Significance
Although fluoride is plentiful in the environment and is commonly used at high concentrations in oral hygiene products, little has been known about how biological systems overcome the toxic effects of this anion. We demonstrate that a protein called FEX in many fungi is essential for cell survival in the presence of high fluoride concentrations. The protein is required for the rapid expulsion of cytoplasmic fluoride, indicating that many eukaryotic species that carry FEX genes likely avoid fluoride toxicity by purging cellular fluoride.
Keywords: toxicity resistance, environmental toxin, ion transport, dual membrane topology
Abstract
Fluorine is an abundant element and is toxic to organisms from bacteria to humans, but the mechanisms by which eukaryotes resist fluoride toxicity are unknown. The Escherichia coli gene crcB was recently shown to be regulated by a fluoride-responsive riboswitch, implicating it in fluoride response. There are >8,000 crcB homologs across all domains of life, indicating that it has an important role in biology. Here we demonstrate that eukaryotic homologs [renamed FEX (fluoride exporter)] function in fluoride export. FEX KOs in three eukaryotic model organisms, Neurospora crassa, Saccharomyces cerevisiae, and Candida albicans, are highly sensitized to fluoride (>200-fold) but not to other halides. Some of these KO strains are unable to grow in fluoride concentrations found in tap water. Using the radioactive isotope of fluoride, 18F, we developed an assay to measure the intracellular fluoride concentration and show that the FEX deletion strains accumulate fluoride in excess of the external concentration, providing direct evidence of FEX function in fluoride efflux. In addition, they are more sensitive to lower pH in the presence of fluoride. These results demonstrate that eukaryotic FEX genes encode a previously unrecognized class of fluoride exporter necessary for survival in standard environmental conditions.
Halides have long been known to be important to biology. Chloride and iodide have been well studied for their effects on organisms (1–5), but less is understood about the biological importance of fluoride, the smallest and most electronegative anion in this series. The size and reactivity of fluoride give it unique chemical and biochemical properties, but the mechanisms by which cells respond to this halide are incompletely characterized.
Fluoride is ubiquitous in the environment, where it is found in soil, water, and air (6, 7), and it is a highly abundant element in the earth’s crust (0.32 g/kg) (8–10). The distribution of this halide in soil and water is variable depending on location. Fluoride concentrations in soil can range from ten to thousands of parts per million (ppm) (10). In natural water sources, the concentrations range from <25 μM to >100 mM (<0.5 to >2,000 ppm; 1 ppm ∼ 55 μM) depending if the water is in contact with high levels of fluoride-containing minerals (6, 7, 11). In groundwater specifically, fluoride concentrations are among the highest of any anion (7, 12).
At high concentration, fluoride has toxic effects on bacteria, fungi, plants, and animals. More than a century ago, fluoride was used as an antimicrobial agent (6). The antimicrobial effects of this ion have continued to be elucidated over the last decades, with an emphasis on the bacteria that cause dental caries (13–15). Fluoride is also toxic to eukaryotic organisms. For example, at the beginning of the 20th century, it was noted that baker's yeast, Saccharomyces cerevisiae, was killed in 1% sodium fluoride (about 250 mM) (16). Inhibitory growth effects have also been observed for other species of fungi, including several pathogens (17, 18).
The mechanisms by which fluoride is toxic to these different species are complex and incompletely understood. It is thought that a partial cause of fluoride toxicity is through enzyme inhibition and interactions with important cations in the cell, such as Mg2+ and Ca2+ (12, 19, 20). Crystal structures of the enzyme enolase in complex with fluoride show that the inhibition is due to the formation of a magnesium-fluoride-phosphate complex (19). Another well-known example of the interaction of fluoride with enzymes is the ability of fluoride complexes with aluminum and beryllium to act as phosphate mimics. These complexes affect the activity of phosphoryl transfer enzymes, a large and important group of macromolecules (12, 21).
Because of its ubiquitous presence in the environment and its toxic effects, organisms must have evolved mechanisms of resistance to fluoride. However, little is known about these strategies and pathways. Some new clues emerged with the discovery of a class of regulatory RNAs, or riboswitches, that bind to fluoride and regulate the expression of genes in response to this anion in eubacteria and archaea (22, 23). Riboswitches are metabolite or ion-sensing structured RNA motifs typically located in the noncoding regions of certain mRNAs. They control the expression of adjoining protein-coding regions through several different mechanisms including transcription termination, translation blocking, and alternative splicing (24–27). One of the genes most commonly associated with fluoride-sensing riboswitches is the crcB gene. This gene was originally identified in Escherichia coli and had been implicated previously in chromosome condensation and camphor resistance (28, 29). It encodes a small protein (127 amino acids) that has four predicted transmembrane domains. There is evidence that CrcB functions as a dimer with dual membrane topology, similar to several small multidrug transporters (30).
Due to its association with the fluoride-binding riboswitch, it was hypothesized that crcB may be involved in fluoride resistance, specifically fluoride efflux. An E. coli strain lacking crcB is 200-fold more sensitive to fluoride than the WT strain (22). Additionally, this strain appears to have a higher intracellular concentration of fluoride than WT, and the growth of this strain can be rescued on high fluoride media by the addition of the eriCF gene (22), a variant of eriC that encodes a F−/H+ antiporter (31). Because eriCF can substitute for crcB, it is likely that these proteins have some level of functional equivalence, namely the transportation of fluoride ions.
