom) demonstrating equal protein loading.
We confirmed the 5′ bias in EZH2–RNA contacts at the genome-wide level by mapping the RCSs identified above to all mouse transcripts and plotting their distribution over the body of the 250 genes that exhibited the strongest signal, and thus were more likely to reflect true in vivo interactions (Fig. 2c). This bias was not an artefact of RCS identification, as it was readily visible also when we mapped the raw CLIP tags to the gene models in a similar fashion (Supplementary Fig. 2d). In both cases (RCS or raw CLIP tag mapping), the bias became more obvious as we increased the stringency of our bioinformatic cutoff (Supplementary Fig. 3). Both CLIP tags containing the T>C mutation, diagnostic of RNA–protein crosslinks, and those not containing the mutation, more likely to be contaminants, peaked at the TSS (i.e. at the first exon) and to a lesser extent at the TTS (i.e. at the last exon), but the T>C tags were more broadly enriched over the 5′ of the transcript body (Fig. 2d), confirming that bona fide CLIP tags crosslinked to EZH2 also originated from introns. Indeed, EZH2 CLIP tags mapped equally well to the beginning of the first exon or of the first intron of protein-coding genes (Fig. 2e), lending further support to the conclusion that EZH2 establishes these contacts with RNA during transcription and before further mRNA processing can occur. To verify that the observed 5′ bias was not due to misannotation of the 3′ portions of some of these genes, we analyzed the distribution of conventional RNA-seq, which is expected to display a 3′ bias due to oligo-dT purification. Indeed, conventional RNA-seq displayed the expected 3′ bias (Fig. 2c, Supplementary Fig. 2d), and clear discrimination between exons and introns (Fig. 2e) demonstrating that the 5′ bias displayed by the CLIP tags was not an artefact of our bioinformatic analyses. Chasing the 4-SU pulse with non-crosslinkable uridine led to a fading of the EZH2-associated PAR-CLIP signal (Fig. 2f), further supporting the conclusion that EZH2 bound preferentially to nascent RNAs.
EZH2-bound RNAs originate from PRC2lo genes
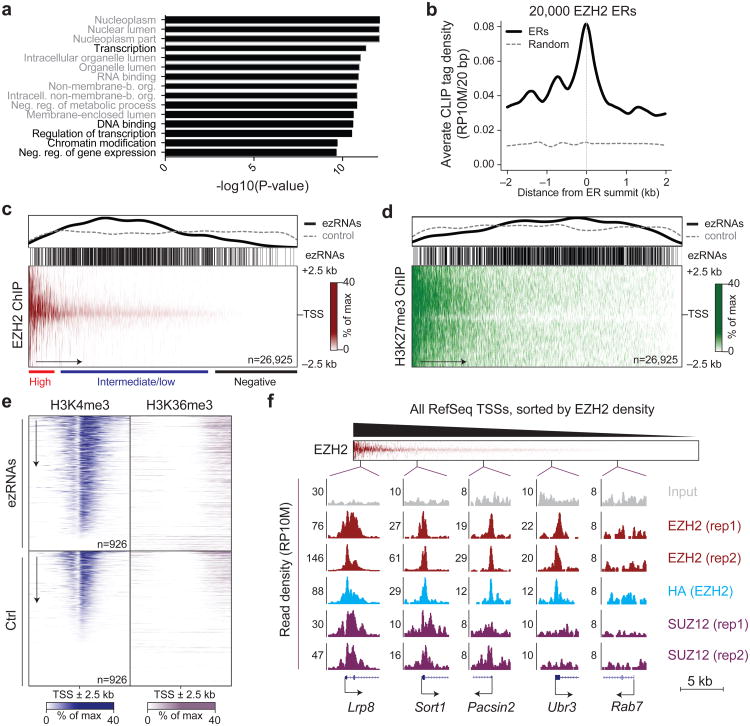
Because CLIP tags mapped equally well to introns and exons (Fig. 2e), we defined a moderately conservative set of EZH2-bound nascent RNAs (henceforth referred to as ezRNAs), based on the presence of at least 3 separate RCSs in their introns. Using this parameter, we identified 1,108 transcripts originating from 784 genes (Supplementary Table 2). Genes encoding ezRNAs were enriched in GO terms suggestive of transcriptional control and nuclear processes, including “chromatin modification” and “regulation of transcription” (Fig. 3a, Supplementary Table 3).
Figure 3.
