Significance
The Mre11/Rad50/Nbs1 (MRN) complex recognizes double-strand breaks in eukaryotic cells, initiating DNA repair and signaling cell cycle checkpoint arrest. Several enzymes are known to be stimulated at sites of breaks through the actions of MRN, including nucleases and the ATM protein kinase. Here we investigated the interactions between MRN and DNA ends using a single-molecule approach that allows us to measure the unwinding of the helix while MRN is bound. We found that MRN promotes local separation of the DNA strands at the end of the helix (15–20 base pairs) in an ATP-dependent manner. A mutation in Rad50 that blocks this ATP-dependent opening also blocks DNA end processing in human cells, confirming the importance of duplex opening for MRN function.
Keywords: DNA structure, DNA–protein interaction
Abstract
The Mre11/Rad50/Nbs1 (MRN) complex initiates and coordinates DNA repair and signaling events at double-strand breaks. The interaction between MRN and DNA ends is critical for the recruitment of DNA-processing enzymes, end tethering, and activation of the ATM protein kinase. Here we visualized MRN binding to duplex DNA molecules using single-molecule FRET, and found that MRN unwinds 15–20 base pairs at the end of the duplex, holding the branched structure open for minutes at a time in an ATP-dependent reaction. A Rad50 catalytic domain mutant that is specifically deficient in this ATP-dependent opening is impaired in DNA end resection in vitro and in resection-dependent repair of breaks in human cells, demonstrating the importance of MRN-generated single strands in the repair of DNA breaks.
The Mre11/Rad50/Nbs1 (MRN) complex initiates DNA repair and signaling responses to DNA double-strand breaks (DSBs) in eukaryotic cells (1). The Mre11 and Rad50 components of the complex are conserved in all organisms and encode an endonuclease and a structural maintenance of chromosomes family ATPase, respectively, which regulate the initial processing of DSB ends, recruitment of exonucleases and endonucleases, stimulation of nonhomologous end-joining, and homologous recombination (2–4). In addition, the MRN complex in eukaryotes controls the activation of the ataxia-telangiectasia-mutated (ATM) kinase at sites of DNA breaks, initiating a signaling cascade that triggers cell cycle checkpoint activation (5, 6). All of these diverse activities require high-affinity association of Mre11/Rad50 complexes with linear DNA ends, one of the earliest events known to occur at DSBs in eukaryotes in vivo (7, 8).
A DNA end can be distinguished from unbroken DNA by the presence of the end itself, but the discontinuity also allows transient opening and distortion of the strands. We previously hypothesized that the binding of MRN to duplex DNA may involve limited unwinding of the end of the helix, based on the observations that recombinant human MRN can dissociate short duplexes (9); both human MRN and yeast Mre11/Rad50/Xrs2 (MRX) complexes recruit Exo1, which binds preferentially to branched DNA structures (10, 11); and ATM activation is regulated by strand opening when the ATP-dependent activities of MRN are compromised (12). MRN has never been directly observed to open duplexes, however, and its interactions with DNA have only been inferred from gel-based ensemble assays and microscopy, which cannot interrogate local changes in DNA structure.
Results
To examine the mechanisms and dynamics of MRN associations with DNA, we used single-molecule total internal reflection fluorescence microscopy to observe DNA duplex opening by the MRN complex in real time. DNA molecules containing Cy3 donor and Cy5 acceptor fluorophores on opposite strands were attached to a slide through a biotin-streptavidin linkage (Fig. 1A), and FRET between the dyes was used to monitor strand opening.
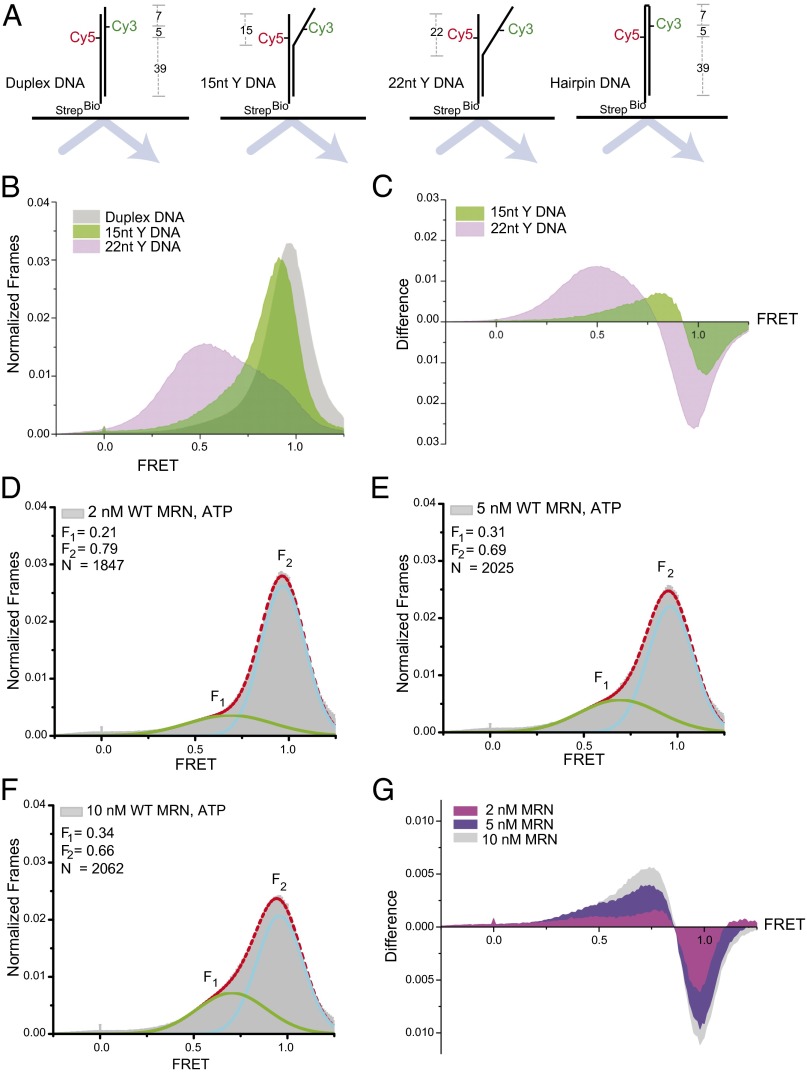
Fig. 1.
