Significance
Cells have evolved mechanisms to silence foreign DNA to prevent the expression of foreign genes within them. In mammalian cells, this involves the assembly of heterochromatin on foreign DNAs such as viral or transfected DNA. Herpesviruses have evolved strategies to counteract these host mechanisms to express their own genes. Herein we demonstrate that the nuclear DNA sensor IFN-inducible protein 16 (IFI16) is involved in the host silencing response to foreign DNA. IFI16 promotes the assembly of heterochromatin on herpesviral DNA and inhibition of viral immediate–early gene expression and replication. This silencing effect is also observed on transfected plasmid DNAs. Therefore, our results indicate that IFI16 is broadly involved in the cellular response to foreign DNA.
Keywords: DNA sensing, intrinsic resistance, host restriction, cellular response
Abstract
Mammalian cells have evolved mechanisms to silence foreign DNA introduced by viruses or by transfection. Upon herpesviral infection of cells, the viral genome is chromatinized in an attempt by the host cell to restrict expression of the viral genome. HSV ICP0 acts to counter host-intrinsic and innate responses to viral infection. We have found that nuclear interferon (IFN)-inducible protein 16 (IFI16) acts as a restriction factor against ICP0-null herpes simplex virus 1 (HSV-1) to limit viral replication and immediate–early gene expression. IFI16 promoted the addition of heterochromatin marks and the reduction of euchromatin marks on viral chromatin. IFI16 also restricted the expression of plasmid DNAs introduced by transfection but did not restrict SV40 DNA introduced into the cellular nucleus in the form of nucleosomal chromatin by viral infection. These results argue that IFI16 restricts unchromatinized DNA when it enters the cell nucleus by promoting the loading of nucleosomes and the addition of heterochromatin marks. Furthermore, these results indicate that IFI16 provides a broad surveillance role against viral and transfected DNA by promoting restriction of gene expression from the exogenous DNA and inducing innate immune responses.
A common intrinsic cellular response to foreign DNA is the restriction of its expression. Whereas bacteria have evolved restriction–modification systems to recognize and degrade DNA that is foreign to them (1), mammalian cells attempt to silence foreign DNA introduced into them via transfection by assembling chromatin and heterochromatin on that DNA (2) and/or by ligating the introduced DNA molecules together and integrating them into cellular heterochromatin (3). Similarly, nucleosome-free viral genomic DNA, such as that associated with herpesvirus infection, is rapidly chromatinized by host cell mechanisms when it enters the host cell nucleus (4–6). However, although the mechanisms of recognition of foreign DNA are well defined for bacteria, less is understood about the sensing and restriction in mammalian cells.
DNA deposited in the nucleus by transfection or viral infection is sensed by mechanisms that lead to a number of cellular responses including rapid chromatinization, initiation of innate responses, the recruitment of repressive nuclear domain 10 (ND10) components, and the activation of DNA damage responses. In the context of viral infection, the constitutively expressed proteins that mediate these responses are collectively categorized as intrinsic resistance factors, the defining characteristic of which is their ability to act immediately to counteract infection. As such, these proteins are a first line of defense against virus infection, acting before innate immunity and the induction of interferons and proinflammatory cytokines.
Much of our understanding of responses to foreign DNA comes from studies of nuclear replicating DNA viruses, which have evolved strategies to counteract or subvert intrinsic resistance pathways to promote efficient viral gene expression. In particular, HSV-1 counteracts these responses in part through the expression of the infected cell protein 0 (ICP0) immediate–early (IE) protein. The importance of counteracting this intrinsic response is documented by reports that ICP0-null viruses are significantly attenuated for viral replication (7, 8), particularly in primary human fibroblasts (8, 9).
During HSV-1 infection, ND10 components accumulate de novo in the nucleus at sites near incoming viral DNA, and this is associated with their ability to restrict viral gene expression (10). However, depletion of ND10 by simultaneous knockdown of the three major ND10 components, promyelocytic leukemia protein (PML), Sp100, and hDaxx, does not completely rescue the replication of an ICP0-null virus (11), indicating that additional mechanisms are involved in the intrinsic resistance to HSV-1. Recently, it was reported that the IFN-inducible protein 16 (IFI16) DNA sensor restricts human cytomegalovirus (HCMV) replication in human fibroblasts by inhibiting viral early gene transcription (12). IFI16 is an AIM2-like receptor of innate sensing molecules and a member of the PYHIN family because it contains a pyrin domain, a member of the death domain superfamily of signaling modules, and two DNA-binding HIN-200 (hematopoietic IFN-inducible nuclear proteins with a 200-aa repeat) domains (13). Although IFI16 is known to promote IFN-regulatory factor 3 (IRF-3) (14–16) and inflammasome (17, 18) signaling in response to herpesvirus infection, the previously reported IFI16-dependent restriction of HCMV was independent of IFN-β, indicating IFI16 may act as an intrinsic resistance factor in addition to categorization as an innate pattern recognition receptor. Furthermore, the association of IFI16 with viral DNA in the infected cell nucleus (14) and our identification of IFI16 as a target of ICP0-mediated degradation (15) prompted us to evaluate IFI16’s activity as an intrinsic resistance factor to foreign nuclear DNA. Notably, in the present study, we demonstrate that IFI16 represses herpesviral and transfected DNA expression and is responsible for a portion of the defect in ICP0-null virus replication. Furthermore, we provide evidence that inhibition of IFI16 by depletion or ICP0-mediated degradation results in changes in viral chromatin structure, implicating the role of this protein in epigenetic regulation of HSV gene expression.
Results
Reduction of IFI16 Enhances ICP0-Null HSV Replication in Normal Human Foreskin Fibroblasts.
