Significance
Genome-wide hypomethylation is frequently observed in cancers. In this study, we showed that genome-wide hypomethylation analysis in plasma using shotgun massively parallel bisulfite sequencing is a powerful general approach for the detection of multiple types of cancers. This approach is particularly attractive because high sensitivity and specificity can be achieved using low sequence depth, which is practical diagnostically. This approach can also be used for monitoring patients following treatment. The same sequencing data can be further used for detecting cancer-associated copy number aberrations at no additional costs. One could thus combine plasma hypomethylation and copy number analyses in a synergistic manner for further enhancing detection sensitivity or specificity.
Keywords: epigenomics, epigenetics, next-generation sequencing, tumor markers, global hypomethylation
Abstract
We explored the detection of genome-wide hypomethylation in plasma using shotgun massively parallel bisulfite sequencing as a marker for cancer. Tumor-associated copy number aberrations (CNAs) could also be observed from the bisulfite DNA sequencing data. Hypomethylation and CNAs were detected in the plasma DNA of patients with hepatocellular carcinoma, breast cancer, lung cancer, nasopharyngeal cancer, smooth muscle sarcoma, and neuroendocrine tumor. For the detection of nonmetastatic cancer cases, plasma hypomethylation gave a sensitivity and specificity of 74% and 94%, respectively, when a mean of 93 million reads per case were obtained. Reducing the sequencing depth to 10 million reads per case was found to have no adverse effect on the sensitivity and specificity for cancer detection, giving respective figures of 68% and 94%. This characteristic thus indicates that analysis of plasma hypomethylation by this sequencing-based method may be a relatively cost-effective approach for cancer detection. We also demonstrated that plasma hypomethylation had utility for monitoring hepatocellular carcinoma patients following tumor resection and for detecting residual disease. Plasma hypomethylation can be combined with plasma CNA analysis for further enhancement of the detection sensitivity or specificity using different diagnostic algorithms. Using the detection of at least one type of aberration to define an abnormality, a sensitivity of 87% could be achieved with a specificity of 88%. These developments have thus expanded the applications of plasma DNA analysis for cancer detection and monitoring.
There is much recent interest in the biology and diagnostic applications of cell-free DNA in the plasma of human subjects. In particular, tumor-associated DNA has been detected in the plasma of cancer patients (1) and fetus-derived DNA has been found in the plasma of pregnant women (2). The finding of these types of circulating nucleic acids has implications for the detection of cancer and noninvasive prenatal testing, respectively. There are also many similarities between the two phenomena, with the detection of tumor-associated genetic (3), epigenetic (4), and RNA (5, 6) markers closely mirroring the analogous markers from the fetus in maternal plasma (7–9).
The advent of massively parallel sequencing has allowed fetal chromosomal aneuploidies, such as trisomy 21, to be robustly detected from maternal plasma (10–13). We and others have used a similar approach for detecting tumor-associated copy number aberrations (CNAs) (14–16) and single-nucleotide variants (14, 17) from the plasma of cancer patients. One important determinant of the sensitivity of such an approach for detecting cancer for a given amount of sequencing is the fraction of the genome exhibiting molecular aberrations that are being searched for in the plasma sample. This resembles the situation in noninvasive prenatal testing when deeper sequencing is needed for detecting subchromosomal copy number changes (18, 19) than compared with detecting an aneuploidy involving an entire chromosome (e.g., trisomy 21) (10–13).
We reason that one would be able to develop a highly efficient approach for detecting tumor-associated genomic aberrations in plasma using massively parallel sequencing if one could target a molecular alteration that is pervasive across the genome. DNA hypomethylation is known to be a genome-wide change that is present in most cancer types (20–22). Here, we describe our results for cancer detection using shotgun massively parallel bisulfite sequencing of plasma DNA to look for tumor-associated hypomethylation.
Results
Analysis of Genome-Wide Hypomethylation in the Plasma of Hepatocellular Carcinoma Patients.
Plasma DNA samples obtained from 26 hepatocellular carcinoma (HCC) patients and 32 healthy subjects were bisulfite converted and analyzed by massively parallel sequencing. Twenty-five of the HCC patients were of Barcelona Clinic Liver Cancer (BCLC) stage A, whereas the remaining patient was of stage B. Twenty-two HCC patients had chronic hepatitis B infection and one had hepatitis C. The 32 healthy subjects were randomly divided into two groups, each consisting of 16 subjects. One group (the reference group) was used to determine the normal range and the other (the test group) was assessed alongside the cancer cases for determining the specificity of the method. A mean of 163 million reads (range: 49 million to 232 million) were obtained per case, using one lane of an Illumina HiSeq 2000 sequencer. On average, 78.4% of the raw sequenced reads were uniquely alignable to the human reference genome. Amongst these reads, a mean of 73.3% were nonduplicated reads. Following the alignment step and filtering of duplicated reads, a mean of 93 million reads per case was used for downstream analyses.