MRN opens DNA duplexes. (A) Schematic of the DNA molecules used in this study. The duplex DNA contains Cy3 and Cy5 located 5 bp apart in the duplex, which has a 4-nt 3′ overhang. The 15- and 22-nt Y substrates contain noncomplementary ends that replace 15 and 22 bp, as shown, and the hairpin DNA contains a 4-nt hairpin and a 51-bp stem. (B) FRET histograms of immobilized constructs in the absence of MRN. (C) Difference plot showing the FRET histogram from the duplex subtracted from those of the DNAs with noncomplementary ends (Y DNAs) (D–F) FRET histograms and fits of the duplex construct in the presence of increasing concentrations of MRN. F1 and F2 peaks were fit with a Gaussian function (green and blue lines as indicated), and N molecules were analyzed. (G) Difference plot showing a histogram for the DNA duplex in the absence of MRN subtracted from data in the presence of various concentrations of MRN.
We first measured FRET in the absence of MRN with a fully paired duplex and with duplexes containing noncomplementary ends of 15 or 22 nt (Y structures) to mimic an unwound duplex end (Fig. 1A). The fully paired duplex exhibited a single peak with a high FRET value, whereas the 15- and 22-nt Y structures were present in lower-FRET states, with the 22-nt unwound molecules occupying a range of conformations with different FRET values (Fig. 1B). A difference plot, with the distribution from the fully paired duplex subtracted from the distributions of each of the Y structures, illustrates the increase in low-FRET states and the loss of high-FRET states as a consequence of the noncomplementary ends (Fig. 1C).
When recombinant human MRN was added to the fully paired duplex in the presence of ATP, we observed a subpopulation of molecules (F1) with lower FRET values, centered at 0.69–0.71, that was increasingly prominent as the MRN concentration was increased (Fig. 1 D–F). The difference plot in Fig. 1G shows that the increase in low-FRET values on MRN binding is reminiscent of the shift to lower FRET values observed with the Y structures, with the center of the peak intermediate between the positions of the 15- and 22-nt Y structures. The decrease in FRET values on MRN addition was not related to DNA degradation, as demonstrated by the return of the high-FRET form after removal of MRN with proteinase K (Fig. S1). The simplest interpretation of these results is that MRN disrupts ∼15–20 base pairs, unwinding the DNA locally on binding to the end of the duplex.
The Rad50 catalytic head domains bind two ATP molecules in trans with another Rad50 molecule (13, 14). This conformation is important for DNA end binding, DNA tethering, and ATM activation (15). We found that the maximal decrease in FRET value requires ATP (Fig. 2A) and all components of the complex, including Nbs1 (Fig. 2B), consistent with previous observations that Nbs1 is essential for the ATP-dependent function of human MRN in vitro (9, 16). The Mre11 protein by itself did not induce low-FRET states in the DNA duplex (Fig. 2C). Likewise, MRN did not induce a low-FRET state for a DNA in which the ends were connected to form a hairpin (Figs. 1A and 2D), indicating that the low-FRET state for the base-paired duplex results from opening of the end on MRN binding rather than from a change in the environment of the dyes.
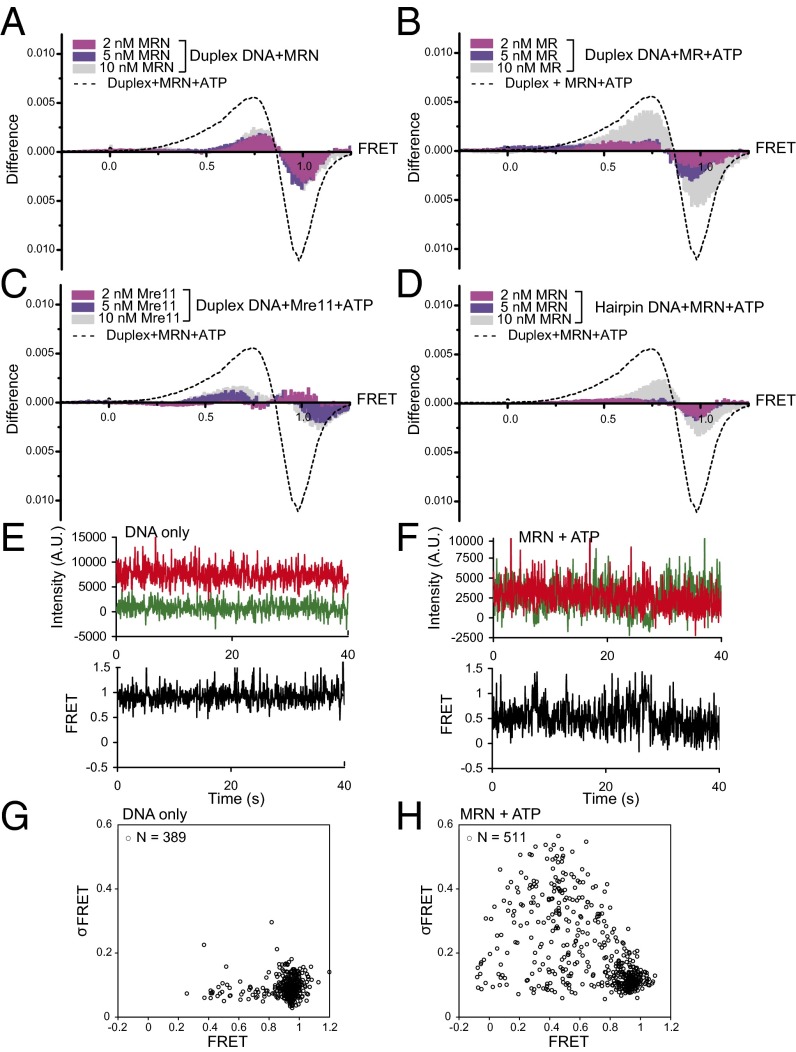
Fig. 2.