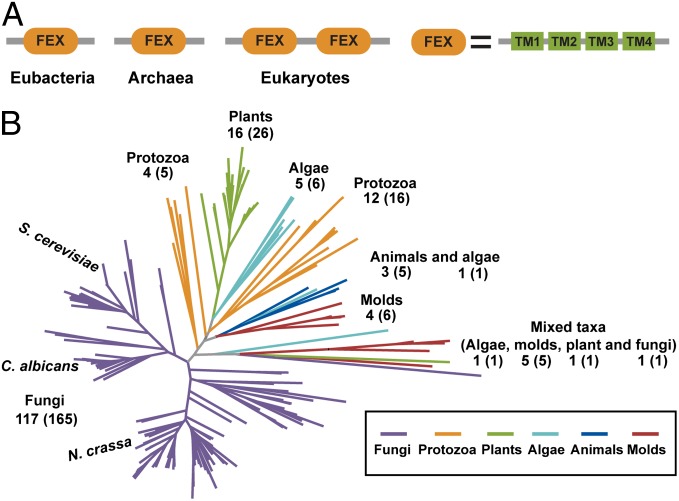
Although no fluoride riboswitches have been identified in eukaryotes, many of these organisms have crcB homologs. There are more than 8,000 genes in the UniProt database annotated as containing at least one crcB domain (32). Although the majority of identified sequences are in prokaryotic organisms, there are a significant number in eukaryotic (>200; Fig. 1) and archaeal (>200) species as well. Many of the homologs contain at least four transmembrane domains and are classified in a transporter superfamily in Pfam (30, 33).
Fig. 1.
FEX proteins architecture and distribution. (A) Eubacterial, archaeal, and eukaryotic FEX domain arrangments. TM represents transmembrane domain. (B) Phylogenetic tree for eukaryotic FEX proteins.
To determine whether the eukaryotic homologs of crcB are involved in fluoride homeostasis, we used genetic approaches in three organisms: the filamentous fungus Neurospora crassa, the budding yeast S. cerevisiae, and the pathogen Candida albicans. Here we demonstrate that CrcB proteins play a role in fluoride resistance in eukaryotes and we provide direct evidence that they are involved in removing fluoride from the cell. This is the only known example of a gene identified to function in fluoride efflux in eukaryotes. Based on these results, we propose renaming these genes FEX, for fluoride exporters, and will refer to them using this nomenclature.
Results
Conserved Domains and Phylogenetic Tree for Eukaryotic FEX Proteins.
Examination of FEX homologs in eubacteria, archaea, and eukaryotes reveals clear similarities and differences in domain organization. Most of the archaeal and bacterial genes encode proteins containing a single conserved FEX domain, whereas most eukaryotic homologs encode two conserved FEX domains in a single polypeptide (Fig. 1A). There is evidence that the CrcB protein from E. coli forms a dimer, suggesting that two CrcB domains may be needed for biological function (30). If this is the case, most prokaryotic FEX proteins would be predicted to form dimers, whereas most eukaryotic homologs would be predicted to function as monomers, with the two necessary FEX domains present in the same polypeptide.
Eukaryotic FEX proteins segregate into several distinct groups based on phylogeny and are found in a wide variety of organisms, including fungi, algae, molds, protozoans, plants, and animals (Fig. 1B). Fungal FEX genes comprise the two largest groups with the homologs from yeast species closely related to each other but distinct from those found in filamentous fungi. Examples are also present in agriculturally important plants such as rice, corn, and grapes, as well as the model plant species Arabidopsis thaliana. Additionally, FEX genes are present in several animal species, such as Oikopleura dioica (a tunicate) and Ciona intestinalis (a sea squirt). The widespread distribution of this protein suggests an important biological function for FEX.
Deletion of FEX Causes Fluoride Sensitivity in Eukaryotes.
To test the role of eukaryotic FEX proteins in fluoride resistance, we selected three important model organisms that would sample phylogenetic diversity but are also easily genetically manipulated. We deleted the FEX gene(s) in three organisms, N. crassa, S. cerevisiae, and C. albicans, and tested each knockout (KO) strain for growth on fluoride containing media.
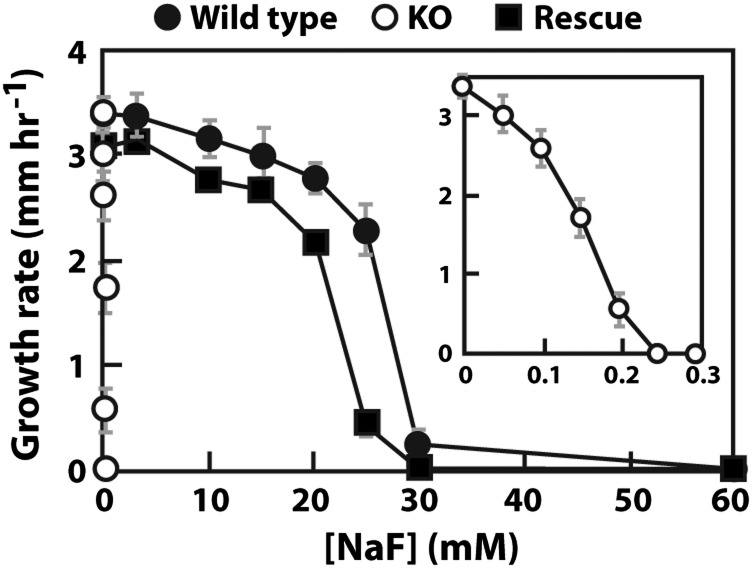
N. crassa is a haploid organism that contains one FEX gene (NCU06262, chromosome III; fex-1) encoding a 526 amino acid (aa) protein with two conserved FEX domains. This gene was deleted, and growth was assessed on both liquid and solid media with increasing amounts of fluoride. When both strains were cultured on solid agar media without fluoride, the KO strain had a similar growth rate compared with WT (Table S1). However, on both solid and liquid media containing fluoride, the growth of the KO organism was inhibited at an ∼200-fold lower concentration than WT (Fig. 2 and Fig. S1). WT N. crassa experienced a reduction in the number of spores, and the amount of mycelium as the fluoride concentration was increased (Fig. S1).When a copy of the fex-1 gene was reintroduced into the genome at the his-3 locus, the resulting rescue strain fully recovered the ability to tolerate high fluoride concentrations (Fig. 2, Figs. S1 and S2, and Table S1).
Fig. 2.