EZH2-bound nascent RNAs originate from PRC2+ and H3K27me3- promoters. (a) Bar plot for the –log10 of the P-value of the top 15 most enriched GO terms in genes producing ezRNAs, as determined by the hypergeometric distribution. (b) Density profile of raw CLIP tags (after duplicate removal and repeat masking) relative to the top 20,000 EZH2 ChIP enriched regions (ERs). As a control, the density of the same CLIP tags was calculated on 20,000 random genomic regions of comparable size (dashed gray line). (c) The heatmap (bottom) shows EZH2 densities at all unique RefSeq TSSs ± 2.5 kb, sorted by EZH2 occupancy. The distribution of ezRNA-producing TSSs within this heatmap is indicated by individual bars (middle) and a density plot (top, black line) and compared to their density over a randomly permutated heatmap (top, dashed gray line). Data was from 2 biological replicates. (d) Same as c but for H3K27me3. The heatmap shows the average of 3 biological replicates. (e) Heatmaps for H3K4me3 and H3K36me3 occupancy comparing the 1,108 genes producing ezRNAs and a random gene set. Data were obtained from GSM590111 and GSM59011931. (f) PRC2 occupancy in 5 kb windows at TSSs from different position along the EZH2 gradient in the heatmap (top). The y axis represents reads per 10 million mapped (RP10M) and the scale is indicated by the number to the left of each profile. In all cases the input was scaled at the highest magnification for meaningful comparison.
Having observed EZH2 peaks at the promoters of visually inspected ezRNA+ genes (Fig. 2b, Supplementary Fig. 2c), we wished to determine whether this was a general feature of RNAs crosslinked to EZH2, despite the fact that, at least based on GO enrichment, ezRNA+ genes constituted a set distinct from traditional Polycomb target genes30. Indeed, EZH2 CLIP-tags tended to accumulate near chromatin regions enriched for EZH2 (Fig. 3b); however, plotting the distribution of ezRNAs on a heatmap of all RefSeq TSSs sorted by PRC2 occupancy revealed that they were missing not only from regions with no detectable PRC2 (Fig. 3c), as expected, but also from regions with very high PRC2 levels (Fig. 3c), and originated, for the most part, from TSSs with intermediate levels of PRC2, which, to our surprise, comprised a majority of all annotated TSSs (Fig. 3c). On the other hand, when the TSSs were sorted by H3K27me3 density, the ezRNA-producing TSSs clustered in the rightmost section of the heatmap, in regions depleted for this histone mark (Fig. 3d). We independently validated this observation by analyzing a different ChIP-seq dataset31, generated in the same cell line and similar culture conditions. Consistent with our findings above, ezRNAs mapped to genes with intermediate levels of EZH2 and low levels of H3K27me3 also in this dataset (Supplementary Fig. 4a,b). Despite the presence of PRC2, genes giving rise to ezRNAs were transcriptionally active, as demonstrated by the enrichment of H3K4me3 and H3K36me3 compared to a random gene set of equal size (Fig. 3e), and were by and large distinct from bivalent genes31,32 (Supplementary Fig. 4c,d).
The presence of EZH2 at such a large proportion of genes, including actively transcribed ones, was unexpected; therefore, we sought confirmatory evidence for this finding. Consistent with the results above, SUZ12, another core subunit of PRC2, was also found at a majority of TSSs, (Supplementary Fig. 4e), although the overall enrichment with this antibody was considerably lower (Fig. 3f). Moreover, the ChIP-seq profiles obtained with HA antibodies in N3–EZH2-expressing mouse ESCs were indistinguishable from those obtained with EZH2 antibodies in the parental cell line (Supplementary Fig. 1c,d), and in both cases peaks of considerable intensity were easily discernible in the promoter regions of genes that would be traditionally considered PRC2-negative (Fig. 3f).
In summary, the results above show that ezRNAs originate from a subset of genes that, although occupied by intermediate levels of PRC2, are actively transcribed in mouse ESCs and devoid of H3K27me3.