The MRN duplex opening is ATP-dependent and requires a DNA end. (A) Difference plots as in Fig. 1 showing the effects of ATP on MRN-induced low-FRET states with Cy3/Cy5 duplex DNA. The dotted curve represents the difference plot from Fig. 1G, the effect of 10 nM MRN on FRET values in the presence of ATP. (B and C) Difference plots as in A but with MR and Mre11 protein, respectively. (D) Difference plot as in A but with hairpin substrate (Fig. 1A). (E and F) Representative FRET traces for individual molecules of immobilized DNA with DNA only (E) or with MRN and ATP (F). (G and H) Average FRET values for Cy3/Cy5 duplex DNA molecules incubated alone (G) or with MRN and ATP (H), with the SD of the FRET value over the collection period (σFRET) plotted on the y-axis.
Evaluation of individual time traces show that nearly all of the molecules that displayed reduced FRET values did not transition back to the high-FRET state during the observation period (40 s), suggesting a long lifetime for MRN association. Representative traces for DNA only and MRN-bound DNA are shown in Fig. 2 E and F. Transition density plots derived from the time traces did not reveal discrete states within these reduced FRET values (Fig. S2); however, the FRET values of molecules bound by MRN varied substantially over time, as reflected in an increased standard deviation (SD) relative to molecules that remained in the high-FRET state (Fig. 2 G and H). This latter group exhibited behavior indistinguishable from that of the DNA in the absence of MRN, and its relative population decreased with increasing MRN concentration (Fig. 1 D–F), suggesting that at least some of these molecules were not bound to MRN. For the molecules with gave lower FRET values, the increased variability of the FRET value may reflect dynamics in the extent of unwinding induced by MRN and/or in the orientation of the dyes within the protein–DNA complex.
To specifically examine the population of DNA molecules bound to MRN, we immobilized the MRN complex to a chelated-copper PEGylated slide through the 6X His tag and flowed the Cy3/Cy5-labeled duplex over the slide. After washing away unbound DNA, we assessed the FRET values of the DNA that was retained by binding to the immobilized MRN (Fig. 3A). In the absence of ATP, quantitation of the FRET values showed a prominent peak centered at 0.95 (Fig. 3B), similar to the behavior of biotinylated DNA bound to the slide without MRN. In contrast, the addition of ATP with the DNA generated an additional population of molecules in a lower-FRET state (Fig. 3C), with an average FRET value of 0.61 (shown in a difference plot in Fig. 3D). This FRET value is similar to that of MRN-bound, immobilized DNA, indicating that the extent of DNA opening is not affected by the configuration of the surface immobilization.
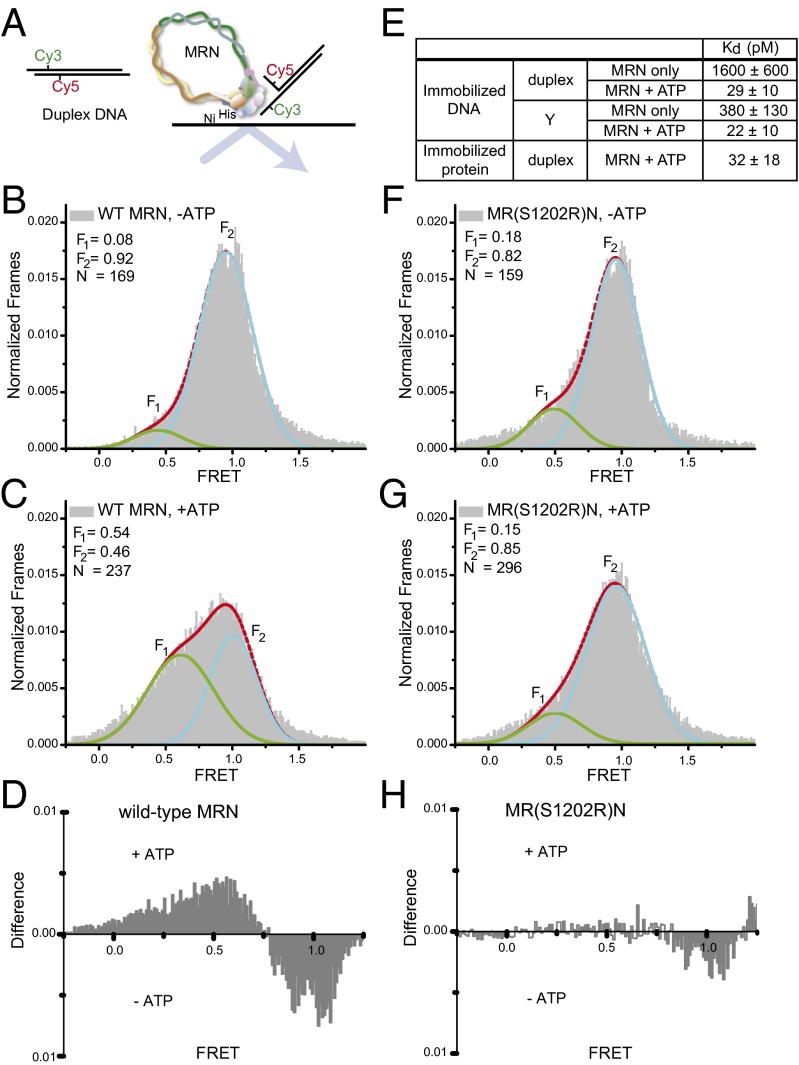
Fig. 3.