Sensitivity of the N. crassa fex-1 KO strain to NaF by race tube assay. All strains are cultured on solid agar media with different concentrations of NaF. (Inset) Curve for the KO strain. The results are from three independent repeats. Gray bars represent SDs.
S. cerevisiae contains two ORFs, YOR390W (FEX1) and YPL279C (FEX2), that each encodes a protein of 375 aa. Both of these proteins are composed of two FEX domains connected by a linker. These proteins, as well as the sequences upstream and downstream, are nearly identical, suggesting a gene duplication event. Both genes were previously uncharacterized and were not included in the systematic yeast deletion library and were therefore not implicated in yeast fluoride biology, despite a large-scale screen for mutants that had growth defects in millimolar sodium fluoride (34).
To test whether these crcB homologs are involved in fluoride resistance in yeast, we made single deletion strains as well as a strain in which both ORFs are deleted. The single deletion of either FEX1 or FEX2 does not significantly increase fluoride sensitivity, suggesting a redundancy of function for the two proteins. Single KOs grow only slightly worse than the WT in media containing millimolar quantities of fluoride (Fig. S3). For the WT strain, the IC50 for fluoride is 70 mM and the minimum inhibitory concentration (MIC) is ∼120 mM. In the case of the single deletion mutants, the IC50 for fluoride is ∼55–60 mM and the MIC is about 110 mM (Table S1).
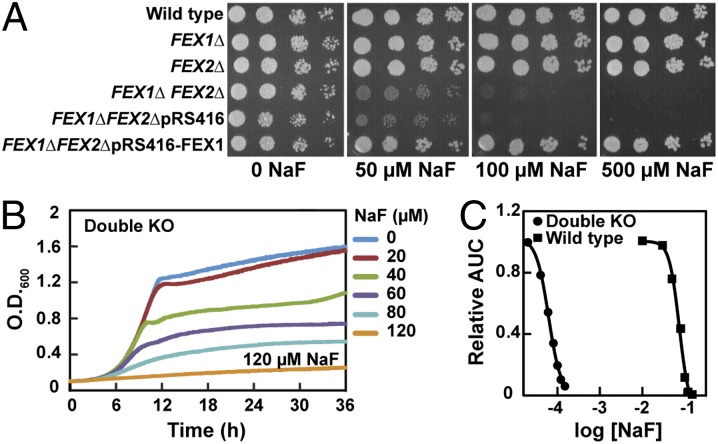
In contrast to the single KOs, deletion of both FEX genes results in a greater than 1,000-fold increase in fluoride sensitivity (Fig. 3 and Table S1). The growth of the double KO is significantly affected at micromolar concentrations of fluoride in both solid and liquid culture. For this double deletion strain, the IC50 for fluoride is ∼60 μM and the MIC is 120 μM. To determine if this phenotype is unique to fluoride, we tested the WT and double KO strains with increasing concentrations of the other sodium halide salts NaCl, NaBr, and NaI. We observed no change in sensitivity to these halides even at concentrations that completely inhibited growth (Fig. S4).
Fig. 3.
Fluoride sensitivity of the S. cerevisiae double KO strain. (A) Serial dilution assay of strains grown on YPD + NaF at the concentrations stated. All plates were grown at 30 °C and imaged after 3 d of growth. (B) Liquid growth assay of the double KO strain grown in YPD + NaF at the concentrations stated. Cultures were grown at 30 °C. (C) Representative IC50 curves for wild-type S. cerevisiae and the double KO strain. AUC is area under the growth curve.
To determine whether the large increase in fluoride sensitivity observed in the double KO was due solely to the deletion of the FEX proteins, we sought to restore fluoride resistance by supplying FEX1 extrachromosomally. A centromeric plasmid (maintained at one copy per cell) containing FEX1 was transformed into the double KO and this strain was assayed for growth in the presence of increasing amounts of fluoride. The transformed strain behaves similarly to the single deletion mutant with an IC50 >900-fold greater than the double KO (Table S1). This observation establishes that the change in fluoride tolerance observed in the double KO is entirely due to the deletion of the FEX genes.
The pathogenic yeast C. albicans is a diploid organism that has one FEX gene (CaO19.7095, chromosome 7; FEX1) encoding a 390 aa protein with two conserved FEX domains. We generated a strain in which one allele is deleted (referred to as the heterozygous KO) and one in which both alleles are deleted (referred to as the homozygous KO). The fluoride sensitivity of all strains was tested in liquid media growth assays.
Similarly to S. cerevisiae, the heterozygous KO does not have a significant effect on fluoride sensitivity, whereas the growth of the homozygous KO is compromised at concentrations of fluoride several orders of magnitude lower than for WT (Fig. 4 and Fig. S5). The WT strain has an IC50 for fluoride of 140 mM and an MIC of 175 mM (Table S1). The heterozygous KO has inhibition values very similar to WT, whereas the homozygous KO has an IC50 of 100 μM and an MIC of 0.5 mM, both >300-fold lower. Neither C. albicans KO strain shows an increased sensitivity to Cl− compared with WT (Fig. S6). A rescue strain was constructed by integrating a WT copy of FEX1 into the ribosomal protein S10 (RPS10) locus. The reintroduction of FEX1 at this alternate locus restores the ability to resist high concentration of NaF similarly to WT, although this strain grew more slowly than WT for unknown reasons (Fig. S5).
Fig. 4.
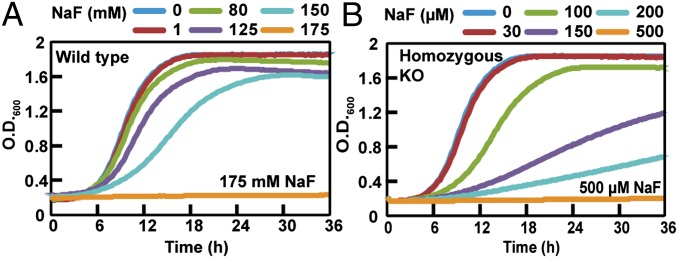
Sensitivity of the C. albicans FEX1 KO strain to NaF. Growth curves of (A) WT and (B) homozygous KO.