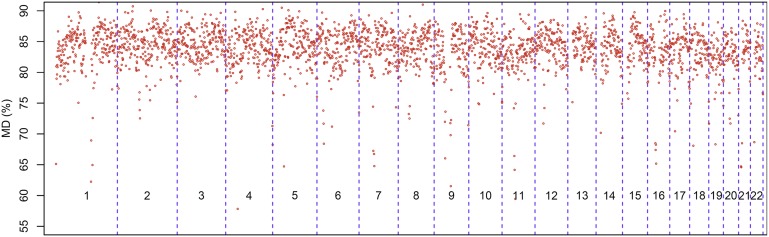
We first assessed the degree of variation in the plasma methylation profile between healthy individuals. We therefore determined the methylation density (MD) for each 1-Mb region, referred to as a bin, among the 32 healthy subjects (Fig. 1). We observed that the mean methylation densities varied from bin to bin. However, within the same bin, the interindividual variation across the 32 healthy controls was relatively small. Indeed, 95% of the bins had a coefficient of variation (CV) across the 32 healthy controls of ≤1.8%. Based on this observation, we proposed to compare the plasma methylation profile of cancer cases and healthy controls between bins of equivalent genomic regions. We defined a bin of a test case as hypomethylated if its mean MD was 3 SDs or more below the mean of the corresponding bin of the 16 healthy subjects in the reference group. We then determined the number of bins within the genome that was hypomethylated and expressed the result as a percentage of the total number of bins analyzed within the genome.
Fig. 1.
Variations of plasma methylation density (MD) of the 32 healthy subjects for each 1 Mb according to the genomic coordinates. The numbers within each box represent the chromosome number.
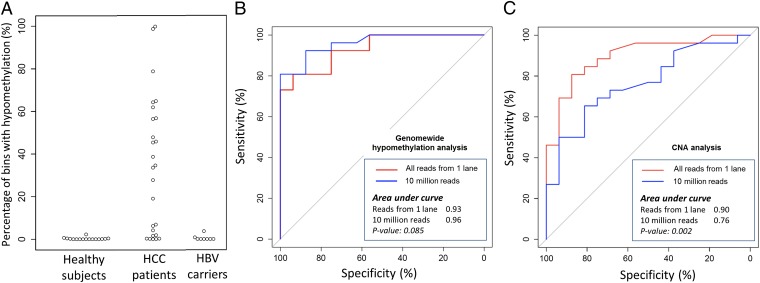
In the plasma samples of the HCC patients, a median of 34.1% [interquartile range (IQR): 2.5–56.7%] of the bins showed hypomethylation (Table 1). For the 16 healthy control subjects in the test group, a median of 0% (IQR: 0–0.26%) of the bins showed hypomethylation (Table S1). The hypomethylation patterns of one representative HCC patient and one healthy control subject are shown in Fig. 2. The diagnostic performance of this assay for detecting HCC is illustrated in Fig. 3A and has been further explored by receiver operating characteristic (ROC) curve analysis (Fig. 3B). For the latter analysis, the area under the curve (AUC) was 0.93 [95% confidence interval (CI): 0.87–1.00]. From the ROC curve, a cutoff of 1.1% of bins showing hypomethylation gave a sensitivity of 81% and a specificity of 94% for the detection of HCC. The selection of the cutoff of 1.1% was based on the point closest to the top left corner of the ROC curve. In the subsequent analyses in this report, we used this cutoff to define a positive result for plasma hypomethylation analysis.
Table 1.
Plasma hypomethylation and CNA analyses for the 26 HCC patients
| Case no. | BCLC stage | Tumor(s) largest dimension,* cm | HBV/HCV | Methylation analysis† | CNA analysis† | Classification based on the “OR” algorithm | Classification based on the “AND” algorithm |
| Percentage of bins with significant hypomethylation, % | Percentage of bins with copy number gains or losses, % | ||||||
| TBR36 | A | 16 | HBV | 98.7 | 25.3 | Positive | Positive |
| TBR34 | A | 13 | None | 64.3 | 57.0 | Positive | Positive |
| HOT198 | A | 12 | None | 99.8 | 45.9 | Positive | Positive |
| HOT205 | A | 6 | HBV | 1.7 | 1.4 | Positive | Positive |
| HOT192 | A | 5.5 | HBV | 47.9 | 14.6 | Positive | Positive |
| HOT227 | A | 5.5 | HBV | 56.8 | 36.7 | Positive | Positive |
| HOT238 | A | 5.5 | HBV | 27.7 | 16.8 | Positive | Positive |
| HOT197 | B | 5.3 | HCV | 4.2 | 0.3 | Positive | Negative |
| HOT156 | A | 5 | HBV | 34.6 | 14.8 | Positive | Positive |
| HOT236 | A | 3.7 | HBV | 38.7 | 0.9 | Positive | Positive |
| HOT170 | A | 3.5 | HBV | 45.4 | 14.2 | Positive | Positive |
| HOT229 | A | 3.1 | HBV | 0.3 | 0.7 | Positive | Negative |
| HOT233 | A | 3 | HBV | 0.3 | 3.8 | Positive | Negative |
| HOT240 | A | 2.8 | HBV | 1.9 | 8.0 | Positive | Positive |
| HOT222 | A | 2.6 | HBV | 61.9 | 2.1 | Positive | Positive |
| HOT162 | A | 2.5 | HBV | 45.8 | 25.0 | Positive | Positive |
| HOT172 | A | 2.5 | HBV | 33.7 | 11.3 | Positive | Positive |
| HOT164 | A | 2.3 | HBV | 56.4 | 2.5 | Positive | Positive |
| HOT215 | A | 2.3 | None | 64.9 | 6.6 | Positive | Positive |
| HOT207 | A | 2.1 | HBV | 6.9 | 0.9 | Positive | Positive |
| HOT224 | A | 2 | HBV | 0.3 | 0.2 | Negative | Negative |
| HOT159 | A | 1.5 | HBV | 6.3 | 0.4 | Positive | Negative |
| HOT204 | A | 1.5 | HBV | 0.1 | 1.9 | Positive | Negative |
| HOT151 | A | 1.5 | HBV | 0.1 | 0.1 | Negative | Negative |
| HOT208 | A | 1.2 | HBV | 19.1 | 2.5 | Positive | Positive |
| HOT167 | A | 1 | HBV | 78.8 | 0.6 | Positive | Negative |
HBV, hepatitis B virus infection; HCV, hepatitis C virus infection.