(A) Diagram of duplex DNA substrate and binding to immobilized MRN. (B and C) FRET histograms for duplex DNA bound to immobilized WT MRN in the absence (B) or presence (C) of ATP. (D) Difference plot showing the effect of ATP on FRET level for DNA bound to WT MRN. (E) Kd values for MRN–DNA binding, measured by immobilized DNA and mOrange MRN binding (Upper) or by immobilized mOrange MRN protein and duplex binding (Lower). (F and G) FRET histograms for duplex DNA bound to MR(S1202R)N in the absence (F) and presence (G) of ATP. (H) Difference plot showing the effect of ATP on FRET level for DNA bound to MR(S1202R)N.
As expected, the fraction of the DNA that populated this low-FRET state (54%) was larger than that seen in the experiments with immobilized DNA, because here only the DNA bound to MRN is retained on the surface. Nevertheless, 46% of the DNA molecules displayed high FRET values, equivalent to those of the free duplex. This high-FRET population most likely consists of DNA bound to MRN through its unlabeled end and/or DNA bound to individual MRN heterotrimers, which appear not to give efficient DNA unwinding (see below).
MRN is thought to be one of the first complexes to arrive at a DSB site in vivo (17–20), yet it has been difficult to quantitatively measure its DNA binding in ensemble assays owing to its large size and the formation of multiple complexes in gel mobility shift assays (16). To monitor MRN binding to immobilized DNA, we expressed MRN as a fusion of mOrange to the Mre11 protein. Using this system, we measured an equilibrium dissociation constant for duplex DNA of 29 ± 10 pM in the presence of ATP (Fig. 3E and Fig. S3). An alternative configuration with labeled DNA and immobilized MRN gave the same value within error (standard deviation). In the absence of ATP, titration of mOrange MRN into reactions with immobilized DNA gave a Kd value of ∼1.6 nM, indicating that MRN binds ATP and DNA cooperatively. Interestingly, MRN bound to a Y-shaped DNA with 15-nt single-stranded regions (Fig. 1A) with relatively high affinity even in the absence of ATP (Kd = 380 pM), suggesting that a key role of ATP in this cooperative binding is to promote a conformational change in MRN that promotes unwinding at the DNA end.
To measure the dissociation kinetics of mOrange MRN from duplex DNA, we used a laser pulsing method to extend the observation time beyond the lifetime imposed by dye photobleaching. In either configuration, with immobilized protein or immobilized DNA, we found that MRN complexes are extremely long-lived, with a dissociation rate constant of ∼5 × 10−3 s–1 (Fig. S4).
To explore the roles of the nucleotide in DNA unwinding by MRN, we also performed the immobilized DNA FRET experiments with the nonhydrolyzable nucleotide AMP-PNP. These experiments showed no apparent unwinding with adenyl imidodiphosphate (AMP-PNP) (Fig. S5), consistent with unwinding requiring the hydrolysis of ATP. However, it is also possible that AMP-PNP does not efficiently induce the changes seen with ATP. Thus, the failure of AMP-PNP to mimic the effects of ATP instead may be related to differences in the interactions of the nucleotides with MRN.
Considering the evidence for oligomeric MRN complexes provided by atomic force microscopy, gel mobility shift, and analytical ultracentrifugation experiments (15, 21), we investigated whether the MRN complexes in our experiments are monomeric or oligomeric. For this, we analyzed immobilized mOrange-MRN as well as mOrange-MRN bound to immobilized DNA for multistep photobleaching, which we took as an indication that oligomeric forms of the complex were present (Fig. S6). Using this criterion, we found that the percentage of immobilized DNA molecules bound by such oligomeric MRN complexes increases with protein concentration to a maximum of ∼30% at 3 nM MRN (Fig. S7). In addition, ∼35% of MRN complexes bound to DNA are oligomeric at concentrations of 1–3 nM (Fig. S7). Thus, oligomeric MRN complexes form under these conditions and are active in binding DNA. It is possible that these oligomeric complexes are responsible for the ∼30% of low-FRET molecules observed in the experiments with immobilized DNA (Fig. 1), and that this may explain why the binding of MRN to duplex DNA occurs in the 30 pM range (Fig. 3), yet the unwinding of DNA duplexes by MRN requires concentrations of 1–10 nM (Fig. 1).
We previously showed that a mutation in the conserved signature motif of Rad50 (S1202R) compromises the ATP-stimulated activities of MRN (22). The MR(S1202R)N complex is unable to stimulate ATM kinase activity in vitro, yet the deficiency of the mutant complex can be suppressed using a pre-unwound DNA structure with a heterologous sequence at one end (12), suggesting that this mutant may be specifically deficient in DNA unwinding. To test this hypothesis directly, we immobilized the MR(S1202R)N complex and found that the FRET values of the DNA bound to the complex were similar to those of the WT complex in the absence of ATP, but the mutant complex did not exhibit the shift to lower FRET values in the presence of ATP (Fig. 3 F–H). Thus, the S1202R mutation in the Rad50 C-terminal catalytic domain blocks the ATP-dependent unwinding of DNA ends.