Given that deletion of FEX in all three organisms produced a similar fluoride-sensitive phenotype, we performed the rest of the analyses only on the two yeasts. These unicellular organisms were more amenable to analysis than the filamentous species.
Intracellular Fluoride Concentration Increases in Organisms Without FEX.
If FEX genes encode proteins that remove fluoride from cells, FEX KO cells should have higher levels of fluoride than the WT strain. To quantitatively determine the amount of fluoride in WT and KO organisms, we measured the intracellular fluoride concentration using radioactive 18F (35, 36).
We first applied this method to measure the concentration of fluoride in WT S. cerevisiae. Yeast cells were incubated with a mixture of nonradioactive fluoride and a trace concentration of 18F. Any fluoride not taken up by the cells was washed away, and the counts per minute (cpm) were measured for the cell suspension. We additionally determined the volume of the WT and double KO cells, allowing us to calculate the concentration of fluoride inside the yeast cells. The measured intracellular volume of 38 ± 5 fL (mean ± SD) matches cell volumes reported previously (37).
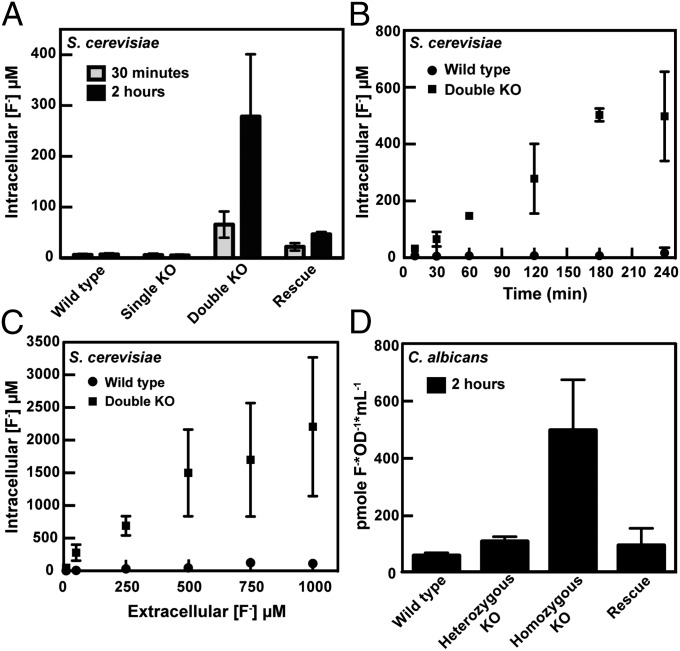
WT S. cerevisiae are able to maintain a low intracellular fluoride concentration over a range of external fluoride concentrations. When incubated in 50 μM external fluoride, the intracellular concentration remains below 10 μM (Fig. 5 A and B). There is essentially no change in the intracellular fluoride concentration for at least 4 h, which is the upper time limit of the assay due to the short half-life of the 18F isotope (109 min). When the external fluoride concentration was increased up to 1 mM, the internal fluoride concentration remains ∼10-fold below the external concentration (Fig. 5C). This result implies that even in millimolar amounts of fluoride, WT S. cerevisiae are able to maintain a low intracellular fluoride concentration. Consistent with the results from growth assays, single KO cells have levels of fluoride approximately equal to WT (Fig. 5A).
Fig. 5.
Intracellular fluoride concentrations in S. cerevisiae and C. albicans. Values graphed are the average of at least three measurements ± the SD. (A) Intracellular concentration of fluoride for S. cerevisiae strains after incubation with 50 μM NaF. (B) Intracellular fluoride concentration as a function of time for WT and double KO S. cerevisiae. Cells were incubated with 50 μM NaF. (C) Intracellular fluoride concentration as a function of external fluoride concentration for WT and double KO S. cerevisiae. All samples were incubated for 2 h before scintillation counting. (D) Intracellular fluoride concentration of C. albicans strains. Cells were incubated with 500 μM fluoride for 2 h.
In contrast, the intracellular fluoride concentration of double KO S. cerevisiae cells rapidly equilibrates and then exceeds the external fluoride concentration. After only 30 min of incubation, the double KO strain contains >10-fold more fluoride than the WT strain under the same conditions (Fig. 5A). After 2 h, the internal concentration rises to 5-fold higher than the external concentration and ∼40-fold higher than the WT strain (Fig. 5A). When cells were incubated with 50 μM NaF, we observe a nearly linear increase in the amount of fluoride inside the double KO cells with time (Fig. 5B). The intracellular concentration also increases as the external concentration of fluoride is increased. After 2 h in the presence of varying concentrations of fluoride, the amount of fluoride inside the double KO cells is between two- and fivefold higher than the external concentration (Fig. 5C).
When FEX1 is supplied to double KO cells on a plasmid, low intracellular levels of fluoride are restored (Fig. 5A). After 30 min of incubation with 50 μM fluoride, the rescue strain has an intracellular fluoride concentration within three- to fourfold of the WT cells. Even after 2 h of incubation, the concentration of fluoride inside these cells is significantly lower than in the double KO, despite a small increase compared with WT. This slight increase is most likely due to the fact that the cells must be grown in nonselective media before the assay, and this plasmid is known to be lost at a low percentage for each generation grown without selection (38). It is also possible that differences in expression of FEX1 between the plasmid and chromosomal locus may exist and contribute to the discrepancies observed.
The same trends were observed in C. albicans, where the homozygous KO accumulates much higher fluoride in the cell than the WT or heterozygous KO strains (Fig. 5D). Accurate concentrations were not determined due to difficulty in measuring the internal volume of C. albicans. However, we observed that the homozygous KO cells accumulate ∼10-fold more fluoride than WT cells when both are incubated with 0.5 mM fluoride. When FEX1 was reintroduced into a different chromosomal locus, the intracellular concentration of fluoride was equivalent to WT (Fig. 5D). These results suggest that FEX proteins play an important role in removing fluoride from the cell and establish that FEX is necessary and sufficient to lower the internal fluoride concentration (Fig. 5).