Genes producing ezRNAs have lower H3K27me3 levels in ESCs
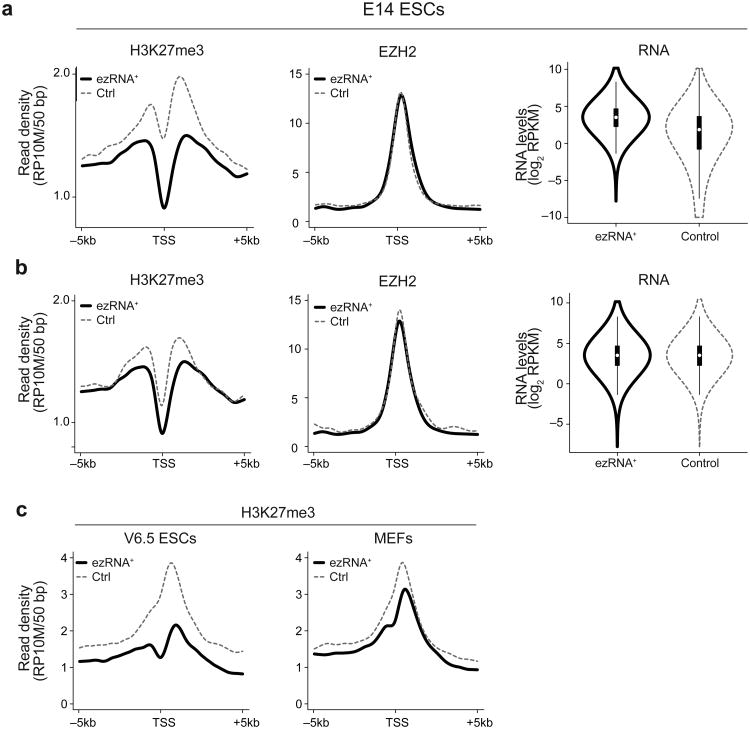
Because genes producing ezRNAs were decorated with chromatin marks consistent with productive transcription (Fig. 3e) and their distribution correlated negatively with H3K27me3 density at a genome-wide level (Fig. 3d), we hypothesized that, despite the presence of PRC2, their promoters might be devoid of H3K27me3. Indeed, when we compared the set of genes producing ezRNAs to a control gene set, matched gene-by-gene for the amount of PRC2 at their promoter, we found that the latter had higher amounts of H3K27me3 (Fig. 4a) despite equal amounts of PRC2 (Fig. 4a). Consistent with the lower levels of H3K27me3, these genes were expressed at higher levels than the control set, as shown by an average increase in RNA-seq RPKM values (Fig. 4a). When we matched the control by equalizing RPKMs rather than PRC2 levels, the difference in H3K27me3 was reduced but still noticeable (Fig. 4b), suggesting that differences in RNA transcription alone are not sufficient to explain the changes in H3K27me3 levels and that contacts between EZH2 and nascent RNAs may influence the function of PRC2 on chromatin. Differences in the average H3K27me3 profiles were not due to a few outliers because regression curves fitted through the individual points had different slopes, confirming that in general ezRNA+ genes contained less H3K27me3 than expected based on PRC2 levels alone (Supplementary Fig. 5a). The differences in H3K27me3 at these genes was also observed in ChIP-seq profiles generated in different laboratories and even using different ESC lines and different culture conditions31 (Supplementary Fig. 5b).
Figure 4.
Decreased levels of H3K27me3 at ezRNA+ genes. (a) Density profiles for H3K27me3 in E14 mESCs (left panel) spanning the promoters of 1,108 ezRNA+ transcripts (black line) or an equally sized control set of promoters (gray dashed line) matched individually for EZH2 occupancy (middle panel). Log-converted RPKM values for the two sets of transcripts are shown in the violin plot (right panel). The plots show the average of 3 (H3K27me3), or 2 (EZH2, RNA-seq) biological replicates. (b) Same as a, but control genes were selected by equalizing RPKM levels on a transcript-by-transcript basis. (c) H3K27me3 density plot calculated as in a for V6.5 ESCs (left) and MEFs (right). Data were obtained from GSE1224132.
Finally, we wished to determine the fate of genes producing ezRNAs in committed cells that had lost the pluripotent characteristics of ESCs. To this end, we compared the H3K27me3 distribution on genes producing ezRNAs in a different pluripotent mESC line (V6.5) and in differentiated mouse embryonic fibroblasts (MEFs), using datasets from Mikkelsen et al.32. The difference in H3K27me3 densities on ezRNAs genes versus an EZH2-matched control was pronounced in the pluripotent V6.5 ESCs (Fig. 4c), but it was much decreased in MEFs (Fig. 4c), suggesting that upon differentiation, several of the genes that produced ezRNAs in ESCs can become fully repressed by acquiring H3K27me3. Presumably, repression caused decreased synthesis of ezRNAs and consequent increased deposition of H3K27me3 by PRC2, but only EZH2 PAR-CLIP in MEFs could confirm this hypothesis. Regardless, the accumulation of H3K27me3 at ezRNA+ genes proves that the lack of PRC2 activity at those genes in ESCs was not due to any intrinsic property of their promoters, but rather it was a consequence of dynamic processes, specific to ESCs, likely involving the production and recognition of ezRNAs.