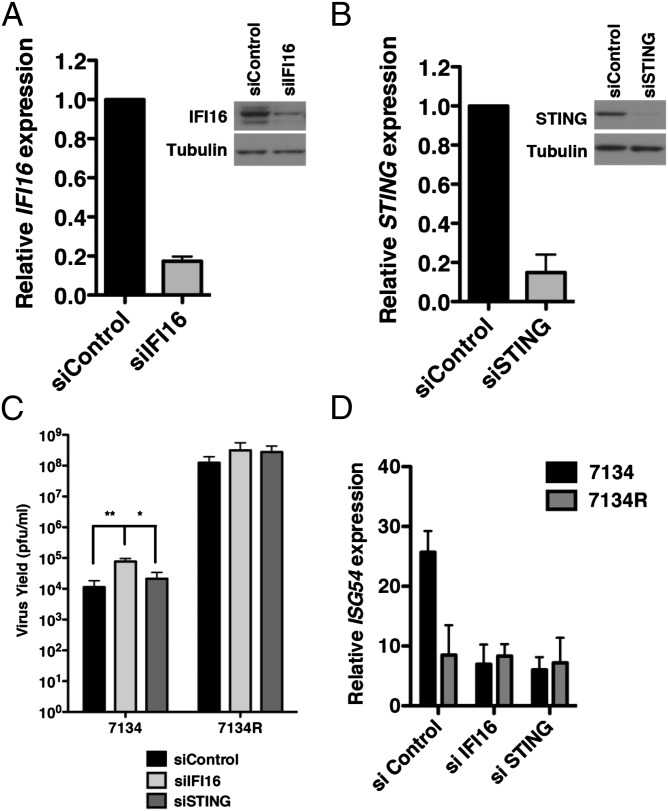
To determine whether IFI16 plays a role in the reduced replication of ICP0-null viruses, we treated normal human foreskin fibroblasts (HFFs) with a pool of siRNAs specific for IFI16 to reduce IFI16 expression or with a nontargeting control siRNA (siControl) pool. We observed a significant decrease in the expression of IFI16 at the mRNA and protein level at 72 h posttransfection (hpt; Fig. 1A), and knockdown was robust through 120 hpt. siRNA-treated cells were subsequently infected with an ICP0-null virus (7134) or its corresponding rescued virus (7134R) at a low multiplicity of infection (MOI; 0.1 pfu per cell), and virus yields were determined by plaque assay. At 48 h postinfection (hpi), we observed a ∼4-log defect in 7134 virus replication compared with 7134R in control-treated cells (Fig. 1C), consistent with previous studies (10). Interestingly, whereas we observed a minimal increase in 7134R virus yield in IFI16 siRNA-treated cells, replication of the 7134 virus was significantly increased (average of sevenfold) upon depletion of IFI16. These results suggested that IFI16 restricts HSV-1 replication in the absence of ICP0 and that the IFI16 protein likely accounts for a portion of the attenuated ICP0-null phenotype seen in human fibroblasts.
Fig. 1.
IFI16 negatively regulates the replication of an ICP0-null virus. (A) IFI16 and (B) STING transcript and protein levels were decreased following transfection of HFF with nontarget control, IFI16-specific, or STING-specific siRNAs. (C) IFI16 depletion resulted in an increase in ICP0-null virus replication relative to cells transfected with control siRNAs. siRNA-transfected cells were infected with HSV-1 ICP0-null (7134) or a rescued virus (7134R) at an MOI of 0.1 and harvested at 48 hpi, and virus yield was determined by plaque assay on U2OS cells. (D) IFI16 or STING depletion decreased IFN-β transcript levels in response to ICP0-null virus infection. siRNA-transfected cells were infected with HSV-1 ICP0-null virus at an MOI of 10, and RNA was harvested at 6 hpi. Results are an average of three (A and B), four (C), or two (D) independent experiments, and error bars represent the SE of means (*P < 0.05 and **P < 0.01, Student t test).
Fibroblasts infected with ICP0-null viruses show enhanced expression of type I interferons and IFN-stimulated genes (ISGs) compared with WT viruses (19) (Fig. 1D). The induction of these antiviral genes are, at least in part, a result of nuclear sensing of viral DNA by IFI16 (15). Therefore, it was possible that the increased replication of 7134 observed in our virus yield assay could be a result of the down-regulation of IFI16-induced antiviral gene expression, as observed by quantitative reverse transcription polymerase chain reaction (qRT-PCR; Fig. 1D). We therefore examined the involvement of these innate pathways in ICP0-null virus replication by examining virus yields in the absence of the stimulator of IFN genes (STING) adaptor. STING is an adaptor in intracellular DNA sensing, and decreased expression of STING greatly inhibits IRF-3 activation and type I IFN induction in response to HSV in different cell types (20, 21) and in our system (15). Depletion of STING (Fig. 1B), while inhibiting ISG54 expression (Fig. 1D), resulted in only 1.8- and 2.2-fold increases (Fig. 1C) in 7134 and 7134R virus replication, respectively, indicating that signaling events downstream of STING, including IRF-3 activation, are not involved in the intrinsic resistance to ICP0-null viruses. Together, these results argued that IFI16 inhibits viral replication independently of STING and its role in IRF-3 signaling.
IFI16 is also implicated in activation of inflammasome signaling in HSV-infected HFF cells (18). However, we did not observe a detectable increase in cleavage of IL-1β in HFF following infection with the 7134 virus (Fig. S1A). We also did not observe a difference in basal cleavage of IL-1β when IFI16 was depleted in these cells (Fig. S1A). These results indicated that the inflammasome is not likely involved in the observed restriction.
Depletion of IFI16 Enhances HSV-1 Immediate-Early Gene Expression.