Cytotoxic Effects of Fluoride on FEX KO Organisms.
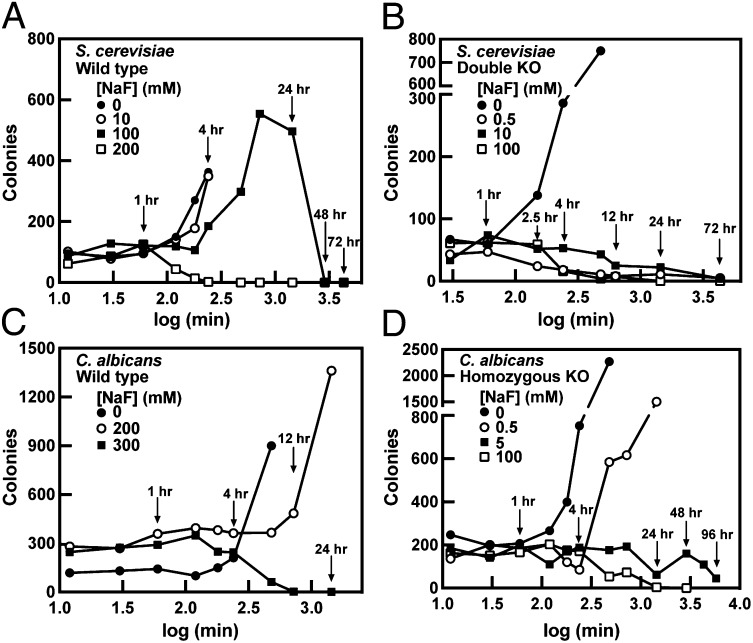
Deleting FEX in eukaryotic organisms causes an increase in both sensitivity to fluoride and intracellular fluoride concentration. We next determined whether this increase in intracellular fluoride in FEX KO yeast results in cell death at lower external fluoride concentration or shorter exposure time. To assess the time- and concentration-dependent killing of cells by fluoride, overnight growths of WT and double KO cells were diluted into media containing fluoride. Periodically, a small volume of this culture was removed, and the cells were washed and plated. The number of colonies was counted after several days of growth.
For WT S. cerevisiae, 200 mM fluoride is required to kill the total cell population within 24 h of exposure. Some growth is observed even at 100 mM fluoride, and at least 48–72 h of incubation is required for cell death at this concentration (Fig. 6A). Although 100 mM is slightly below the MIC for the WT strain, more growth was observed in this assay than in the liquid culture growth assays described above, most likely due to differences in conditions between these two experiments (amount of oxygen, shaking speeds, etc.). In contrast to the WT strain, when double KO cells are incubated with 100 mM fluoride, the number of colonies decreases sharply after 4–8 h, and all cells are dead at 24 h of exposure (Fig. 6B). When double KO cells are incubated with 0.5 mM fluoride, we observe a decrease in the number of colonies after only 2.5 h. However, after 72 h, a few colonies are still able to grow. Curiously, at 10 mM fluoride, the number of colonies does not substantially decrease until ∼12–24 h of exposure, and some cells are still alive after 72 h.
Fig. 6.
Effects of NaF on the growth and survival of S. cerevisiae and C. albicans. Data were measured in triplicate but only one data set is shown as an example. Colonies formed by (A) WT S. cerevisiae, (B) S. cerevisiae double KO, (C) WT C. albicans, and (D) C. albicans homozygous KO cultures.
For WT C. albicans, 300 mM fluoride is required to kill the yeast within 24 h (Fig. 6C); 200 mM fluoride (close to the MIC) inhibits the growth for the first 12 h only but cells continue to multiply after that. The result is similar to what was observed for S. cerevisiae, and we expect that this is due to differences in how the cells were cultured between solid agar plate and liquid medium assays. In the case of the homozygous KO, 100 mM fluoride is required to kill the cells within 24 h of exposure (Fig. 6D). Similarly to S. cerevisiae at lower fluoride concentrations, such as 5 mM here, the C. albicans homozygous KO is inhibited in growth but could survive up to 120 h.
These data suggest that although the inhibitory concentrations of fluoride are dramatically different between WT cells and those in which FEX has been deleted, the concentrations that cause cell death after 24 h are similar. Lower concentrations of fluoride could kill cells lacking a functional FEX, but longer exposure times were needed.
Effect of pH on the Growth of C. albicans with Fluoride.
The cellular permeability of HF is >1,000,000-fold greater than F− (39) due to the fact that HF is neutral and can therefore diffuse through the lipid bilayer. In contrast, F− is repelled by the large Born charging energy that results from the low dielectric environment of the membrane and the small radius of the F− anion (40). The pKa of HF is 3.2 (41), so a lower extracellular pH results in a higher ratio of HF to F−, and therefore a greater amount of fluoride able to enter cells (6, 42). Under low pH conditions, cells lacking FEX would be predicted to accumulate more fluoride and experience greater toxic effects than the same KO cells at higher pH. To test this hypothesis, we measured the growth of different C. albicans strains as a function of pH, both in the absence and presence of NaF. As expected, the growth of the WT, heterozygous KO, and rescue strains of C. albicans is unaffected by pH (Fig. S7). However, even in the absence of added NaF, the growth of homozygous KO cells is inhibited below pH 5. We believe that this effect is due to the presence of residual F− in the media [according to our measurement, YPD (yeast extract, peptone, dextrose) contains 7.3 ± 0.5 μM fluoride]. When a known amount of fluoride (100 μM) is added to the media, the growth of the homozygous KO is inhibited at pH 6.5 or lower, indicating that these cells are more sensitive to pH in the presence of F−.