Denotes that the tumors have been sorted in descending order of size.
For hypomethylation and CNA analyses, a positive result would be classified if the percentage of bins showing hypomethylation and CNA was >1.1% and >0.68%, respectively.
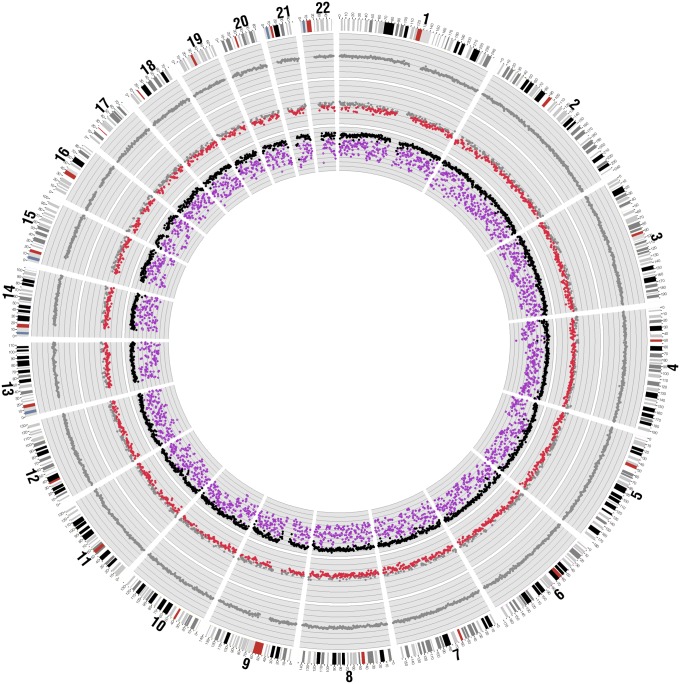
Fig. 2.
Methylation analyses for a representative HCC patient (HOT215) and a healthy control subject. For the HCC patient, the methylation densities of each 1-Mb bin for the tumor tissue (purple) and buffy coat (black) are shown in the inner ring. For the MD in tissue, the range shown is from 0% (innermost) to 100% (outermost) and the distance between two lines is 10%. The methylation z scores of the patient's plasma are shown in the middle ring. For the healthy control subject, the methylation z scores of the plasma are shown in the outer ring. For the plasma analysis, the red and gray dots represent bins with and without hypomethylation, respectively.
Fig. 3.
Percentage of bins showing hypomethylation in HCC patients and chronic hepatitis B virus (HBV) carriers with cirrhosis (A). ROC curves for HCC patient detection using hypomethylation (B) and CNA (C) analyses. The P values shown are for the comparisons between using all sequenced reads from one lane and using 10 million reads.
Among the HCC cases, tumor DNA and buffy coat DNA were available in 15 cases. Massively parallel bisulfite sequencing was performed for these samples. All of the HCC tumors showed significant hypomethylation compared with the corresponding buffy coat samples. The sequencing results in one representative case are presented in Fig. 2. A median of 98.8% (IQR: 97.1–99.9%) of the 1-Mb bins had a lower MD in the tumor tissues compared with the corresponding buffy coat samples, thus demonstrating the pervasiveness of hypomethylation as a genomic target for cancer detection.
The pervasiveness of hypomethylation across the genome would potentially allow it to be effectively detected at a reduced depth of sequencing. Hence, to explore the effect of reducing the amount of sequencing on the diagnostic performance, 10 million reads were randomly sampled from the massively parallel bisulfite sequencing data from each plasma sample. The hypomethylation patterns using one lane (i.e., mean of 93 million reads) and 10 million reads for all cases are shown in Fig. S1. The diagnostic performance of using 10 million reads per sample was further explored by ROC curve analysis (Fig. 3B). The AUC was 0.96 (95% CI: 0.91–1.00). There was no statistically significant difference (P = 0.085, DeLong test) between the AUCs of the ROC curves generated using all sequenced reads from one lane (i.e., a mean of 93 million reads) and 10 million reads.
Analysis of CNAs Using Bisulfite-Converted Plasma DNA from HCC Patients.
We next proceeded to compare the performance of plasma hypomethylation as a tumor marker with the plasma-based detection of CNAs previously described (14–16). To explore whether we could obtain information on CNAs using the massively parallel bisulfite sequencing data, we performed copy number analysis for two HCC patients (TBR34 and TBR36). We compared the CNAs detected in tumor tissues and those in the plasma of the corresponding patients obtained using bisulfite and nonbisulfite converted DNA. There was a high concordance between the CNAs detected in the tumor DNA and the plasma DNA samples (Fig. S2A). There was also a strong correlation between the two sets of plasma DNA results that were obtained from DNA with and without bisulfite treatment (r = 0.83 and 0.89 for TBR34 and TBR36, respectively, Pearson correlation; Fig. S2B).