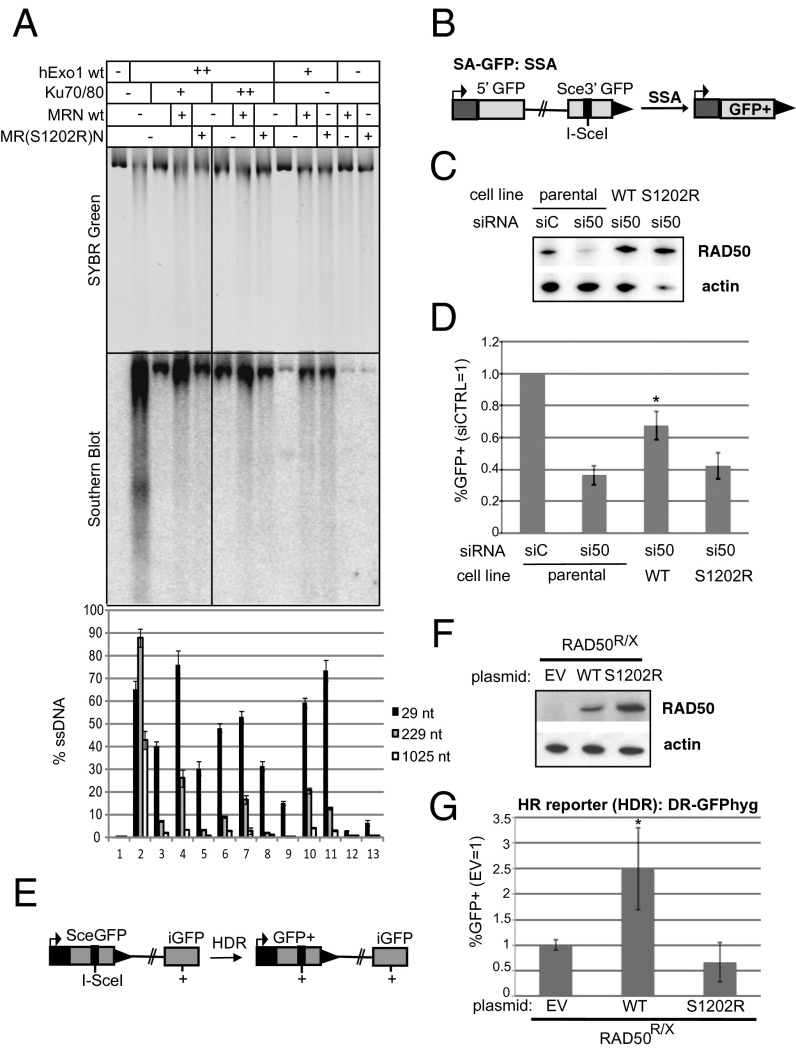
To determine whether the ATP-dependent DNA unwinding is important for DSB processing by MRN, we performed a reconstituted resection assay composed of recombinant MRN, human Exo1, and human Ku70/80 as described previously (11). Exo1 is a 5′ to 3′ exonuclease as well as a 5′ flap endonuclease that is recruited to DNA ends through the action of MRN(X) (10). Exo1 is inhibited by the end-binding heterodimer Ku70/80, but this inhibition can be blocked by MRN(X) in vitro (10, 11), consistent with genetic observations in vivo in Saccharomyces cerevisiae (23–25). Linearized plasmid DNA was incubated with human Ku70/80 and Exo1 and either WT or MR(S1202R)N complex (Fig. 4A). Resection was monitored by DNA staining with SYBR Green by Southern blot analysis using a strand-specific RNA probe for the 3′ strand at one end, and by quantitative PCR using a restriction enzyme to distinguish between single-stranded and double-stranded DNA (10). We found that Exo1 activity is blocked by Ku70/80 and relieved by WT MRN, but the MR(S1202R)N complex is deficient in this process (Fig. 4A). For instance, measuring resection at a site located 229 nt from the break, WT MRN stimulated Exo1-mediated degradation to 7.5- to 8-fold higher levels compared with reactions with the MR(S1202R)N mutant complex (lanes 4, 5, 7, and 8). In fact, 5′ strand degradation in the presence of the MR(S1202R)N mutant MRN was indistinguishable from the reaction without MRN by any of the methods used to assess resection. In contrast, there was little difference between the WT and mutant MRN complexes in the stimulation of Exo1 without the presence of Ku (lanes 10 and 11); thus, we conclude that the ATP-driven opening of DNA ends by MRN specifically contributes to the removal or displacement of Ku70/80. The equivalent MR(S1205R)X complex in budding yeast also shows a deficiency in resection of HO endonuclease-induced breaks in vivo (22).
Fig. 4.
RAD50-S1202R is deficient in promoting resection. (A) Resection assays on linearized plasmid DNA performed in vitro with purified Exo1, Ku, WT MRN, and MR(S1202R)N complexes as indicated. Reaction products were separated by agarose gel electrophoresis and analyzed by SYBR Green staining (Top), Southern blot analysis with an RNA probe specific for the 3′ strand at one end (Middle), and quantitative PCR (Bottom), as described previously (11). (B) Diagram of the SA-GFP reporter integrated into U2OS cells. The SSA product between two tandem GFP fragments restores a GFP+ cassette and causes a 2.7-kb deletion. (C) Immunoblot analysis of clonal stable cell lines harboring RAD50-WT and RAD50-S1202R expression cassettes resistant to an siRNA directed against endogenous RAD50 (siRAD50#1/si50). Shown are RAD50 and actin immunoblot signals from the parental cell line (U2OS SA-GFP) treated with a nontargeting siRNA (siCTRL/siC) and the parental cell line and RAD50-WT and RAD50-S1202R cell lines treated with siRAD50#1. (D) Cell lines used in B were treated with siRNA before transient expression of I-SceI and subsequent analysis of GFP+ cells by FACS. Shown is the frequency of SSA for siRAD50#1-treated samples relative to parallel siCTRL-treated samples for each cell line. *P < 0.0001 vs. parental siRAD50#1-treated; n = 12. Error bars represent SD. (E) Diagram of the DR-GFPhyg reporter for homology-directed repair (HDR) (27), which was integrated into cells derived from a Rad50-deficient human patient (RAD50R/X) (28) between the tandem GFP fragments restores a GFP+ cassette. (F) Immunoblot signals of RAD50R/X cells transfected with RAD50 WT, RAD50-S1202R (S1202R), or EV. (G) RAD50R/X cells with DR-GFPhyg were transfected with expression vectors for I-SceI along with RAD50 expression vectors or EV as indicated, and GFP+ cells were subsequently analyzed by FACS. The frequency of HDR (GFP+ cells) normalized to parallel EV transfections is shown. *P < 0.01 vs. EV and S1202R; n = 4. Error bars represent SD.