Discussion
FEX genes are widespread across all three domains of life, suggesting that these proteins have an important function in diverse organisms. Deleting crcB in E. coli makes the cells more sensitive to fluoride, likely due to the accumulation of fluoride in the cell as indicated by a reporter assay (22). Here we showed that deleting FEX genes in three model eukaryotic organisms produces a fluoride sensitive phenotype, and KO cells become 200–1,000 times more sensitive to fluoride than their corresponding WT strain. These data demonstrate the importance of FEX in resistance to fluoride toxicity. We also provided direct evidence that cells lacking FEX have higher intracellular fluoride concentrations relative to their WT counterparts, directly implicating FEX in fluoride efflux.
The FEX protein product is predicted to be a multipass transmembrane protein with homology to a variety of membrane proteins responsible for transporting different substrates. Combined with our data, this suggests that the FEX gene encodes a fluoride channel or transporter, although we cannot rule out the possibility that FEX functions as a required accessory element to another protein that itself exports fluoride. However, a recent report shows conclusively that bacterial FEX homologs are highly selective fluoride channels that discriminate against Cl− by a factor of >10,000-fold (43). These channels reconstituted in lipid bilayers exist primarily in an open state and export F− via passive electrodiffusion by using the negative membrane potential. Because of the large degree of homology that exists between bacterial and fungal FEX genes, the eukaryotic FEX proteins are likely to also function as selective F− channels. Furthermore, the resting membrane voltage of fungi is normally highly negative (about −200 mV) (44, 45), which would provide a sufficient thermodynamic gradient to expel F− by the same mechanism. The results from both eubacteria and eukaryotes indicate that FEX proteins have a conserved function in maintaining low levels of intracellular fluoride across multiple domains of life and that these proteins promote the survival of cells under fluoride stress.
The conservation of FEX proteins across many species implies that a large number of organisms encounter fluoride stress and require the ability to remove this anion. Environmental fluoride concentrations vary from low micromolar to mid millimolar, suggesting that to survive, organisms must be tolerant to these levels (6, 46). It is possible that different species may be resistant to different concentrations of this ion. Indeed, we observe differences in the MIC for the different eukaryotic species that we have studied here, with C. albicans being the most tolerant to fluoride and N. crassa being the least. A previous study reports the MIC for C. albicans to be even higher, suggesting that different strains within species may vary in their level of resistance (18). Wide variation is observed in the fluoride tolerance of higher eukaryotes. For instance, the model organism Caenorhabditis elegans experiences toxic effects at low to submillimolar concentrations (47), whereas some plants (such as the cotton plant) can tolerate up to 4,000 ppm (about 220 mM) NaF (48). Relative to the WT strains, all of the FEX deletion mutants that we made in this study exhibited increased sensitivity to fluoride by several orders of magnitude, meaning they experienced growth inhibition at low to mid-micromolar fluoride. These data indicate that many organisms would not be able to grow in the absence of FEX in many environmental conditions. In fact, the S. cerevisiae FEX double KO shows growth inhibition at the level of fluoride supplementation found in some municipal water supplies (∼50 μM) (7). Even at these low micromolar amounts, our data using 18F as a reporter for intracellular fluoride concentration indicate that over time the internal fluoride concentration can reach much higher levels as fluoride accumulates.
Lower pH aggravates fluoride stress, presumably by facilitating the influx of fluoride into cells as the conjugate acid HF. This fact may also explain why most bacteria and fungi contain FEX genes. In many situations, the external pH is lower than the intracellular pH, which would promote the accumulation of fluoride in the cell. For instance, tap, distilled, or deionized water all have a pH of ∼5. For environmental water sources where the pH is also near 5, it would be difficult for cells to survive without genes encoding fluoride export proteins.
To date, no homologs of FEX have been identified in many higher eukaryotes, including in humans and other mammals. However, the toxic effects of fluoride on these species have been well documented. These reports along with our data indicate that most organisms likely have evolved mechanisms to combat fluoride toxicity. Although the proteins involved in fluoride response in mammals are largely unknown, the fact that FEX does not appear to be involved could be exploited for therapeutic purposes. Because humans and many animals do not have the FEX protein, whereas many pathogens and parasites do, a drug targeting FEX used in combination with fluoride might be promising as a novel therapeutic (49, 50). For example, when an antifungal reagent (Amphotericin) was combined with a small amount of NaF, the efficacy of the inhibition was greatly improved (49). If inhibitors targeting FEX were developed, such a combination would be predicted to have broad activity against diverse pathogens and parasites while being relatively nontoxic to human or animal cells that do not contain the FEX protein.
Materials and Methods
Strains and Growth Assays.
Strains used in this study are listed in Table S2, plasmids used are listed in Table S3, and primers used are listed in Table S4. FEX deletions in N. crassa, S. cerevisiae, and C. albicans were made using previously published methods. For a detailed description, see SI Materials and Methods. N. crassa growth assays were performed in both liquid and solid (race tubes) minimal media with and without NaF. S. cerevisiae growth assays were performed on solid agar plates and also in liquid culture using standard procedures. C. albicans growth assays were done in liquid culture using standard procedures. For a detailed description, see SI Materials and Methods.
18F Assays.
Measurements of internal fluoride concentrations in S. cerevisiae were made using yeast grown to ∼1 OD600; 675 μL of this cell suspension was mixed with 75 μL of 10× NaF solution containing a mixture of 18F and nonradioactive NaF. A variable amount of 18F was added to the 10× solutions to reduce the range of specific activities for the samples: 0.013 μCi/μL for the 10 and 50 μM samples, 0.026 μCi/μL for the 250 μM samples, 0.04 μCi/μL for the 500 μM samples, 0.053 μCi/μL for the 750 μM samples, and 0.067 μCi/μL for the 1,000 μM samples. After addition of NaF, the cells were incubated at room temperature for variable time. The media were removed, and the cells were washed four times by vacuum filtration. Cells were resuspended in 200 μL YPD, and the radiation intensity was measured by scintillation counting using a 18F measurement protocol. Each sample was counted for 1 min, and the decay of 18F during counting was accounted for by using the half-life of 109 min. One hundred microliters of a 1:1,000 dilution of each 10× stock was counted alongside the samples to determine the specific activity of the 10× mixture in moles per count. This value was used to determine the moles of NaF in the cells by multiplying the counts of each cell sample by the moles per count value obtained. Samples without 18F were prepared in an identical fashion and used to determine the OD600 of the cells after incubation and washing. The OD600 of the samples was used to determine the volume of the cells in each sample as described below. The concentration of NaF in the cells was determined by dividing the moles of NaF by the cell volume.