We then carried out CNA analysis using the massively parallel bisulfite sequencing results from the plasma of the 26 HCC patients. All sequenced reads from one lane were used for each case. The reference range for plasma CNA analysis was determined using the massively parallel bisulfite sequencing results from the plasma of the 16 healthy reference subjects mentioned before. A bin is considered as showing copy number gain or loss as previously described (14) (Materials and Methods). We then determined the proportion of bins across the genome that showed copy number gains or losses. We next used ROC curve analysis to determine the best cutoff for discriminating HCC patients and controls based on proportion of bins with CNA. A cutoff of 0.68% bins showing CNAs, which was the point closest to the top left corner of the ROC curve, was selected. At this cutoff, HCC could be detected at a sensitivity of 81% and a specificity of 88%. In terms of the diagnostic performance of plasma CNA analysis, the AUC of the ROC curve was 0.90 (95% CI: 0.81–0.97; Fig. 3C). For samples positive for both hypomethylation and CNA analyses, the percentages of bins showing aberrations were significantly higher for hypomethylation (P < 0.01, Wilcoxon sign-rank test) (Table 1).
To explore the effect of reducing the amount of sequencing on the diagnostic performance of plasma CNA analysis, 10 million reads were randomly sampled from the massively parallel bisulfite sequencing data from each plasma sample. The diagnostic performance of using 10 million reads per sample was further explored by ROC curve analysis (Fig. 3C). The AUC dropped to 0.76 (95% CI: 0.62–0.91; P = 0.002, DeLong test).
Combination of Hypomethylation and CNA Analyses in HCC Patients.
For the detection of HCC, the plasma hypomethylation and CNA analyses could be combined using either an “OR” algorithm or an “AND” algorithm. In the former, a plasma sample would be called positive if either hypomethylation or CNAs were observed. In the latter, a plasma sample would be called positive if both hypomethylation and CNAs were observed. The diagnostic performances of these algorithms are shown in Table 1. The sensitivity and specificity are 92% and 88%, respectively, for the “OR” algorithm. However, the sensitivity and specificity are 69% and 94%, respectively, for the “AND” algorithm.
Patients With Chronic Hepatitis B and Cirrhosis but Without HCC.
We further analyzed plasma samples from eight patients with chronic hepatitis B infection and cirrhosis, but without HCC (Fig. 3A, Table 2, and Fig. S1). For sequencing depth of one lane and 10 million reads, seven of the eight patients had negative plasma hypomethylation test results and all eight had negative plasma CNA test results. The remaining patient (HBV8) had a positive result for plasma hypomethylation analysis for sequencing depths of one lane and 10 million reads (Table 2). The respective proportions of bins showing plasma hypomethylation were 3.7% and 3.5%. This patient had an ultrasound scan 4 mo before blood sampling, which did not show evidence of HCC.
Table 2.
Plasma hypomethylation and CNA analyses for the eight patients with chronic hepatitis B and cirrhosis
| Case no. | Percentage of bins with hypomethylation, % | Percentage of bins with CNA, % |
| HBV1 | 0 | 0.2 |
| HBV2 | 0 | 0.08 |
| HBV3 | 0 | 0.04 |
| HBV4 | 0 | 0.15 |
| HBV5 | 0 | 0.11 |
| HBV6 | 0.8 | 0.49 |
| HBV7 | 0.04 | 0.2 |
| HBV8 | 3.7 | 0.2 |
Serial Analysis of Plasma Hypomethylation and CNA from HCC Patients.
In two HCC patients, TBR34 and TBR36, apart from the presurgery plasma samples, additional plasma samples were obtained at multiple time points following surgical resection. Analyses for hypomethylation and CNAs were performed for these plasma samples (Fig. 4).
Fig. 4.
Serial analysis for plasma methylation and CNA for cases TBR34 (A) and TBR36 (B). For methylation analysis, the innermost ring shows the MD of the buffy coat (black) and tumor tissues (purple). For CNA analysis, the innermost ring shows the CNA detected in the tumor tissues. For both types of analyses, from the second innermost ring outward, the plasma results for different time points are shown. (A) For TBR34, the plasma samples were taken before surgery, at 3 d and 2 mo after tumor resection, respectively. (B) For TBR36, the plasma samples were taken before surgery, at 3 d, 3 mo, 6 mo, and 12 mo after tumor resection, respectively.
For TBR34, the percentages of bins showing hypomethylation and CNAs were 64.3% and 57%, respectively, before operation. At 3 d following tumor resection, the percentages of bins showing hypomethylation and CNAs were 75.5% and 11.7%, respectively (Table S2). Although the percentage of bins showing hypomethylation increased slightly, the magnitude of hypomethylation showed a significant reduction. Before treatment, 15.9% of the bins had MD more than 10 SDs below the mean of the control subjects. At 3 d after tumor resection, the degree of hypomethylation was reduced in the plasma, with none of the bins having MD more than 10 SDs below the mean of the controls. Of note, the percentages of bins showing hypomethylation and CNAs remained elevated at 38.8% and 14.6%, respectively, at 2 mo after operation. This patient was later diagnosed with having multifocal tumor deposits (previously unknown at the time of surgery) in the remaining nonresected liver at 3 mo and was noted to have multiple lung metastases at 4 mo after the operation. The patient died of metastatic disease at 8 mo after the operation.
For TBR36, before operation, the percentage of bins showing hypomethylation and CNAs were 98.7% and 25.3%, respectively. At 3 d following tumor resection, the percentages of bins showing hypomethylation and CNAs were reduced to 6.3% and 6.3%, respectively. Both changes almost completely disappeared at 3 mo after tumor resection and remained undetectable at 12 mo after the operation. The patient remained in clinical remission at 20 mo after tumor resection.