To test whether the Rad50 S1202R mutant also shows a deficiency in promoting resection in human cells, we used I-SceI–based DSB reporter systems for homologous recombination (HR), in which HR products are marked by GFP+. First, we used osteosarcoma U2OS cells with an HR reporter for single-strand annealing (SSA; Fig. 4B) (26), with which we established clonal cell lines with RAD50-WT and RAD50-S1202R expression cassettes with silent mutations to resist an RAD50 siRNA. Thus, these cell lines maintain an RAD50 immunoblot signal after siRAD50 treatment; RAD50 levels in parental cells were reduced by more than fourfold (Fig. 4C). These cell lines were treated with siRAD50, I-SceI was transiently expressed, and the frequency of SSA was quantified (%GFP+ cells normalized to parallel siCTRL-treated samples; Fig. 4B). We found that the RAD50-WT cell line showed a significantly higher frequency of SSA compared with the parental cell line (1.8-fold; P < 0.0001; Fig. 4D), whereas the RAD50-S1202R cell line did not.
Next, we integrated a reporter for homology-directed repair (HDR; Fig. 4E) (27) into a human patient cell line deficient in RAD50 (RAD50R/X) (28). With these cells, I-SceI was transiently expressed with an empty vector (EV), RAD50-WT, or RAD50-S1202R, and the frequency of HDR was quantified (%GFP+ cells normalized to I-SceI with the EV; RAD50-WT and S1202R expression in RAD50R/X confirmed by immunoblot analysis; Fig. 4F). We found that RAD50-WT expression caused a significant increase in HDR (2.5-fold; P < 0.01; Fig. 4G), but RAD50-S1202R had no effect.
These findings indicate that RAD50-S1202R is deficient for promoting HR. Considering that the MR(S1202R)N mutant binds to DNA but specifically fails to produce ATP-dependent opening of the duplexes (Fig. 3), these results collectively argue that this opening event is critical for resection of DNA ends in human cells.
Discussion
In this work, we have shown that ATP-driven association of MRN with DNA ends promotes a dramatic change in DNA structure such that ∼15–20 bp of duplex is destabilized and held open by the complex. The dependence on ATP in strand opening by MRN is consistent with previous observations of nucleotide-stimulated strand displacement (9) and our analysis of MRN effects on the ATM protein kinase (12). Activation of ATM is also sensitive to the S1202R mutation, which can be suppressed by heterologous, unwound strands at a DNA end. In addition, we recently showed that the ATP-bound (but not hydrolyzed) form is the form of MRN that activates ATM in vitro (15).
Structural analysis of Rad50 from archaea and bacteria has shown that two Rad50 “heads,” composed of N-terminal and C-terminal DNA-binding domains, dimerize in the presence of ATP (13, 14, 29). Mre11 binds underneath the dimerized domains in this configuration, blocking the nuclease active sites of Mre11 (14, 29). In contrast, Mre11/Rad50 catalytic domains in the absence of ATP are in an open, extended structure (30). No structural information on the location of DNA in either configuration is available, but our finding that ATP is required for efficient DNA unwinding suggests that it is the closed, ATP-bound dimeric structure that engages and unwinds the DNA strands at a break. Under the conditions of these experiments, the steady-state ATP hydrolysis by MRN is much slower than the DNA opening. Considered together with results indicating that ATP binding induces conformational changes in MRN (29, 30), these results most simply suggest that it is bound ATP that promotes formation of the closed structure and leads to local DNA unwinding.
All of the experiments reported in this paper were performed in the presence of magnesium ions, which support only a low level of Mre11 nuclease activity. The nuclease function of Mre11 does not affect the measurement of unwinding by FRET, as demonstrated by the recovery of a high-FRET state after removal of bound proteins. Many of the known biological functions of MRN have been reconstituted in magnesium and are similar to observations in human cells, including stimulation of DNA end resection by Exo1 and Dna2 (10, 11, 31, 32) and activation of ATM in the presence of DNA DSBs (12).
The single-molecule assays used in this study also allowed us to determine the binding constants for MRN association with DNA ends, which yielded an estimate of ∼30 pM in the presence of ATP. This binding is much tighter than has been estimated from gel-based assays (16), possibly because the complexes observed in the gel mobility shift assays are formed primarily from oligomerized proteins, as also observed visually by scanning force microscopy (21). We infer that higher concentrations of MRN are required to form these oligomeric complexes than are required for a single functional unit of MRN to bind to a DNA end. Our results indicate that these oligomeric complexes are capable of opening DNA and are linked to the biological functions of MRN, thus providing insight into the strength of the functional interaction of MRN with DNA ends. It is possible that the larger oligomers also play important roles in DNA end tethering and functions of MRN related to the DNA damage-induced foci of repair proteins.
In addition to ATM, MRN recruits specific DNA-processing enzymes to sites of DNA DSBs. We have shown that unwound single strands at these ends promote the binding of yeast and human Exo1 to DNA ends (10, 11), suggesting that the modulation of DNA structure at ends is critical for MRN regulation of DNA end processing. Based on these results, we propose that MRN acts as a gatekeeper at a DNA break, opening the strands and recruiting specific enzymes while restricting access of other complexes to ensure the coordinated assembly of repair and signaling activities at the break site.
Materials and Methods
Protein Expression and Purification.