Discussion
By performing PAR-CLIP-seq for EZH2 in undifferentiated mouse ESCs (Fig. 1), we have identified a family of nascent transcripts that bind to EZH2 while they are being transcribed and before they are spliced and processed into mature mRNAs (Fig. 2). The majority of these nascent transcripts originate from transcriptionally active regions containing intermediate or low levels of PRC2 (Fig. 3c), which we detected in multiple biological replicates and using different antibodies (Fig. 3f), as well as in datasets generated by other groups (Supplementary Fig. 4). A peculiar characteristic of these ezRNA-producing genes is the lack of H3K27me3, despite measurable levels of PRC2 (Fig. 4), which hints at the possibility that PRC2 might function as RNA “sensor” (see below).
One conclusion of our study is that PRC2, at least in mouse ESCs, binds to a majority of promoters. In addition to regions where PRC2 is entrenched and large amounts of H3K27me3 accumulate (Fig. 3c), low but detectable levels of EZH2 are present at a majority of TSSs (Fig. 3c, Fig. 3f), including many that have no detectable H3K27me3. The PRC2hi regions correlate with strong repression and contain the gene targets that have traditionally been associated with PcG function in ESCs30, including developmental regulators such as the Hox genes. At these genes heterochromatin has already been nucleated and is likely propagated by the known positive feedback cycles affecting PRC28. On the other hand, the PRC2lo promoters appear to be transcriptionally competent if not completely activated, and this is in contrast with the traditional view that PRC2 enforces epigenetic repression. What might be the reason for this constitutive and broad TSS occupancy and how can we explain that these genes are transcribed despite the presence of PRC2?
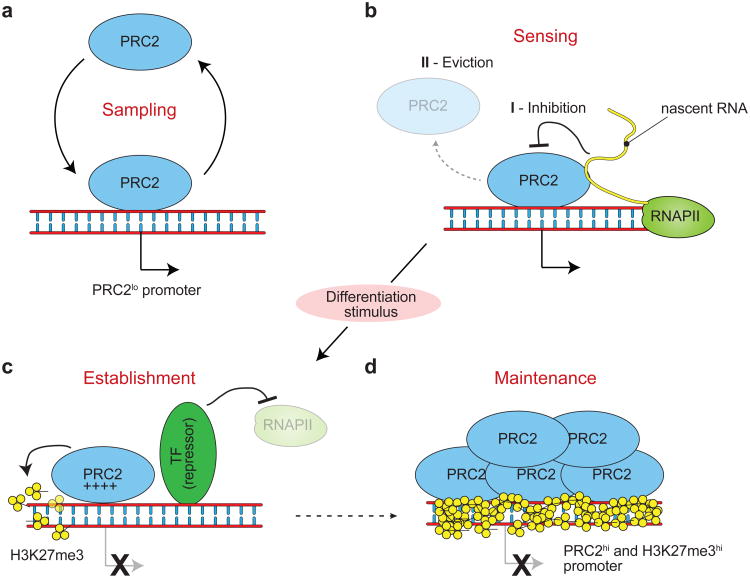
Considering that the function of the PcG axis is that of maintaining—not establishing— transcriptional repression33, the results presented above suggest a model wherein PRC2 exerts its epigenetic function by sensing the activation states of promoters through contacts with nascent RNAs (Fig. 5). The first step of this model is “promoter sampling”, that is the ability of PRC2 to interact with a majority of TSSs, at least in pluripotent mouse ESCs, using low affinity or low frequency binding events (Fig. 5a), possibly directed by GC-rich sequences34, other protein factors22,35,36, and even ncRNAs14,16,18 (S.K., R.B., unpublished). This broad promoter surveillance gives PRC2 the opportunity to survey the activation state of a large number of genes. If the surveyed gene is active and transcription is ongoing, PRC2 “senses” this by interacting with the nascent RNA, presumably still tethered to RNA polymerase II, and this interaction causes a decreased deposition of H3K27me3, allowing chromatin to stay open and the promoter to remain active. Whether and how exactly ezRNA–EZH2 contacts directly cause decreased H3K27me3 are open questions that will require further investigation. It is tempting to speculate that RNA contacts directly inhibit the enzymatic activity of EZH2 (Fig. 5b, I), but we can also envision more complex scenarios, in which one or more components of the PRC2 complex might be evicted from chromatin through interactions with or competition from ezRNAs (Fig. 5b, II).