Hypomethylation and CNA Analyses in Patients with Other Cancer Types.
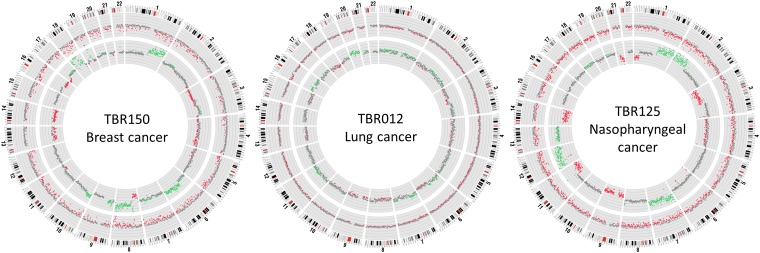
To show that our approach could be applied to cancer types other than HCC, we analyzed another 20 cancer samples including breast cancer, lung cancer, nasopharyngeal cancer, smooth muscle sarcoma, and neuroendocrine cancer. Patients with metastatic stages of all studied cancers had the highest percentages of bins showing plasma hypomethylation and CNAs among patients of the same cancer types (Table 3). Among the 20 patients evaluated, 15 (75%) had positive plasma hypomethylation results and 14 (70%) had plasma CNAs. Combined hypomethylation and CNA analyses using the “OR” algorithm was able to detect 17 (i.e., sensitivity of 85%) patients at a specificity of 88% (Table 3). Combined hypomethylation and CNA analysis using the “AND” algorithm was able to detect 12 (i.e., sensitivity of 60%) patients at a specificity of 94%. Representative results are shown in Fig. 5.
Table 3.
Plasma hypomethylation and CNA analyses for multiple types of cancers
| Cancer type | Case no. | Staging*/tumor max dimension | Methylation analysis† | CNA analysis† | Classification based on the “OR” algorithm | Classification based on the “AND” algorithm |
| Percentage of bins with significant hypomethylation, % | Percentage of bins with copy no. gains or losses, % | |||||
| Breast cancer | TBR150 | Metastatic | 30.3 | 36.9 | Positive | Positive |
| TBR126 | T2N1M0 | 28.8 | 16.6 | Positive | Positive | |
| 4.5 cm | ||||||
| TBR127 | T2N1M0 | 6.8 | 0.4 | Positive | Negative | |
| 2.5 cm | ||||||
| TBR128 | T2N0M0 | 0.3 | 22.5 | Positive | Negative | |
| 2 cm | ||||||
| TBR111 | T1N0M0 | 0.3 | 1.7 | Positive | Negative | |
| 0.9 cm | ||||||
| Smooth muscle sarcoma | TBR051 | Metastatic | 7.5 | 59.4 | Positive | Positive |
| Neuroendocrine tumor | TBR052 | Metastatic | 100 | 80.3 | Positive | Positive |
| Lung cancer | TBR164 | Metastatic | 94.6 | 37.8 | Positive | Positive |
| TBR012 | Metastatic | 19.9 | 22.8 | Positive | Positive | |
| TBR014 | Metastatic | 5.9 | 6.8 | Positive | Positive | |
| TBR177 | T2N1M0 | 3.0 | 0.2 | Positive | Negative | |
| 3.9 cm | ||||||
| Nasopharyngeal cancer | TBR031 | Metastatic | 6.1 | 15.3 | Positive | Positive |
| TBR125 | Metastatic | 49.7 | 42.3 | Positive | Positive | |
| TBR124 | T4N1 | 0.0 | 0.5 | Negative | Negative | |
| TBR123 | T1N2M0 | 20.8 | 0.8 | Positive | Positive | |
| TBR062 | T3N1M0 | 0.0 | 0.3 | Negative | Negative | |
| TBR108 | T3N1M0 | 0.0 | 0.3 | Negative | Negative | |
| TBR107 | T3N0M0 | 1.9 | 2.6 | Positive | Positive | |
| TBR106 | T1N1M0 | 2.6 | 1.2 | Positive | Positive | |
| TBR099 | T1N0M0 | 16.2 | 0.4 | Positive | Negative |
According to Union for International Cancer Control/AJCC 2010 Cancer Staging Manual, 7th Ed. (31).
For methylation and CNA analyses, a positive result would be scored if the percentages of bins showing hypomethylation and CNA were >1.1% and 0.68%, respectively.
Fig. 5.
Plasma hypomethylation and CNA analyses for three representative patients suffering from breast, lung, and nasopharyngeal cancers. The inner ring and outer ring show the CNA and methylation z scores, respectively. Each dot represents a 1-Mb bin. For CNA analysis, the green, red, and gray dots represent bins with chromosome gain, loss, and normal chromosome dosage, respectively. For methylation analysis, the red and gray dots represent bins with and without hypomethylation, respectively. The distance between two parallel lines represents a z-score difference of 5.