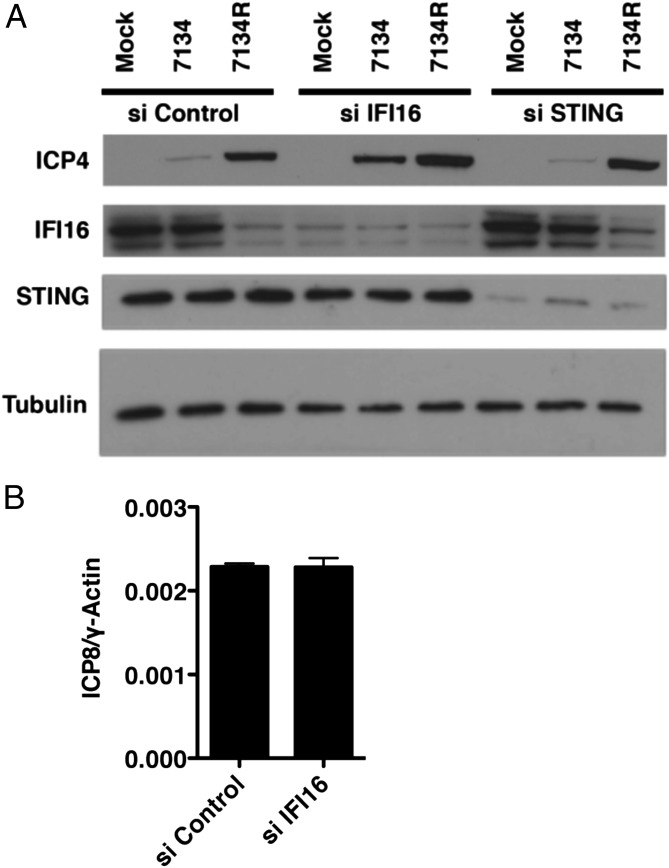
The previous experiments showed that IFI16 can restrict ICP0-null virus replication; however, it was unclear what stage in the viral lifecycle was inhibited by IFI16. To investigate the mechanism(s) of IFI16-mediated inhibition of viral replication, we examined the expression of the viral ICP4 essential IE transactivator during infection of siRNA-treated cells. HFFs were transfected with IFI16-specific, STING-specific, or nontarget control siRNAs, infected with 7134 or 7134R viruses, and, at 6 hpi, whole cell lysates were harvested. Western blot analysis revealed an increase in the expression of ICP4 in 7134-infected IFI16-depleted cells relative to siControl-treated cells (Fig. 2A), but not in STING-depleted cells, consistent with the increase in viral replication observed in Fig. 1C. Furthermore, we observed a minimal increase in ICP4 expression during infection with 7134R virus (Fig. 2A), consistent with ICP0 overcoming IFI16-mediated inhibition by promoting the proteasomal degradation of IFI16 (15). The IFI16-dependent decrease in gene expression was not caused by differences in the initial accumulation of viral DNA in the nucleus as we observed equal amounts of nuclear viral DNA during infection with d109 virus, which expresses no viral gene products (22), in the presence or absence of IFI16 (Fig. 2B). Together, these results argued that IFI16 inhibits HSV-1 replication early during infection at the stage of immediate–early gene expression.
Fig. 2.
Reducing IFI16 protein levels increases the expression of a viral immediate–early protein. (A) Immunoblot examining the levels of the HSV-1 ICP4 immediate–early protein in HFF cells treated with nontargeting control, IFI16, or STING siRNA. Treated cells were mock-infected or infected with an ICP0-null (7134) or rescued virus (7134R) at an MOI of 1. Lysates were prepared at 6 hpi and subjected to Western blot analysis. The cellular tubulin gene was used as a recovery and loading control. (B) IFI16-depleted cells were infected with HSV-1 d109 virus at an MOI of 1, and cells were fractionated at 2 hpi. Total DNA was prepared from the nuclear fraction and quantified. The viral ICP8 gene was normalized to the cellular γ-actin control.
Overexpression of IFI16 Reduces ICP0-Null Virus Gene Expression and Replication.
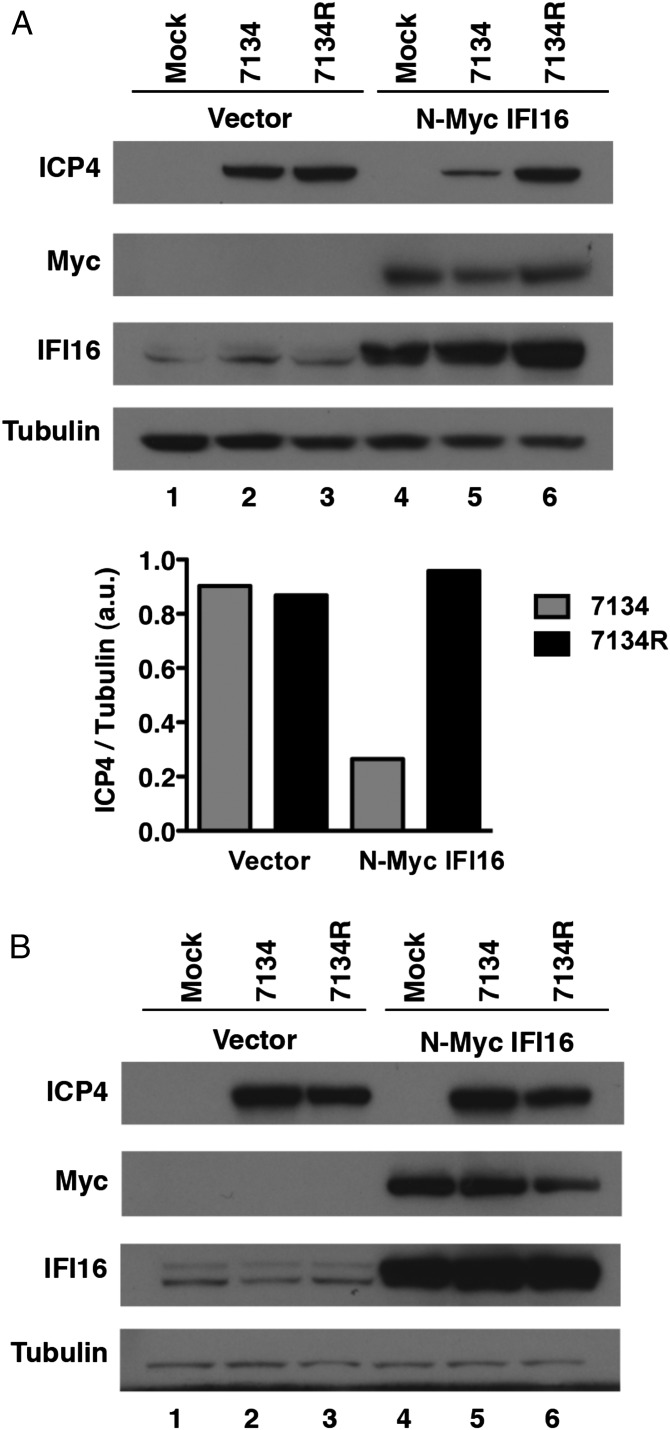
The experiments described here earlier showed that IFI16 is required for a cellular mechanism that decreases IE gene expression by an ICP0-null virus. To determine if IFI16 is sufficient for the silencing effect, we tested the effect of exogenous expression of IFI16 in cells that are defective for IFI16 signaling. Both U2OS cells and HEK293 cells show defects in IFI16-induced innate responses (see Fig. 4A) (14). Therefore, expression of exogenous IFI16 can be used in these cells to test its function. Furthermore, HSV-1 mutants deficient in functional ICP0 are grown in U2OS cells as a result of an intrinsic ability of these cells to complement ICP0-null virus infection. To test if exogenous IFI16 could repress viral gene expression in U2OS cells, we transfected the cells with an N-Myc IFI16-expressing plasmid followed by infection with 7134 and 7134R viruses at low and high MOI. At 6 hpi, we observed a 3.5-fold decrease in ICP4 protein levels in cells expressing N-Myc IFI16 and infected with the 7134 virus at an MOI of 0.1 (Fig. 3A), indicating that exogenous IFI16 can restrict viral gene expression under these conditions. At high MOI, 7134 showed no decrease in ICP4 protein levels, (Fig. 3B, lanes 2 vs. 5), consistent with the MOI-dependent phenotype of ICP0 mutant viruses. Interestingly, we observed no degradation of endogenous IFI16 in U2OS cells infected with the 7134R virus even during a high-MOI infection (i.e., MOI of 10; Fig. 3B, lanes 1 vs. 3), although some loss of exogenous N-Myc IFI16 was apparent (Fig. 3B, lanes 4 vs. 6). We speculate that U2OS cells express a defective IFI16 that cannot be activated for signaling and is not degraded by ICP0.