The human MRN complex was expressed and purified from insect cells as described previously (33) with modifications. After binding and elution from nickel-nitrilotriacetic acid resin, the protein was bound to a 1-mL Hitrap SP column (GE Healthcare) in buffer A [25 mM Tris (pH 8.0), 10% glycerol, 1 mM DTT] and eluted with buffer A containing 0.5 M NaCl before purification on anti-Flag antibody resin as described. The MR(S1202R)N mutant complex (12) was prepared similarly. Expression and purification of Ku and Exo1 were done as described previously (11). The mOrange gene was amplifed by PCR from a template containing the ORF of mOrange (gift from M. Modesti, Centre National de la Recherche Scientifique, Marseille, France) and cloned into the C terminus of the full-length human Mre11 gene in pTP109 with XbaI/KpnI sites to generate pTP1987, a pFastBac1 baculovirus transfer plasmid (Invitrogen) containing the Mre11-mOrange fusion cassette. The plasmid was used to make a bacmid (pTP1973) according to the manufacturer’s instructions (Invitrogen), and the resulting virus was used instead of WT Mre11 to make M(mOrange)RN complex in Sf21 insect cells as described previously (33).
Purification of the M(mOrange)RN complex was performed as for human MRN, but after SP ion exchange, 0.5 mL of the peak fraction was loaded onto a Superose 6 10/300 GL gel filtration column (GE Healthcare) in buffer A. Peak fractions were collected and aliquoted at −80 °C for future use. For the immobilized MRN assays, purified MR complex was used with the addition of purified Nbs1 at an equimolar ratio. We observed no differences between the activity of MRN purified as a coexpressed complex and MR + Nbs1.
Oligonucleotides.
In immobilized DNA experiments, the fully annealed duplex was composed of TP2451 (5′-CTTGCATG[iCy5]CCTCAGCTATTCCGGATTATTCATACCGTTCCCATGTA-biotin-3′) annealed to TP2450 (5′-TGGGAACGGTATGAATAATCCGGAATAGCTGAGGCATGC[iCy3]AAGTCTA-3′). The 15-nt and 22-nt Y structures were composed of TP2451 annealed to TP2972 (5′-TGGGAACGGTATGAATAATCCGGAATACAACATTGCCAT[iCy3]GCATCTA-3′) and TP2973 (5′-TGGGAACGGTATGAATAATCGACTCACCAACATTGCCAT[iCy3]GCATCTA-3′), respectively. The hairpin DNA substrate was made by ligation of TP2591 (5′-TGGGAACGGTATGAATAATCCGGAAT-3′) to TP2592 (5′-ATTATTCATACCGTTCCCATGTA-biotin) to TP2586 (5′-AGCTGAGGCATGC/iCy3/AAGTCTATATATAGACTTGCATG/iCy5/CCTCAGCTATTCCGG-3′), and the ligated substrate was gel-purified in a denaturing sequencing gel before use.
Another fully annealed duplex was composed of TP2450 annealed to TP3343 (5′-CTTGCATG[iCy5]CCTCAGCTATTCCGGATTATTCATACCGTTCCCATGTA-3′). All Cy3 and Cy5 oligonucleotides were purchased from Integrated DNA Technologies.
Immobilized DNA Single-Molecule Experiments.
DNA duplexes, 3′ biotinylated, were immobilized through biotin-streptavidin linkages to streptavidin molecules conjugated to the surface of a quartz slide passivated with methoxy polyethylene glycol (mPEG). The molecules were imaged by prism-type total internal reflection microscopy on an Olympus IX-71 microscope with a cooled I-PentaMAX IIC CCD (Princeton Instruments). An oxygen-scavenging system (OSS) consisting of 0.6 mM Trolox, 8% glucose, 0.05 mg/mL catalase, and 0.1 mg/mL glucose oxidase was used to slow photobleaching. For FRET experiments, the duplexes were directly excited with a 532-nm laser (Crystalaser), and a 637-nm laser (Coherent) was used to confirm the presence of the Cy5 dye for each duplex after 50 s of monitoring FRET by excitation of Cy3. The data were acquired at 10–25 frames/s. The final solution conditions were 25 mM 3-(N-morpholino)propanesulfonic acid (MOPS; pH 7.0), 60 mM NaCl, 5 mM Mg2+, 1 mM DTT, and a nucleotide concentration of 1 mM when applicable.
Immobilized Protein Single-Molecule Experiments.
MRN was immobilized through a 6X His tag to chelated copper mPEG quartz slides (MicroSurfaces). DNA duplexes with either a single Cy5 (for the colocalization experiments with immobilized mOrange-MRN) or dual-labeled with Cy3 and Cy5 (for FRET experiments) were added to the flow chamber, with the OSS described in the preceding section.
Single-Molecule Measurements of Binding Equilibrium and Kinetics.
For equilibrium measurements, the immobilized species (Cy5-labeled DNA substrates or mOrange-MRN) were incubated with free mOrange-MRN or Cy5-labeled DNA, respectively, at concentrations ranging from 1 pM to 3 nM in the flow chamber for 10 min before data collection. Based on the measured rate constant for dissociation (below), this incubation was sufficient for equilibration at all concentrations of protein and DNA. The fraction of the immobilized species colocalized with dye-labeled molecules from the solution was measured for several fields of view at each concentration. Kinetics were measured by allowing binding of mOrange-MRN to immobilized Cy5-labeled DNA or binding of Cy5-labeled DNA to immobilized mOrange-MRN, and then washing out the unbound, dye-labeled molecules. The process of dissociation of dye-labeled MRN or DNA was performed for 16 min, with repeated cycles of unshuttering the excitation laser for 2 s and then shuttering it for 28 s.
In Vitro Resection Assays.