Figure 5.
PRC2 senses transcriptional activity at ESC promoters. (a) In ESCs, PRC2 binds to a majority of TSSs in mouse ESCs, independent of their transcriptional state, sampling their activity. (b) Regulation of PRC2 activity occurs during the “sensing” phase, when PRC2 binds to nascent ezRNAs that somehow impede the deposition of H3K27me3 on chromatin. This could happen either by (I) inhibition of PRC2 activity, possibly dependent on additional in vivo factors, or (II) eviction of PRC2 from chromatin or of other factors required for H3K27me3 deposition. This allows the cell to continue expressing genes for which activating transcription factors are present, despite the constant presence of PRC2. (c) During differentiation, lineage specific transcriptional repressors are upregulated and silence transcriptional activity at selected target genes. Lacking the inhibitory ezRNAs, PRC2 initiates deposition of H3K27me3, establishing the nucleating conditions for the formation of facultative heterochromatin. (d) By the time the differentiation program is completed, a positive feedback loop of H3K27me3 deposition, EED binding, PRC2 recruitment, and more H3K27me3 deposition gives rise to stable and self-perpetuating facultative heterochromatin, which maintains transcriptional repression even after the original transcription factor ceases to be expressed8.
Whatever the mechanism may be, PRC2 responds to the activated state of ezRNA+ promoters by not depositing H3K27me3, which allows the gene to remain active; however, the Polycomb axis is better known for maintenance of repression, not activation. How does this fit our model? We propose that when the cell detects a new stimulus, for example during differentiation, DNA-binding transcriptional repressors are expressed de novo or are otherwise activated, enter the nucleus, bind to chromatin, and silence their target genes, abolishing the production of nascent RNAs at these loci. Without inhibitory ezRNAs, PRC2 complexes that are still surveying these loci deposit increased amounts of H3K27me3, which establish repressed chromatin structures (Fig. 5c). As H3K27me3 marks accumulate, a tipping point is reached when heterochromatic structures are maintained in an epigenetic manner8, even in absence of the original transcriptional repressor (Fig. 5d).
Although the physical contacts between RNA and EZH2 reported herein are highly suggestive of a direct regulatory mechanism, we cannot exclude that other molecular features associated with ongoing transcription may also contribute to the attenuation of PRC2 activity. For example, others and we recently showed that, in certain contexts, the presence of H3K4me3 inhibits the enzymatic activity of PRC237,38. This regulatory mechanism may function in addition to or in cooperation with the recognition of ezRNAs.
Our results and the model presented above are consistent with the genetic evidence that PRC2 does not repress transcription de novo but rather senses loci that are already silenced, and maintain their repressed state over time and cell division39,40. In Drosophila, PRC2 binds to the Polycomb responsive element of Ultrabithorax independent of its activation state, but H3K27me3 only spreads to the promoter and body of the gene in imaginal discs that repress Ultrabithorax41. In fact, transient activation of a reporter locus controlled by Polycomb is sufficient to switch its epigenetic state from repressed to active42, which fits well with our proposed model in which PRC2 is inhibited by active transcription.
Although we focused our attention on the 784 top ezRNA+ candidates, to our knowledge this is the first report of genome-wide PAR-CLIP for a non-canonical RNA binding protein (the second counting Guil et al.29, who performed CLIP without 4-SU), and we must consider the possibility that other nascent RNAs that we cannot detect with the current protocol might bind to EZH2 in vivo. In fact, according to our model, binding to RNA is likely to be very promiscuous in vivo, because the presence of RNA, not necessarily specific sequences or structures, would suffice to deliver a regulatory signal to PRC2. This would help explain the large number of EZH2-interacting RNA species recovered by previous studies17,18, but it does not exclude that other RNAs, particularly lncRNAs, may have evolved to further exploit this property of EZH2 in the context of complex regulatory scenarios such as that of X chromosome inactivation43 or imprinting16,18.