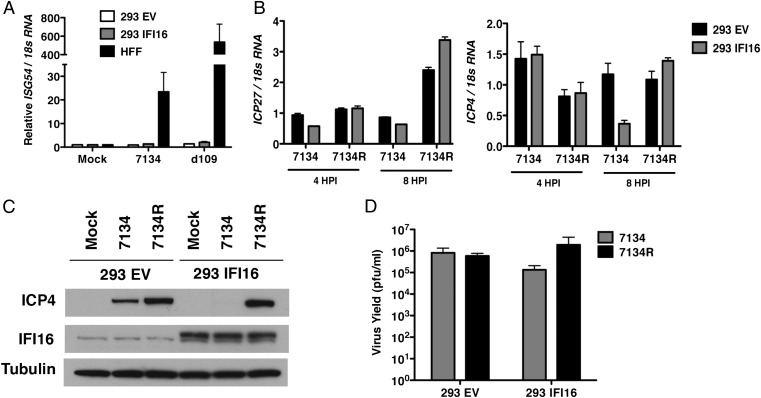
Fig. 4.
Stable expression of IFI16 in HEK293 cells inhibits viral gene expression and replication independently of IRF-3 activation. (A) Control HEK293 cells (293 EV), HEK293 cells stably expressing IFI16 (293 IFI16), or HFFs were infected with 7134 or d109 viruses. Total cell-associated RNA was harvested at 6 hpi and prepared for qRT-PCR. ISG54 transcripts were normalized to 18S RNA and made relative to mock-infected cells. (B) Total RNA was harvested at 4 or 8 hpi and analyzed by qRT-PCR for ICP4 and ICP27 mRNA. Transcripts were normalized to 18S RNA. (C) Cell lysates were harvested at 6 hpi and analyzed by Western blot for ICP4, IFI16, and Tubulin levels. (D) Virus yields at 24 hpi were determined by plaque assay on U2OS cells. Cells were infected with an MOI of 10 for A and an MOI of 0.1 for B–D.
Fig. 3.
Overexpression of IFI16 reduces HSV-1 gene expression and replication. U2OS cells transfected with an empty vector control or an N-terminal Myc-tagged IFI16 construct were infected with ICP0-null (7134) or rescued virus (7134R) at an MOI of 0.1 (A, Upper) or an MOI of 10 (B). Cell lysates were harvested at 6 hpi and analyzed for ICP4, IFI16, and Tubulin protein levels. (A, Lower) ICP4 and Tubulin band intensities from A (Upper) were quantified by ImageJ analysis and compared with a standard curve.
We next examined whether this decrease in gene expression corresponded to a similar decrease in virus replication. Attempts at producing a stable IFI16-expressing U2OS cell line have thus far been unsuccessful; therefore, we examined viral gene expression and replication in HEK293 cells stably expressing IFI16 (293 IFI6). Infection of these cells with 7134 virus resulted in a reduction in ICP4 and ICP27 mRNA and protein levels (Fig. 4 B and C) and virus yields (Fig. 4D) relative to vector control cells. Interestingly, overexpression of IFI16 in HEK293 cells did not result in a rescue of these cells to induce ISG54 expression in response to HSV infection (Fig. 4A). Together these results provided further evidence of IFI16’s activity as an intrinsic resistance factor and indicate that, even though expression of IFI16 is sufficient to restrict viral gene expression, it is not sufficient to induce an innate immune response in these cells, further arguing that these activities of IFI16 are separable. Furthermore, our results suggested that endogenous IFI16 in U2OS cells is nonfunctional and accounts for at least a portion of this cell line’s permissivity for ICP0-null virus replication.
Depletion of IFI16 Does Not Detectably Affect PML Recruitment to Viral Genomes.