The cell volumes were determined using the cell displacement method and inulin as previously described (35). Thirty mL of ∼20 OD600 cells were pelleted and resuspended in 200 μL water with 10,000 counts/μL 14C purified inulin. The volume of the resuspension was determined, and the OD600 of the cells was measured. The increase in volume over 200 μL was due to both the cellular volume and the extra cellular volume. To determine the extracellular volume, the cells were filtered through a 0.45-μm filter by centrifugation. The radiation intensity of the filtrate and the cells were measured by scintillation counting. The radiation intensity of the filtrate is the specific activity of the extracellular liquid and was used to determine the extracellular volume from the radiation intensity of the cells. The cell volume was determined by subtracting the extracellular volume from the total volume increase above 200 μL. The OD600 and volume of the cells were used to determine the cell volume per OD600 × mL. This analysis was done four times for S. cerevisiae to determine a value of 0.00038 μL (cell volume)/μL (sample volume) × OD600 (sample). This value and the OD600 and volume of the NaF-incubated cells were used to determine the internal volume of the cells.
The internal NaF concentrations in C. albicans were determined by growing cells to a density of 2 OD600; 10× NaF solutions were added to the cells at similar specific activities as the S. cerevisiae samples. The picomoles of NaF were obtained using the same method as describe for the S. cerevisiae samples. Because of the challenge of using 14C inulin with a pathogenic fungus, the volume of the cells was not specifically determined. Because of this, the amount of fluoride in the C. albicans samples is reported in (picomoles F−)/[OD600(sample) × mL (sample)].
Fluoride Toxicity Assays.
A low cell density (about 200 cells) of each strain was incubated with different concentrations of fluoride in test tubes in a total volume of 2 mL YPD shaking at 30 °C. At different time points, 100 μL of this cell culture was removed and spread onto YPD agar plates. For experiments with the double KO strains, the 100 μL of culture was centrifuged and washed twice with 1 mL of YPD before being spread onto agar plates. Colonies were counted after 24–48 h of incubation at 30 °C.
Supplementary Material
Acknowledgments
We thank M. Hochstrasser (Yale University) for the generous gift of the BY4741 and BY4742 strains and resistance cassette plasmids; S. M. Noble (University of California, San Francisco) for the C. albicans SN152 strain and plasmids pSN40, pSN52, and pSN69; the Fungal Genetics Stock Center (Kansas City, MO) for Neurospora strains FGSC#4200 and FGSC#9717 and plasmid pCSN44; K. Ruff for cloning of pRS416-FEX1; D. Spakowicz for help with the phylogenetic tree; and R. Tomko Jr., J. W. Nelson, N. Sudarsan, and D. Hiller for helpful advice. This work was supported by National Institutes of Health Grants DE022340 (to R.B.) and GM022778 (to S.A.S.). Research in the R.B. laboratory is also supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310439110/-/DCSupplemental.
References
- 1.Jentsch TJ, Stein V, Weinreich F, Zdebik AA. Molecular structure and physiological function of chloride channels. Physiol Rev. 2002;82(2):503–568. doi: 10.1152/physrev.00029.2001. [DOI] [PubMed] [Google Scholar]
- 2.Duran C, Thompson CH, Xiao Q, Hartzell HC. Chloride channels: Often enigmatic, rarely predictable. Annu Rev Physiol. 2010;72:95–121. doi: 10.1146/annurev-physiol-021909-135811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards JC, Kahl CR. Chloride channels of intracellular membranes. FEBS Lett. 2010;584(10):2102–2111. doi: 10.1016/j.febslet.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmermann MB, Jooste PL, Pandav CS. Iodine-deficiency disorders. Lancet. 2008;372(9645):1251–1262. doi: 10.1016/S0140-6736(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 5.Zimmermann MB. The role of iodine in human growth and development. Semin Cell Dev Biol. 2011;22(6):645–652. doi: 10.1016/j.semcdb.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein LH, Davison AW. Fluorides in the Environment: Effects on Plants and Animals. Cambridge, MA: CABI; 2004. [Google Scholar]
- 7.Jagtap S, Yenkie MK, Labhsetwar N, Rayalu S. Fluoride in drinking water and defluoridation of water. Chem Rev. 2012;112(4):2454–2466. doi: 10.1021/cr2002855. [DOI] [PubMed] [Google Scholar]
- 8. WHO (1970) Fluorides and human health. Switzerland Monograph Series No. 59 (World Health Organization, Geneva) [PubMed]
- 9.WHO . Fluorine and Fluorides, Environmental Health Criteria 36, IPCS International Programme on Chemical Safety. Geneva: World Health Organization; 1984. [Google Scholar]
- 10.Smith FA, Hodge HC, Dinman BD. Airborne fluorides and man: Part I. CRC Crit Rev Environ Control. 1977;8:293–371. [Google Scholar]
- 11.WHO . Fluoride in Drinking Water. Geneva: World Health Organization; 2006. [Google Scholar]
- 12.Barbier O, Arreola-Mendoza L, Del Razo LM. Molecular mechanisms of fluoride toxicity. Chem Biol Interact. 2010;188(2):319–333. doi: 10.1016/j.cbi.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Lesher RJ, Bender GR, Marquis RE. Bacteriolytic action of fluoride ions. Antimicrob Agents Chemother. 1977;12(3):339–345. doi: 10.1128/aac.12.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maltz M, Emilson CG. Susceptibility of oral bacteria to various fluoride salts. J Dent Res. 1982;61(6):786–790. doi: 10.1177/00220345820610062701. [DOI] [PubMed] [Google Scholar]