In conclusion, we propose that, at least in mammals, PRC2 performs its epigenetic function by sensing the presence of active transcription via interactions with nascent RNAs and curtailing its activity on chromatin in their presence. The increased facility by which the genome of ESCs can be edited will soon offer the opportunity to test this hypothesis experimentally.
Online Methods
Original images of gels, autoradiographs and blots used in this study can be found in Supplementary Figure 6.
Antibodies
For the purpose of CLIP and PAR-CLIP, we used 2–10 μg of anti-HA antibody (Abcam ab9110, lot GR98618-2). For ChIP-seq, we used 2 μg of anti-HA (Abcam ab9110, lot GR98618-2), EZH213, SUZ12 (Cell signaling #3737, lot 2) and H3K27me3 (Millipore #07-449, lot 1999681). For western blotting, we used antibodies against EZH2 (BD Biosciences #612666), SUZ12 (Cell signaling #3737), Beta-tubulin (Abcam ab6046), and HA (Covance MMS-101P), diluted 1:1000.
Cell culture
E14Tg2A.4 mouse ESCs were cultured as described23.
Lentiviral transduction
To produce lentiviruses, viral vector and packaging plasmids were cotransfected in 293T cells with Lipofectamine 2000 (Invitrogen) according to manufacture's instruction. After 48–72 hours, cell culture medium containing lentiviruses was collected and purified by using Fast-Trap Virus Purification and Concentration Kit (Millipore). Lentiviral transduction of mESCs was carried out in the presence of polybrene (5 μg/ml).
Knockdown of Ezh2 and expression of epitope-tagged EZH2
To establish conditional knockdown of Ezh2 in E14 mESCs, pTRIPZ lentiviral inducible shRNAmir targeting human and mouse Ezh2 were purchased from Open Biosystems (Item# RHS4696-99635303). Lentiviruses were prepared as described above. Selection was performed with Puromycin (1 μg/ml) and clones were screened by red fluorescent protein (RFP) expression 3–4 days after doxycycline (1 μg/ml) induction. To replace the endogenous copy with an epitope-tagged Ezh2, we cloned mouse Ezh2 into a pINTA-N3 vector, based on the Tet-On 3G system (Clontech) and encoding for three different N-terminal epitope tags: FLAG, HA, and OneSTrEP (IBA, Germany). The mESC clones were selected with zeocin (50–100 μg/ml). Knockdown of endogenous Ezh2 and simultaneous rescue with epitope-tagged Ezh2 were induced by treating with doxycycline (1 μg/ml) for 72 hours.
ChIP and ChIP-seq
ChIP from mouse ESCs was performed as described44 with minor modification. Briefly, cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Crosslinking was quenched by addition of glycine (pH 7.0) to 200 mM final concentration. Cells were washed three times with ice-cold PBS and harvested in ChIP buffer (50 mM Tris-HCl pH 7.9, 150 mM NaCl, 1% Triron X-100, 0.5% NP-40, 5 mM EDTA pH 8.0, 1 mM PMSF and protease inhibitor (Sigma, cat# P8849)). Samples were sonicated by using Diagenode Bioruptor to generate DNA fragments of ∼400 base pairs for quantitative PCR (qPCR) and ∼200 base pairs for library construction. For the purpose of ChIP-seq, each antibody (2–3 μg) was added into the sheared chromatin (50–200 μg), and incubated in an ultrasonic water bath for 30 min at 4°C. After centrifugation, supernatants were incubated with Dynabeads protein G (Invitrogen) for 30 min at 4°C. Beads were washed twice with ChIP buffer, treated with RNase A at 37°C for 15 min, and washed three more times with ChIP buffer. Immunocomplexes were heated at 65°C for ∼16 hours and incubated with proteinase K for 15 min at 55°C. Immunoprecipitated DNA was extracted by phenol–chloroform and precipitated with ethanol.
Libraries for ChIP-seq were prepared according to manufacturer's instructions (Illumina) and as described45. Briefly, immunoprecipitated DNA was end-repaired using End-It Repair Kit (Epicenter), tailed with a deoxyadenine using Klenow exo- (New England Biolabs), and ligated to custom adapters with LigaFast (Promega). Fragments of 300±50 bp were size-selected and subjected to ligation-mediated PCR amplification (LM-PCR) using Phusion DNA polymerase (NEB M0530). Libraries were quantified by qPCR using primers annealing to the adapter sequence and sequenced on an Illumina HiSeq2000 at the NYU core facility. Custom barcoding was utilized to sequence more than one sample per lane.