During the cellular response to HSV-1 infection, components of ND10 bodies accumulate at sites adjacent to viral DNA (23, 24), which correlates with their involvement in the repression of viral replication (9, 25). This accumulation is observed during infection with ICP0-null viruses, as WT virus infection overcomes ND10-mediated repression by targeting components of these domains for degradation in an ICP0-dependent manner (26). IFI16 has not been identified as an ND10 component, nor does it localize to nuclear foci that would be indicative of this domain in normal HFFs (15, 27). However, given the involvement of IFI16 in the restriction of viral gene expression, we asked whether the protein is involved in the recruitment of ND10 components to viral genomes. To answer this question, we used a technique previously developed for analyzing ND10 accumulation at sites of viral DNA entry into the nucleus. This assay involves imaging cells along the edge of a viral plaque, where incoming viral genomes accumulate asymmetrically in the cell nucleus (10, 28, 29). HFF were treated with control siRNAs or IFI16 siRNAs to decrease IFI16 expression, infected with 7134 (MOI of 1) or 7134R (MOI of 0.001), and fixed at 24 hpi. Infection with 7134 at the higher MOI was necessary to observe plaque formation on HFF as ICP0-null viruses have as much as a 103-fold defect in plaque-forming efficiency on human fibroblasts (30). Viral genome complexes were visualized with an antibody specific for the viral ICP4 immediate–early protein, which has previously been shown to colocalize with viral DNA (10), and PML was used as a marker for ND10. Depletion of IFI16 did not detectably affect PML recruitment to ICP4 foci in 7134-infected cells (Fig. S2B vs. Fig. S2E), and PML was degraded in 7134R-infected cells irrespective of knockdown (Fig. S2C vs. Fig. S2F). Because there appeared to be no defect in PML recruitment in IFI16-depleted cells, the effect of IFI16 may be independent of PML effects.
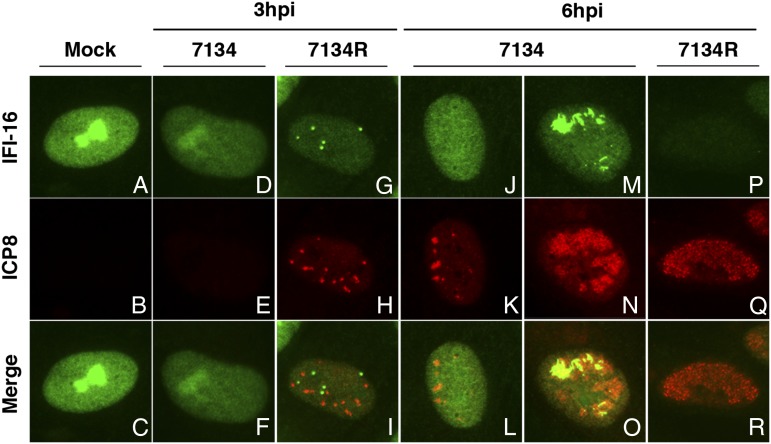
IFI16 Relocalizes to Sites of HSV-1 Viral DNA Synthesis.
In an earlier study, we did not observe a detectable intranuclear relocalization of IFI16 during infection with a replication-defective HSV-1 recombinant virus (15). However, others have reported that endogenous IFI16 colocalizes with viral DNA during infection with KSHV, a γ-herpesvirus, and IFI16 colocalizes with HSV-1 DNA when overexpressed in U2OS cells (14, 17), consistent with the DNA binding activity of this protein (16). It is possible that viral DNA synthesis enhances the relocalization of IFI16 as a result of an increased availability of the viral pathogen-associated molecular pattern (PAMP); therefore, we reinvestigated the localization of IFI16 during infection with replication-competent HSV-1. HFF cells were infected with the 7134 or 7134R viruses at an MOI of 10, fixed, and processed for immunofluorescence at 3 and 6 hpi. Viral replication compartments, sites of viral DNA synthesis (31), were visualized with an antibody specific for the viral ICP8 ssDNA binding protein. In mock-infected HFFs, IFI16 was nuclear and noticeably stained both the nucleoplasm and the nucleolus (Fig. 5A), as observed previously (14, 15, 26). In 7134R-infected cells, we observed the accumulation of IFI16 in nuclear foci and the subsequent loss of IFI16 (Fig. 5 I and R), consistent with our previous study describing the ICP0-dependent loss of IFI16 during HSV-1 infection (15). This pattern of relocalization was not observed in 7134-infected cells. Instead, we initially observed a decrease in nucleolar IFI16 at 3 hpi (Fig. 5D). This loss did not appear to be a result of degradation of the protein, as we did not observe a reduction in the steady-state levels of IFI16 by Western blot (Fig. 2A). Instead, this initial redistribution was most likely a consequence of the viral disruption of cellular nucleoli (32, 33). At 6 hpi, we observed two distinct populations of 7134-infected cells: (i) cells with small replication compartments and diffuse IFI16 staining (Fig. 5L) and (ii) cells with large replication compartments and notable IFI16 accumulation within those compartments (Fig. 5O). These results indicate that, in the absence of ICP0, IFI16 relocalizes to replication compartments possibly as viral DNA synthesis progresses.
Fig. 5.
IFI16 relocalizes to replication compartments during HSV-1 infection. HFF were infected with ICP0-null (7134) or rescued virus (7134R) at an MOI of 10 and fixed at 3 and 6 hpi. Cells were simultaneously stained with mouse anti-IFI16 (Abcam) and rabbit anti-ICP8 antibodies followed by Alexa Fluor 488-conjugated goat anti-mouse and Alexa Fluor 594-conjugated goat-anti rabbit secondary antibodies. Representative immunofluorescence images are shown. (A–C) Mock, (D–F, J–O) 7134, (G–I, P–R) 7134R.