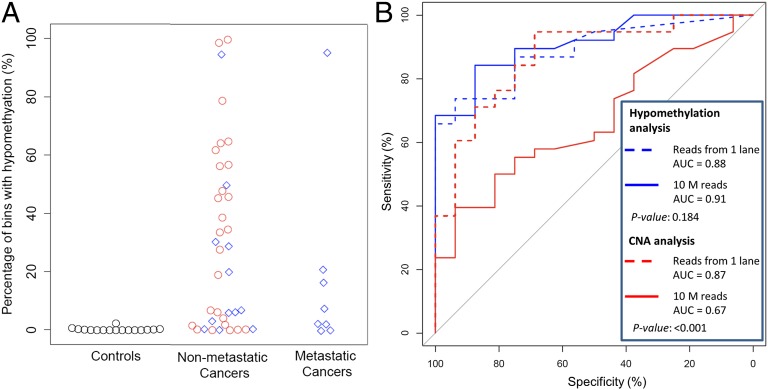
To give an overall view on the diagnostic performances of plasma hypomethylation and plasma CNA analyses for all of the cancer cases analyzed in this study, the results for the methylation analysis for the 16 healthy test cases, the 38 nonmetastatic cancer cases (HCC and other cancers), and the 8 metastatic cancer cases are shown in Fig. 6A. ROC curve analysis was performed for the 38 nonmetastatic cancer cases (Fig. 6B). ROC curves were plotted for analyses performed using all aligned reads from one lane (i.e., a mean of 93 million reads) and for just 10 million reads. For hypomethylation analysis, the diagnostic performance was similar when all sequenced reads in one lane (i.e., a mean of 93 million) and only 10 million reads were analyzed. However, the diagnostic performance for CNA analysis deteriorated when the number of sequenced reads was reduced from a mean of 93 million to 10 million. The diagnostic performances of using hypomethylation alone, CNA alone, and combining hypomethylation and CNA using the “OR” and “AND” algorithms for different sequencing depths are summarized in Table 4.
Fig. 6.
Diagnostic performance for hypomethylation and CNA analyses for patients with metastatic and nonmetastatic cancers. (A) Percentage of bins showing hypomethylation. The red circles and blue rhombuses represent cancer patients with HCC and non-HCC, respectively. (B) ROC curves for the detection of all cases with nonmetastatic cancers using hypomethylation and CNA analyses. The P values were for the comparisons between using all sequenced reads of one lane (i.e., a mean of 93 million reads) and using 10 million reads.
Table 4.
Overall diagnostic performance for plasma-based detection of the 38 nonmetastatic cancers
| No. of sequenced reads | Algorithm | Sensitivity, % | Specificity, % |
| Mean of 93 million | Hypomethylation alone | 74 | 94 |
| CNA alone | 71 | 88 | |
| Hypomethylation OR CNA | 87 | 88 | |
| Hypomethylation AND CNA | 58 | 94 | |
| 10 million | Hypomethylation alone | 68 | 94 |
| CNA alone | 39 | 94 | |
| Hypomethylation OR CNA | 74 | 94 | |
| Hypomethylation AND CNA | 34 | 94 |
Discussion
In this report, we have demonstrated that the detection of genome-wide hypomethylation in plasma using massively parallel bisulfite sequencing is a promising approach for cancer detection. It is noteworthy that we have shown that using the same bisulfite sequencing data, one could also detect tumor-associated CNAs in plasma at no additional cost. This development thus allows genome-wide hypomethylation and CNA analyses to be used synergistically, as will be discussed below. It is also very encouraging that our method can be used in a broad spectrum of cancer types, including HCC, breast cancer, lung cancer, nasopharyngeal cancer, smooth muscle sarcoma, and neuroendocrine cancer.
One important advantage of genome-wide hypomethylation as a tumor marker is that it is a pervasive feature of the genomes of most types of cancer (20–22). Hence, for a given depth of sequencing of plasma DNA, the probability of reads falling in genomic regions exhibiting such an aberration will be higher than that of reads aligning to genomic regions exhibiting a less pervasive change, e.g., CNAs. This characteristic suggests that the detection of genome-wide hypomethylation in plasma would be a relatively cost-effective approach with regard to the amount of sequencing that is needed. Indeed, our data have borne out these predictions. Thus, we have shown that the diagnostic performances of plasma hypomethylation analysis as illustrated by the AUCs of ROC curves are virtually identical for 10 million reads and 93 million reads. At 10 million reads, the diagnostic sensitivity and specificity for all nonmetastatic cancer cases reported here are 68% and 94%, respectively (Table 4). This is a level of sequencing that is feasible in practice, as evidenced by the recent launch of noninvasive prenatal testing for fetal trisomies, which is based on a similar amount of sequencing (10–13). Hence, plasma hypomethylation detection by massively parallel bisulfite sequencing at low sequencing depth (e.g., 10 million reads) has the potential to be used on its own as a relatively cost-effective screen for cancer.
In contrast, the detection of tumor-associated CNAs by plasma DNA sequencing is much more dependent on the depth of sequencing. Hence, the diagnostic sensitivity for cancer drops from 71% to 39% when the number of reads was reduced from a mean of 93 million to 10 million. Thus, at a low sequencing depth, it appears that plasma CNA analysis would not provide much additional diagnostic value over and above plasma hypomethylation analysis. However, at an increased depth of sequencing, such as 93 million reads per case, CNA analysis can be used synergistically with hypomethylation analysis using either the “OR” algorithm or the “AND” algorithm (Table 4). The former would be for applications in which sensitivity is more important than specificity, such as for cancer screening in subjects with relatively high risk or in the monitoring of relapse of patients known to have cancer. The latter, however, would be for applications in which specificity is relatively more important than sensitivity, such as for cancer screening in relatively low-risk populations. A high specificity for such applications would reduce the number of false-positive cases that would need to be followed up.