For this study we analyzed 2 biological replicates for EZH2 from two different E14 clones (parental line and uninduced E14::N3–EZH2; 2 biological replicates for SUZ12 (two separate ChIPs from the E14 parental line); 3 biological replicates for H3K27me3 (two from separate ChIPs from the E14 parental line, 1 from dox-induced E14::N3–EZH2).
CLIP, PAR-CLIP, and PAR-CLIP-seq
For CLIP experiments, ESCs were crosslinked with 400 mJ/cm2 UVC (254 nm) and then processed as described below.
For PAR-CLIP, ESCs were grown in standard conditions and pulsed with 500 μM 4-SU (Sigma) for 2 h. In some experiments the 4-SU was chased with 500 μM uridine. After washing the plates with PBS, RNA–protein crosslinks were generated by irradiating 4-SU-treated cells with 400 mJ/cm2 UVB (312 nm) using a Stratalinker UV crosslinker (Stratagene, CA). These conditions are different than those previously reported46 and were optimized to maximize crosslinking of RNA to EZH2 (Fig. 1b), a non-canonical RNA-binding protein. Although Rabani et al. observed 5′-biased 4-SU incorporation at the Ifih1 gene after a 45 minute pulse47, it was only at the first time point after gene induction, suggesting that the 5′ bias reflected the increased rate of initiation events rather than a predisposition of RNAPII to incorporate 4-SU at the beginning of transcriptional units. Indeed, in the same experiment, later time points showed homogeneous 4-SU incorporation across the body of the same transcript47. Incubation with 4-SU for 2 h was previously shown to label RNA efficiently in a broad range of cells48 and is considerably longer than the half-life of a majority of mature mRNAs and virtually all pre-mRNAs47. These optimized PAR-CLIP conditions may be useful to determine contacts with noncoding RNAs for other non-canonical RNA-binding proteins found in chromatin.
Whole cell extracts were obtained by incubating the cells for 10 min at 37°C in an appropriate volume of CLIP buffer (20 mM HEPES pH 7.4, 5 mM EDTA, 150 mM NaCl, 2% lauryldimethylbetaine) supplemented with protease inhibitors, 20 U/ml Turbo DNase (Life technologies), and 200 U/ml murine RNase inhibitor (New England Biolabs) and lysates were cleared by centrifugation. Prior to immunoprecipitations, epitope-tagged EZH2 was affinity-purified by using Strep-Tactin beads (IBA) and eluted with CLIP buffer containing 2 mM biotin (Sigma). Immunoprecipitations were carried out with ChIP-grade anti-HA antibody (abcam) in the same CLIP buffer for 16 hours at 4°C, after which, when required, the extracts were treated with various concentration of RNAse A + T1 cocktail (Ambion) for 5′ at 37°C. Immunocomplexes were recovered by adding protein G-coupled dynabeads (Life technologies) for 45 min at 4°C. Contaminating DNA was removed by treating the beads with Turbo DNase (2U in 20 μl). Crosslinked RNA was labeled by successive incubation with 5U Antarctic phosphatase (New England Biolabs) and 5U T4 PNK (New England Biolabs) in presence of 10 μCi [γ-32P] ATP (PerkinElmer, MA). Labeled material was resolved on 8% bis-tris gels, transferred to nitrocellulose membranes and exposed to autoradiography films for 1–24 hours.
For PAR-CLIP-seq experiments, 3′-blocked DNA adapter (100 pmol/μl) was ligated to the RNA after dephosphorylation and before 5′ 32P end-labeling by incubating the beads with T4 RNA ligase 1 (New England Biolabs) for 1 hour at 25°C. Labeled material was resolved on 8% bis-tris gels, transferred to nitrocellulose membranes and exposed to autoradiography films for ∼4 hours. Bands of interest were excised and the RNA eluted from the membrane by treating with proteinase K (Roche, 4 mg/ml) for 30′ at 37°C and then proteinase K in presence of 3.5M urea for 30′ at 55°C. After phenol/chloroform extraction, custom designed 5′ RNA adapters were ligated, the products size-selected on polyacrylamide gels, and libraries amplified and sequenced on an Illumina HiSeq 2000 sequencing system.