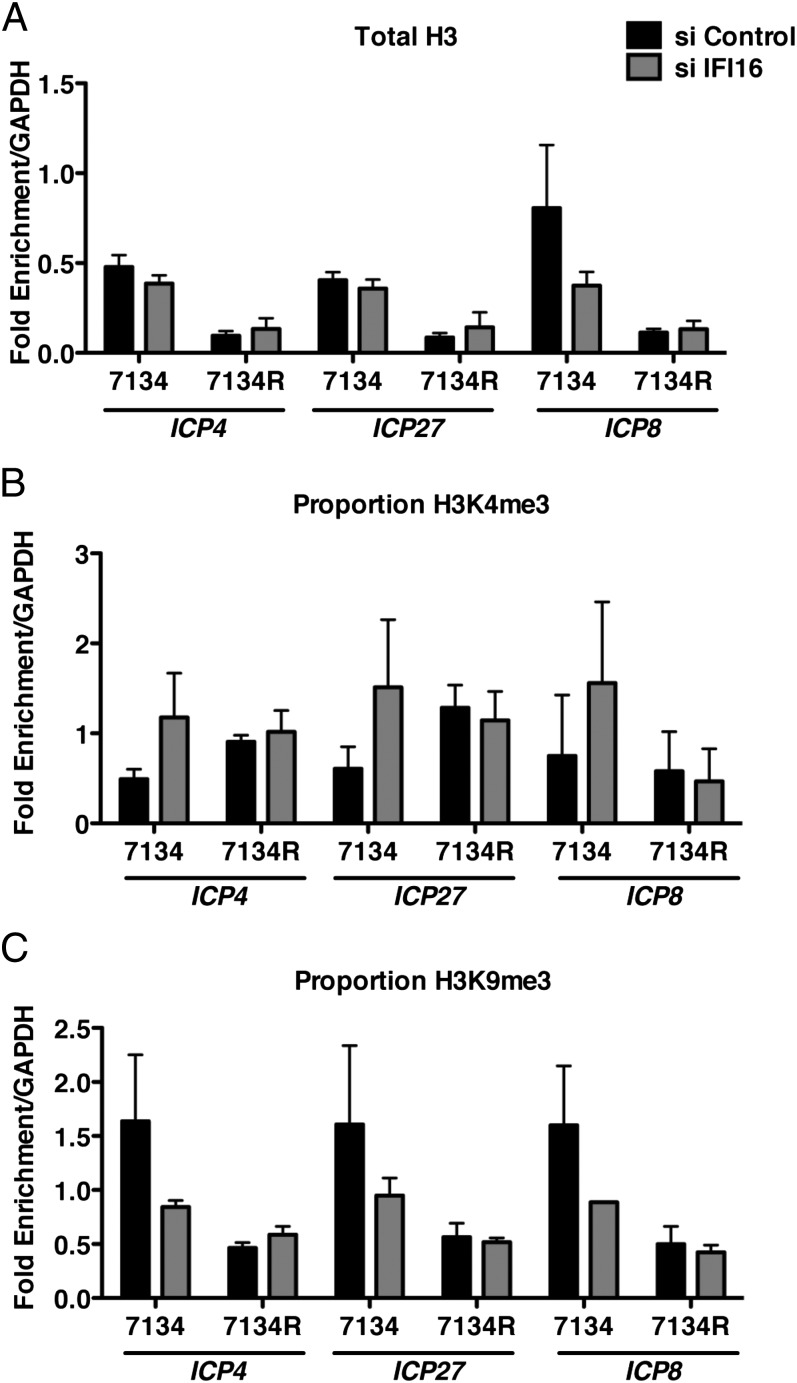
IFI16 Promotes Heterochromatin Association with HSV-1 DNA.
Gene expression is regulated by changes in chromatin structure through the addition of specific modifications to core histones. Modifications such as trimethylation of histone H3 lysine 4 (H3K4me3) are associated with actively transcribing genes, whereas trimethylation of histone H3 lysine 9 (H3K9me3) is a mark of heterochromatin. During HSV-1 infection, ICP0 plays a role in modulating the viral chromatin structure to promote viral gene expression (5, 34, 35), and it has been proposed that ICP0 may facilitate these changes by targeting host proteins involved in silencing of the viral genome (5, 6, 36). To determine whether IFI16 mediates the repressive activity described earlier through changes in HSV-1 chromatin structure, we used chromatin immunoprecipitation (ChIP) to examine histone association with HSV-1 DNA in IFI16-depleted cells. Lysates from formaldehyde-fixed 7134- and 7134R-infected cells were subjected to immunoprecipitation by using antibodies specific for histone H3, H3K4me3, or H3K9me3. The fraction of viral DNA immunoprecipitated was determined by quantitative PCR (qPCR) and normalized to the fraction of cellular GAPDH DNA immunoprecipitated. In control siRNA-treated cells, we observed a significant increase in histone H3 association with the viral ICP4, ICP27, and ICP8 promoters in the absence of ICP0 (Fig. 6A), consistent with our previous results (5). Furthermore, consistent with the reduction in IE gene expression observed in Fig. 2, 7134 IE (ICP4/ICP27) and early (E) (ICP8) gene promoters were associated with decreased H3K4me3 (Fig. 6B) and increased H3K9me3 (Fig. 6C) compared with 7134R promoters. Interestingly, in IFI16-depleted cells, we observed a 7134 virus-specific decrease in H3K9me3 on all promoters examined, as well as an increase in H3K4me3. Together these results argued that IFI16 restricts viral gene expression by promoting heterochromatin association with viral promoters.
Fig. 6.
IFI16 promotes heterochromatin association with viral DNA. Control and IFI16 siRNA-treated HFFs were infected with 7134 and 7134R viruses at an MOI of 1. Cell extracts were prepared at 6 hpi and ChIP was carried out by using antibody specific to (A) histone H3, (B) H3K4me3, (C) and H3K9me3. Immunoprecipitated ICP4, ICP27, and ICP8 promoter sequences were measured by qPCR and viral DNA sequences were normalized to immunoprecipitated GAPDH DNA. Histone marks (B and C) are represented as proportion of the total DNA immunoprecipitated by the H3 antibody.
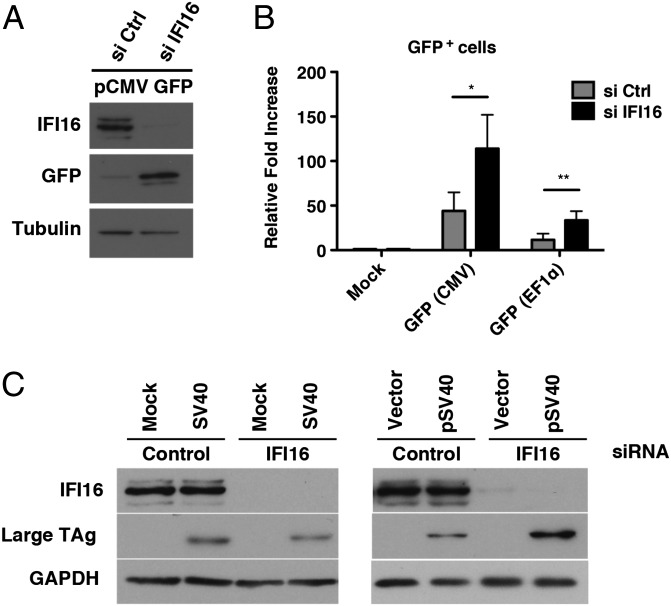
Depletion of IFI16 Enhances Plasmid and SV40 DNA Expression.