The resection assays with plasmid DNA were performed as described previously (10) with the following modifications. The 4.5-kb plasmid pNO1 was linearized with SphI and incubated in reactions containing 25 mM MOPS (pH 7.0), 1 mM DTT, 5 mM MgCl2, and 60 mM NaCl (30 mM in reactions containing Dna2) for 1 h at 37 °C. Each reaction contained 0.135 nM linearized DNA in a total volume of 10 μL. Each reaction was terminated by adding 0.1% SDS and 10 mM EDTA. The reactions were loaded and separated on a 1% nondenaturing agarose gel and further analyzed by SYBR Green (Invitrogen) staining and nondenaturing Southern hybridization as described previously (10).
Homologous Recombination Reporter Assays.
For RAD50 expression vectors, the RAD50-HA-His coding sequence from pTP11 (34) was inserted into an EV (pCAGGS-BSKX) (26). Silent mutations to resist siRAD50#1 (target: 5′gaaacaaacugcagaaugu; mutations: 5′GAAAtAAgtTaCAGAATGT) and the S1202R mutation were introduced. A neomycin resistance expression cassette was inserted upstream of the promoter, linearized plasmids were transfected into the U2OS SA-GFP reporter cell line (26), and single integrant clones were isolated using Geneticin (Invitrogen). To examine SSA, cells were transfected in 24-well dishes with 5 pmol siRAD50#1 or siCTRL#1 (Thermo Scientific) using 1.8 μL of RNAiMAX and the next day were transfected with the I-SceI expression vector pCBASce (0.4 μg) using 1.8 μL of Lipofectamine 2000 (Invitrogen). At 3 d after transfection, GFP+ cells were quantified by FACS analysis (CyAn ADP; Dako) and normalized to the mean value of parallel siCTRL-treated samples; results represent the mean of 12 independent transfections.
To examine HDR, the DR-GFP reporter (26) (hyg version) was transfected into SV40T-transformed fibroblasts from a RAD50-R1093X/X1313Y patient (28). After hygromycin selection (Invitrogen), cells were transfected with 0.4 μg of pCBASce and 0.2 μg of RAD50 expression vectors (lacking an HA-His tag) or an EV, and GFP+ cells were quantified by FACS and normalized to the EV samples. Results represent the mean of four independent transfections. For immunoblot analysis, protein was extracted at 2 d after transfection using NETN buffer as described previously (26), and were probed with RAD50 and actin antibodies (sc-74460; Santa Cruz Biotechnology and A2066; Sigma-Aldrich) and developed using secondary antibody (sc-2004; Santa Cruz Biotechnology) and ECL (GE Healthcare). Statistical analyses were performed using the unpaired t test.
Supplementary Material
Acknowledgments
We thank members of the T.T.P. and R.R. laboratories for helpful comments, Felicia Lopezcolorado for technical assistance, and Mauro Modesti (Centre National de la Recherche Scientifique Marseille, France) for reagents. This work was supported by National Institutes of Health/National Cancer Institute Grants CA094008 (to T.T.P.) and CA120954 (to J.M.S.), and National Institute of General Medical Sciences Grant GM-70456 (to R.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309816110/-/DCSupplemental.
References
- 1.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: The importance of communicating and holding things together. DNA Repair (Amst) 2004;3(8-9):845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85(4):509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 3.Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair (Amst) 2010;9(12):1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mimitou EP, Symington LS. DNA end resection: Many nucleases make light work. DNA Repair (Amst) 2009;8(9):983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26(56):7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 6.Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3(3):155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 7.Lisby M, Rothstein R. Localization of checkpoint and repair proteins in eukaryotes. Biochimie. 2005;87(7):579–589. doi: 10.1016/j.biochi.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Stracker TH, Petrini JH. The MRE11 complex: Starting from the ends. Nat Rev Mol Cell Biol. 2011;12(2):90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13(10):1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17(12):1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang SH, et al. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 2013;32(1):126–139. doi: 10.1038/emboj.2012.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308(5721):551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 13.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101(7):789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 14.Lim HS, Kim JS, Park YB, Gwon GH, Cho Y. Crystal structure of the Mre11-Rad50-ATPγS complex: Understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25(10):1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, et al. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J Biol Chem. 2013;288(18):12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J-H, et al. Regulation of Mre11/Rad50 by Nbs1: Effects on nucleotide-dependent DNA binding and association with ATLD mutant complexes. J Biol Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- 17.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118(6):699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9(6):683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 19.Wen J, Cerosaletti K, Schultz KJ, Wright JA, Concannon P. NBN phosphorylation regulates the accumulation of MRN and ATM at sites of DNA double-strand breaks. Oncogene. 2013;32(37):4448–4456. doi: 10.1038/onc.2012.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14(19):1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jager M, et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8(5):1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 22.Moncalian G, et al. The rad50 signature motif: Essential to ATP binding and biological function. J Mol Biol. 2004;335(4):937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 23.Shim EY, et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29(19):3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14(7):639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 25.Wu D, Topper LM, Wilson TE. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics. 2008;178(3):1237–1249. doi: 10.1534/genetics.107.083535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunn A, Bennardo N, Cheng A, Stark JM. Correct end use during end joining of multiple chromosomal double-strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J Biol Chem. 2011;286(49):42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakanishi K, et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol. 2011;18(4):500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waltes R, et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet. 2009;84(5):605–616. doi: 10.1016/j.ajhg.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Möckel C, Lammens K, Schele A, Hopfner KP. ATP-driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Res. 2012;40(2):914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lammens K, et al. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145(1):54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467(7311):108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467(7311):112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhaskara V, et al. Rad50 adenylate kinase activity regulates DNA tethering by Mre11/Rad50 complexes. Mol Cell. 2007;25(5):647–661. doi: 10.1016/j.molcel.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JH, Paull TT. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006;408:529–539. doi: 10.1016/S0076-6879(06)08033-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.