The diagnostic performance for plasma hypomethylation analysis using massively parallel sequencing appears to be superior to that obtained using PCR-based amplification of specific classes of repetitive elements, e.g., long interspersed nuclear element-1 (LINE-1) (23). One possible explanation for this observation is that, although hypomethylation is pervasive in the tumor genome, it does have some degree of heterogeneity from one genomic region to the next. In fact, we observed that the mean plasma methylation densities of the healthy subjects varied across the genome (Fig. 1). A simple PCR-based assay would not be able to take account of such heterogeneity into its diagnostic algorithm. Such heterogeneity would broaden the range of methylation densities observed among the healthy individuals. A greater magnitude of reduction in the MD would then be needed for a sample to be considered as showing hypomethylation. In contrast, our massively parallel sequencing-based approach divides the genome into 1-Mb bins and measures the methylation densities for such bins individually. We believe that this approach reduces the impact of the variations in the baseline methylation densities across different genomic regions as each region is compared between a test sample and the controls. Our approach therefore has a higher signal-to-noise ratio for the detection of hypomethylation associated with cancer.
With the advent of single-molecule sequencing platforms in which the methylation status of individual DNA molecules can be measured directly without bisulfite conversion (24), it is likely that the approach outlined in this report can be further simplified. The reduction in the number of analytical steps, e.g., omission of the bisulfite conversion step, would potentially further improve the precision of the assay.
Although the data presented in this report are promising, the number of samples that we have studied to date is still relatively small. Thus, it is important that the data should be validated on a much larger cohort of both cancer and noncancer cases, as well as conditions predisposing to cancer, e.g., hepatitis and cirrhosis. In this regard, we are following up the patient with chronic hepatitis and cirrhosis who had a positive plasma hypomethylation test result, to see whether this subject might be at an increased risk of developing HCC in the future. This and other future work would allow us to obtain more precise information on the diagnostic sensitivity and specificity in different subject groups.
Materials and Methods
Sample Collection and Processing.
Cancer patients were recruited from the Department of Surgery and the Department of Clinical Oncology of the Prince of Wales Hospital and the Department of Surgery of the North District Hospital, Hong Kong. Healthy subjects were recruited as controls. All recruited subjects gave informed consent, and the study was approved by the ethics committees of the respective institutions.
Plasma was collected from the blood samples after centrifugation at 1,600 × g for 10 min and 16,000 × g at 10 min. DNA was extracted using the DSP Blood Mini Kit (Qiagen). HCC tumor tissues were collected after tumor resection. DNA was extracted from the buffy coat and tumor tissues using the Blood Mini Kit (Qiagen).
Extracted DNA from tumor tissues and buffy coat was quantified by NanoDrop and plasma DNA was quantitated by real-time PCR targeting the HBB gene coding for β-globin (25) before library preparation.
Sequencing Library Preparation.
For DNA extracted from tumor tissues and buffy coat, 5 µg of DNA with 0.5% (wt/wt) unmethylated lambda DNA (Promega) was fragmented to about 200 bp in length using the Covaris S220 System. DNA libraries were prepared using the Paired-End Sequencing Sample Preparation Kit (Illumina) with methylated adapters according to the manufacturer’s instructions. For DNA extracted from plasma, the fragmentation step was omitted because plasma DNA is already fragmented in nature. For each case, DNA extracted from 4 mL of plasma was used for library construction.
Bisulfite Conversion of Sequencing Libraries.
For bisulfite sequencing (26, 27), the ligated products of the sequencing libraries were purified using AMPure XP magnetic beads (Beckman Coulter) and were then subjected to two rounds of bisulfite conversion by the EpiTect Plus DNA Bisulfite Kit (Qiagen) according to the manufacturer’s instructions. Bisulfite-converted products were amplified with 10 cycles of PCR amplification. The PCR products were then purified using AMPure XP magnetic beads.
DNA clusters either from bisulfite-treated or untreated DNA libraries were generated on a cBot system (Illumina) using a Paired-End Cluster Generation Kit, version 3 (Illumina), before sequencing on a HiSeq2000 system (Illumina) in a 75-bp single-end format.
Sequence Alignment and Identification of Methylated Cytosines.
After base calling, adapter sequences and low-quality bases (i.e., quality score of <5) on the fragment ends were removed. The trimmed reads in FASTQ format were then processed by a methylation data analysis pipeline called Methy-Pipe (27). To align the bisulfite converted sequencing reads, we first performed in silico conversion of all cytosine residues to thymines on the Watson and Crick strands separately using the reference human genome (National Center for Biotechnology Information build 36/hg19). Then we performed in silico conversion of each cytosine to thymine in all of the processed reads and kept the positional information of each converted residue. SOAP2 was used to align the converted reads to the two preconverted reference human genomes (28), with a maximum of two mismatches allowed for each aligned read. Only reads mappable to a unique genomic location were used for downstream analysis. Ambiguous reads mapped to both the Watson and Crick strands and duplicated (clonal) reads were removed.
Methylation Analysis in Plasma and Cancer Tissues.
Cytosine residues in the CpG dinucleotide context were used for downstream methylation analysis. After alignment, the cytosines originally present on the sequenced reads were recovered based on the positional information kept during the in silico conversion. The recovered cytosines among the CpG dinucleotides were scored as methylated. Thymines among the CpG dinucleotides were scored as unmethylated.