We next asked whether this repressive effect of IFI16 was specific to HSV-1 DNA by testing if IFI16 could restrict gene expression from transfected plasmid DNA or during additional DNA virus infections. HFFs were treated with IFI16 or control siRNAs and transfected with a plasmid containing a GFP expression construct under the control of a human CMV promoter (pCMV GFP) at 48 h after siRNA treatment. Whole-cell lysates were harvested at 24 hpt, and GFP protein levels were examined by western blotting. In IFI16-depleted cells, we observed an increase in GFP expression compared with control cells (Fig. 7A). This increase was quantified by measuring GFP-expressing cells by flow cytometry, and we observed a threefold increase in GFP+ cells when IFI16 was depleted (Fig. 7B). To determine whether this response was specific to the viral CMV promoter used to express the GFP plasmid described earlier, we also examined GFP expression from a plasmid under the control of the cellular elongation factor 1 (EF1) promoter (pEF1 GFP). Although the efficiency of expression was lower in control cells transfected with pEF1 GFP compared with pCMV GFP-transfected cells, we observed a similar increase in GFP+ cells upon depletion of IFI16 (Fig. 7B). Together these results suggested that IFI16 can inhibit transfected DNA expression in a promoter-independent manner.
Fig. 7.
Effect of IFI16 on transfected and SV40 DNA. (A) Immunoblot examining GFP and IFI16 expression in pCMV GFP-transfected HFFs treated with nontargeting control or IFI16 siRNA. The cellular tubulin gene was used as a recovery and loading control. (B) Quantification of GFP+ cells in the presence or absence of IFI16 by flow cytometry at 48 hpt. HFFs were transfected with an empty vector plasmid or pCMV GFP or pEF1 GFP at 48 h after siRNA treatment. The results are represented as the fold increase in GFP+ cells compared with their empty vector control. (C) siRNA-transfected HFF were infected with WT SV40 (MOI of 0.1) or transfected with pSV40 (0.5 μg). Cell lysates were prepared at 48 h posttreatment and analyzed by Western blot for TAg protein levels. IFI16 depletion was confirmed by Western analysis, and the cellular GAPDH gene was used as a recovery and loading control (*P < 0.05 and **P < 0.01, Student t test).
To test whether additional DNA viruses might be affected by IFI16, we examined the expression of large T antigen (TAg) during simian virus 40 (SV40) infection or GFP expression from an adenovirus vector (Ad5-GFP) in the presence or absence of IFI16. At 48 hpi, we observed no difference in the accumulation of TAg in IFI16-depleted cells infected with SV40 (MOI of 0.1) compared with control cells (Fig. 7C). Similarly, we observed no increase in GFP+ cells during Ad5-GFP infection (Fig. S3). However, when cells were transfected with a plasmid encoding the WT SV40 genome (pSV40), we observed an increase in the accumulation of TAg in the absence of IFI16 (Fig. 7C), indicating that IFI16 was capable of restricting SV40 gene expression from DNA introduced as a plasmid. The apparent lack of an effect on virion-delivered DNA suggested that the genomes within SV40 and adenovirus virions are resistant to the IFI16 effect. These results suggested that IFI16 may target exogenous DNA not associated with nucleosomes and that the SV40 genome in the virion, which contains nucleosomes, may be resistant to the restriction effect of IFI16. Similarly, the adenoviral genome in the virion is associated with core protein VII (37), which may prevent IFI16 binding.
Discussion
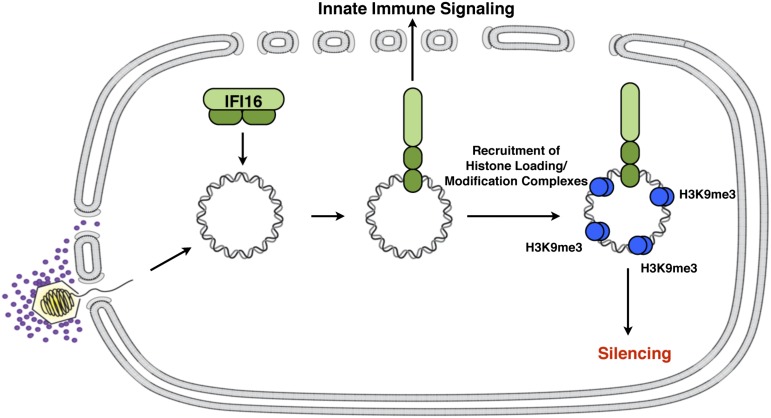
Mammalian cells load chromatin with heterochromatin marks on transfected and viral DNA when these DNAs enter the host cell nucleus, and expression of these exogenous DNAs are silenced by these mechanisms. Viruses have evolved viral gene products that counter these mechanisms. For example, the HSV ICP0 protein counters intrinsic and innate mechanisms (25, 38). In addition, the HCMV IE1 and IE2 gene products counter the cellular epigenetic silencing mechanisms (39). In this study, we have shown that the nuclear IFI16 protein promotes the restriction of HSV-1 IE gene expression in ICP0-null mutant-infected cells and promotes the restriction of transfected plasmid gene expression. This observation is different from the restriction effect previously reported for human CMV, whereby early and late, but not immediate–early, gene expression was inhibited by IFI16 (12). We found that the SV40 genome, when introduced by viral infection, is not restricted by IFI16, suggesting that nucleosomal DNA is not affected by this restriction mechanism. Our results argue for the model shown in Fig. 8 in which nuclear IFI16 in HFFs binds to nonnucleosomal DNA when introduced into the nucleus by transfection or herpesviral infection. IFI16 then undergoes a conformational change upon DNA binding (40) that allows the recruitment of histone modification enzymes that promote heterochromatic modifications on the exogenous DNA, compaction of the chromatin, and silencing of viral IE gene promoters.