RNA-seq
RNA-seq was performed in duplicates in two different E14 subclones. RNA was isolated using TRIzol (Invitrogen). PolyA+ RNA was purified using Oligo(dT)25 Dynabeads (Invitrogen) and converted to cDNA while preserving strand information using the dUTP method49. Libraries were constructed from cDNA with the same protocol as described above for ChIP-seq libraries.
Bioinformatic analyses
In all cases, reads were mapped to the mm9 version of the mouse genome and gene annotations were obtained from ENSEMBL (v.67) or RefSeq (downloaded on July 10, 2013). Except for Fig. 2a, in which all features from ENSEMBL were considered, all other analyses concentrate on the set of 26,925 protein-coding transcripts present in both ENSEMBL and RefSeq database and having unique TSSs and TTSs.
ChIP-seq analysis was performed as described before with modifications45. Sequenced reads from ChIP-seq experiments were mapped with Bowtie using parameters -v2 -m4 --best50. Normalized genome-wide read count densities (typically reads per 10 million mapped, “RP10M”) were computed with samtools51 and visualized on the UCSC genome browser (http://genome.ucsc.edu) after extending the reads to the estimated size of the ChIP fragment (typically 200 nts). Enriched regions (ERs) were identified using MACS 2.0952 using the relevant input as control and default parameters. For the heatmaps shown in Figure 3 and S4 the extended reads were mapped to non-overlapping 25 bp bins spanning 5 kb centered around the TSS and then normalized bin-wise to % of max. Density plots (e.g. Fig. 4) were generated similarly except that the window spanned 10 kb (divided in 50 bp bins) around the TSS and the mean for each given bin in all genes considered was plotted on the y axis. The correlation of binding affinities in Supplementary Fig. 2d was calculated using DiffBind53.
For the initial PAR-CLIP-seq mapping and RCS identification we followed Corcoran et al.27. Briefly, we clipped adapter sequences from PAR-CLIP reads and kept those longer than 17 nts. The resulting reads were collapsed to remove duplicate sequences using the fastx toolkit, then mapped with bowtie -v2 -m40 --best –strata to the mm9 assembly. We pooled the 4 separate replicates and processed them with PARalyzer requiring at least 2 independent T>C conversions, which resulted in the identification of 13,764 RCS. Of these, 2,261 overlapped repetitive elements listed by repeat masker (as downloaded from the UCSC genome browser website on May 29 2013) and were discarded, leaving 11,503 non-repetitive RCSs. For CLIP tag analyses (Fig. 2d, 2e, 3b; Supplementary Fig. 2a, 2b, 2d, 3b), reads were pre-processed in a similar way with the exception that duplicate removal was performed after bowtie mapping.
RNA-seq reads were trimmed to 30 nucleotides, mapped with bowtie -v2 -m40 -best and assigned to gene models using DEGseq54.
GO enrichment analyses were performed with DAVID (http://david.abcc.ncifcrf.gov/) using default parameters55.
Supplementary Material
Acknowledgments
We thank W.W. Tee, L. Vales, and P. Voigt for their critical assessment of the manuscript and the Genome Technology Center at NYU for help with sequencing. This work was supported by grants from the US National Institute of Health (GM-64844 and R37-37120 to D.R.) and the Howard Hughes Medical Institute (to D.R.). R.B. was supported by a Helen Hay Whitney Foundation postdoctoral fellowship and by the Helen L. and Martin S. Kimmel Center for Stem Cell Biology postdoctoral fellow award.
Footnotes
Author Contributions: S.K., J.S., S.S.S., D.R., and R.B. designed and performed experiments, analyzed data, and wrote the manuscript.
Accession codes: Public datasets used in this paper were obtained from series GSE23943 and GSE12241 at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) and from the mouse ENCODE project. All new sequencing data has been deposited to the NCBI GEO as SuperSeries GSE49435.
References
- 1.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 4.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 5.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 6.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 8.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 11.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 12.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 13.Margueron R, et al. Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 17.Khalil A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klattenhoff CA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko S, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciferri C, et al. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corcoran DL, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guil S, et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 30.Boyer L, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 31.Marks H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007 doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 34.Mendenhall EM, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brien GL, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol. 2012;19:1273–1281. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 36.Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Struhl G, Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. The EMBO journal. 1985;4:3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ringrose L, Leonie Ringrose, Renato P, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annual review of genetics. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 41.Papp B, Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes & development. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes & development. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Z, et al. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Molecular cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hafner M, et al. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010 doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolken L, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008;14:1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 55.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.