For methylation analysis, the genome was divided into 1-Mb bins. The MD for each 1 Mb was calculated as the number of methylated cytosines in the context of CpG dinucleotide divided by the total number of cytosines at CpG positions. In this approach, the sensitivity of detecting cancer-associated hypomethylation in plasma would be dependent on the variability of the MD in the 16 reference subjects. A lower variability would result in a better sensitivity for detecting cancer-associated hypomethylation. Thus, we have investigated the CV for MD measurement by focusing on different types of sequences in the genome. As shown in Table S3, the CV for MD analysis was lowest when repeat elements were analyzed. Therefore, in subsequent methylation analyses, the CpGs within repeat elements would be used for MD calculation.
To determine whether the plasma MD of a tested case was normal, the MD was compared with the results of the reference group to calculate the methylation z score (Zmeth) as follows:
 |
where 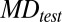 was the MD of the tested case for a particular 1-Mb bin;
was the MD of the tested case for a particular 1-Mb bin; 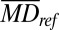 was the mean MD of the reference group for the corresponding bin; and
was the mean MD of the reference group for the corresponding bin; and 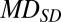 was the SD of the MD of the reference group for the corresponding bin.
was the SD of the MD of the reference group for the corresponding bin.
CNA Analysis.
For CNA analysis, the number of sequenced reads mapping to each 1-Mb bin was determined. Sequenced read density was determined for each bin after correction for GC bias using locally weighted scatter plot smoothing regression as previously described (29). For plasma analysis, the sequenced read density of the tested case was compared with the reference group to calculate the CNA z score (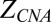 ) as follows:
) as follows:
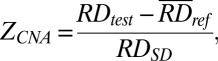 |
where 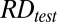 was the sequenced read density of the tested case for a particular 1-Mb bin;
was the sequenced read density of the tested case for a particular 1-Mb bin; 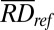 was the mean sequenced read density of the reference group for the corresponding bin; and
was the mean sequenced read density of the reference group for the corresponding bin; and 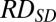 was the SD of the sequenced read density of the reference group for the corresponding bin.
was the SD of the sequenced read density of the reference group for the corresponding bin.
A bin was defined to exhibit CNA if the ZCNA of the bin was less than −3 or greater than 3.
Statistical Analysis for ROC Curves.
The ROC curves were constructed using the open source statistical package pROC software (30). The comparison of the AUC for two ROC curves was performed using the DeLong test.
Supplementary Material
Acknowledgments
This work is supported by the Hong Kong Research Grants Council Theme-Based Research Scheme (T12-CUHK05/10), the S. K. Yee Foundation, and the Innovation and Technology Fund under the State Key Laboratory Programme. Y.M.D.L. is supported by an endowed chair from the Li Ka Shing Foundation.
Footnotes
Conflict of interest statement: K.C.A.C., P.J., C.W.M.C., R.W.K.C., and Y.M.D.L. have filed patent applications on the technology described in this work.
This article is a PNAS Direct Submission.
Data deposition: Sequence data for 84 of the 86 subjects studied in this work who had consented to data archiving have been deposited at the European Genome–Phenome Archive (EGA), www.ebi.ac.uk/ega/, which is hosted by the European Bioinformatics Institute (EBI) (accession no. EGAS00001000566).
See Profile on page 18742.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313995110/-/DCSupplemental.
References
- 1.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 2.Lo YMD, Chiu RWK. Genomic analysis of fetal nucleic acids in maternal blood. Annu Rev Genomics Hum Genet. 2012;13:285–306. doi: 10.1146/annurev-genom-090711-163806. [DOI] [PubMed] [Google Scholar]
- 3.Chen XQ, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2(9):1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- 4.Wong IH, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59(1):71–73. [PubMed] [Google Scholar]
- 5.Lo KW, et al. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin Chem. 1999;45(8 Pt 1):1292–1294. [PubMed] [Google Scholar]
- 6.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5(8):1961–1965. [PubMed] [Google Scholar]
- 7.Lo YMD, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 8.Chim SSC, et al. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc Natl Acad Sci USA. 2005;102(41):14753–14758. doi: 10.1073/pnas.0503335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poon LL, Leung TN, Lau TK, Lo YMD. Presence of fetal RNA in maternal plasma. Clin Chem. 2000;46(11):1832–1834. [PubMed] [Google Scholar]
- 10.Chiu RWK, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105(51):20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu RWK, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palomaki GE, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011;13(11):913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 13.Bianchi DW, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 14.Chan KCA, et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59(1):211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 15.Leary RJ, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitzer E, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5(4):30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 18.Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013;92(2):167–176. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu SCY, et al. Noninvasive prenatal molecular karyotyping from maternal plasma. PLoS One. 2013;8(4):e60968. doi: 10.1371/journal.pone.0060968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 21.Ross JP, Rand KN, Molloy PL. Hypomethylation of repeated DNA sequences in cancer. Epigenomics. 2010;2(2):245–269. doi: 10.2217/epi.10.2. [DOI] [PubMed] [Google Scholar]
- 22.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5(3):223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 23.Tangkijvanich P, et al. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta. 2007;379(1-2):127–133. doi: 10.1016/j.cca.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 24.Korlach J, Turner SW. Going beyond five bases in DNA sequencing. Curr Opin Struct Biol. 2012;22(3):251–261. doi: 10.1016/j.sbi.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Lo YMD, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: Implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62(4):768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lun FMF, et al. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin Chem. 2013;59(11):1583–1594. doi: 10.1373/clinchem.2013.212274. [DOI] [PubMed] [Google Scholar]
- 28.Li R, et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 29.Chen EZ, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One. 2011;6(7):e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robin X, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edge SB, et al. AJCC Cancer Staging Manual. 7th Ed. New York: Springer; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.