- 15.Ochoa-Herrera V, et al. Toxicity of fluoride to microorganisms in biological wastewater treatment systems. Water Res. 2009;43(13):3177–3186. doi: 10.1016/j.watres.2009.04.032. [DOI] [PubMed] [Google Scholar]
- 16.Arthus M, Gavelle J. Action of sodium fluoride on yeast. Compt Rend Soc Biol. 1903;55:1481–1483. [Google Scholar]
- 17.Treshow M. Response of some pathogenic fungi to sodium fluoride. Mycologia. 1965;57:216–221. [PubMed] [Google Scholar]
- 18.Flisfisch S, Meyer J, Meurman JH, Waltimo T. Effects of fluorides on Candida albicans. Oral Dis. 2008;14(4):296–301. doi: 10.1111/j.1601-0825.2007.01385.x. [DOI] [PubMed] [Google Scholar]
- 19.Lebioda L, Zhang E, Lewinski K, Brewer JM. Fluoride inhibition of yeast enolase: crystal structure of the enolase-Mg(2+)-F(-)-Pi complex at 2.6 A resolution. Proteins. 1993;16(3):219–225. doi: 10.1002/prot.340160302. [DOI] [PubMed] [Google Scholar]
- 20.Agalakova NI, Gusev GP. Molecular mechanisms of cytotoxicity and apoptosis induced by inorganic fluoride. ISRN Cell Biol. 2012;2012:1–16. [Google Scholar]
- 21.Li L. The biochemistry and physiology of metallic fluoride: Action, mechanism, and implications. Crit Rev Oral Biol Med. 2003;14(2):100–114. doi: 10.1177/154411130301400204. [DOI] [PubMed] [Google Scholar]
- 22.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335(6065):233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486(7401):85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breaker RR. Riboswitches: From ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4(7):771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43(6):867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4(2):a003566. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu KH, et al. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143(4):1521–1532. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sand O, Gingras M, Beck N, Hall C, Trun N. Phenotypic characterization of overexpression or deletion of the Escherichia coli crcA, cspE and crcB genes. Microbiology. 2003;149(8):2107–2117. doi: 10.1099/mic.0.26363-0. [DOI] [PubMed] [Google Scholar]
- 30.Rapp M, Granseth E, Seppälä S, von Heijne G. Identification and evolution of dual-topology membrane proteins. Nat Struct Mol Biol. 2006;13(2):112–116. doi: 10.1038/nsmb1057. [DOI] [PubMed] [Google Scholar]
- 31.Stockbridge RB, et al. Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proc Natl Acad Sci USA. 2012;109(38):15289–15294. doi: 10.1073/pnas.1210896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UniProt Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40(Database issue):D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drescher M, Suttie JW. Intracellular fluoride in cultured mammalian cells. Proc Soc Exp Biol Med. 1972;139(1):228–230. doi: 10.3181/00379727-139-36115. [DOI] [PubMed] [Google Scholar]
- 36.Quissell DO, Suttie JW. Development of a fluoride-resistant strain of L cells: Membrane and metabolic characteristics. Am J Physiol. 1972;223(3):596–603. doi: 10.1152/ajplegacy.1972.223.3.596. [DOI] [PubMed] [Google Scholar]
- 37.Tyson CB, Lord PG, Wheals AE. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J Bacteriol. 1979;138(1):92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutknecht J, Walter A. Hydrofluoric and nitric acid transport through lipid bilayer membranes. Biochim Biophys Acta. 1981;644(1):153–156. doi: 10.1016/0005-2736(81)90071-7. [DOI] [PubMed] [Google Scholar]
- 40. Hall JE (1981) Membrane Transport, eds Bonting SL, de Pont JJHHM (Elsevier, New York), pp 107–121.
- 41.Ayotte P, Hébert M, Marchand P. Why is hydrofluoric acid a weak acid? J Chem Phys. 2005;123(18):184501. doi: 10.1063/1.2090259. [DOI] [PubMed] [Google Scholar]
- 42.Sharma R, Tsuchiya M, Skobe Z, Tannous BA, Bartlett JD. The acid test of fluoride: How pH modulates toxicity. PLoS ONE. 2010;5(5):e10895. doi: 10.1371/journal.pone.0010895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stockbridge RB, Robertson JL, Kolmakova-Partensky L, Miller C (2013) A family of fluoride-specific ion channels with dual-topology architecture. eLife 2:e01084. [DOI] [PMC free article] [PubMed]
- 44.Vacata V, Kotyk A, Sigler K. Membrane potentials in yeast cells measured by direct and indirect methods. Biochim Biophys Acta. 1981;643(1):265–268. doi: 10.1016/0005-2736(81)90241-8. [DOI] [PubMed] [Google Scholar]
- 45.Ballarin-Denti A, Slayman CL, Kuroda H. Small lipid-soluble cations are not membrane voltage probes for Neurospora or Saccharomyces. Biochim Biophys Acta. 1994;1190(1):43–56. doi: 10.1016/0005-2736(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 46.WHO . Guidelines for Drinking Water Quality. 4th Ed. Geneva: World Health Organization; 2011. [Google Scholar]
- 47.Li Q, et al. Toxicity of sodium fluoride to Caenorhabditis elegans. Biomed Environ Sci. 2012;25(2):216–223. doi: 10.3967/0895-3988.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 48.Jacobson JS, Weinstein LH, McCune DC, Hitchcock AE. The accumulation of fluorine by plants. J Air Pollut Control Assoc. 1966;16(8):412–417. doi: 10.1080/00022470.1966.10468494. [DOI] [PubMed] [Google Scholar]
- 49.Li S, Breaker RR. Fluoride enhances the activity of fungicides that destabilize cell membranes. Bioorg Med Chem Lett. 2012;22(9):3317–3322. doi: 10.1016/j.bmcl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zasloff M, Steinberg WH (1993) US Patent: 5,217,956.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.