Fig. 8.
Model for IFI16 restriction of HSV gene expression. IFI16 binds to nucleosome-free DNA that accumulates in the nucleus. DNA-bound IFI16 undergoes a conformational change releasing the pyrin domain (light green) from an autoinhibited state. Activated IFI16 can signal from the nucleus to the cytoplasm to activate innate immune signaling pathways and recruit chromatin modification complexes that promote H3K9me3 on viral genomes, resulting in gene silencing.
Specificity of IFI16 Restriction.
The ability of IFI16 to restrict various DNA molecules suggests that IFI16 recognizes foreign DNA in a sequence-independent manner. This hypothesis is consistent with the recently described crystal structure of the IFI16 and related AIM2 HIN DNA-binding domains in which the protein makes contacts with sugar-phosphate backbone of DNA (40). DNA binding is then predicted to release the pyrin signaling domain from its intramolecular complex with the HIN domain (40). DNA binding studies with IFI16 have shown a preference for supercoiled and cruciform DNA over relaxed or linear DNA (41), but the basis for discrimination between viral and host DNA remains to be explained. Our observation of the restriction of SV40 DNA introduced by transfection but not by viral infection argues that the target of IFI16 restriction is unchromatinized DNA in that SV40 DNA in the virion is assembled in nucleosomes (42) while transfected SV40 DNA acquires chromatin and becomes supercoiled over 72 h (43). We therefore hypothesize that the mechanism by which IFI16 distinguishes “foreign” DNA from “self” DNA is that self DNA is tightly wrapped in nucleosomes and is not recognized by IFI16 whereas transfected or herpesviral DNA is underchromatinized and IFI16 can bind to these forms of DNA.
The packaging of nucleosome-bound viral DNA by SV40, in addition to promoting transcription, also provides an evasion strategy adopted by the virus, and it is likely that other DNA viruses possess similar mechanisms to shield their genomic DNA from IFI16 detection. Interestingly, in this study, we observed no IFI16-dependent restriction of adenovirus gene expression, which, like herpesviruses, is nucleosome-free upon entry into the nucleus. However, incoming adenovirus DNA is associated with core protein VII, which protects the viral DNA from nuclear DNA damage responses (44). It is therefore possible that, like nucleosomes on SV40 DNA, core protein VII may also protect adenovirus DNA from an IFI16-dependent response during infection.
IFI16 Effector Mechanisms.
We hypothesize that, when IFI16 has bound to these exogenous DNA molecules, as shown by Li et al. (14), it undergoes a conformational change, freeing the pyrin domain and/or other domains to interact with another pyrin domain-containing protein and/or other protein complexes. Our evidence that IFI16 promotes histone H3K9 trimethylation modifications on HSV chromatin while reducing histone H3K4 trimethylation argues that IFI16 recruits histone modification enzymes that add this heterochromatin mark, leading to silencing of the exogenous DNA. Thus, IFI16 is likely part of the chromatinization response to HSV DNA (4, 5, 34). Proteomic studies are under way to identify proteins that associate with IFI16 upon HSV infection.
ND10 components localize near genomic complexes, and this localization is linked to restriction of viral replication by the combined action of ND10 components hDaxx, PML, and Sp100 (11). However, depletion of these three cellular factors does not fully restore ICP0-null mutant virus replication, suggesting that other factors are operative in human cells. Our results raise the possibility that IFI16 is at least part of this additional restriction effect. The relationship of these restriction factors to IFI16 is not well defined, but we observed that PML still localizes to sites near HSV genome complexes in most cells when IFI16 is depleted, so IFI16 appears to have no role in recruiting this protein to viral genome complexes. Further studies are needed to determine if IFI16 and ND10 restriction are fully independent mechanisms of HSV restriction. The mechanism(s) of ND10 restriction of viral replication have not been defined; thus, the effects of these factors on viral chromatin are not known. This work on IFI16 provides a mechanism for its restrictive effect, as well as defining a component of the cellular chromatinization response.
Implications.
Our results may have practical implications for enhancing expression of certain types of vectored DNAs in normal cells. It is well documented that exogenous plasmid DNA can be silenced upon transfection, and that this has been a significant drawback to the use of nonviral vectors in gene therapy (45, 46). Thus, reduction of IFI16 levels or activity may enhance expression of certain types of vectored DNAs in normal cells. We have shown that expression of herpesviral genomes and transfected DNAs is restricted by IFI16, and it will be interesting to test additional vectors, including parvovirus vectors, lentiviral, foamy viral, and other retroviral vectors. IFI16 may also affect integration of vectors as well as expression; thus, there are a number of potential scenarios in which manipulation of IFI16 may increase the efficacy of gene therapy vectors.
The restriction of exogenous gene expression by IFI16 may also explain in part the different properties of different cell lines with regard to permissivity for infection by DNA viruses. The extent to which ICP0-null HSV strains are restricted for replication is cell type-dependent. Human fibroblasts are among the most restrictive cells for ICP0-null virus replication, whereas ICP0 is dispensable for growth in U2OS cells. The cellular mechanisms that impart these differences have not been identified. Hancock et al. (47) reported that heterokaryon formation between U2OS cells and human fibroblasts imparted a dominant restrictive phenotype on U2OS cell nuclei, suggesting that U2OS cells lack a restrictive factor(s) that is functional in fibroblasts (47). Others have examined the levels of known intrinsic resistance factors in U2OS cells (48), and it has been noted that these cells do not express ATRX, a component of ND10; however, the addition of this protein failed to restrict HSV-1 growth (49). Surprisingly, U2OS cells expressed detectable IFI16 protein, which was inconsistent with the permissivity of these cells to ICP0-null virus infection. The ability of HSV-1 to promote the degradation of exogenous, but not endogenous, IFI16 indicates the endogenous protein might be dysfunctional in tumor cell lines and is consistent with reports that transformed cells lack a functional DNA sensing mechanism (50). However, the downstream factors involved in restricting viral gene expression appear to be functional, as exogenous IFI16 was able to restrict HSV-1 gene expression. We therefore speculate that the permissivity of cells to ICP0-null virus infection may, in part, rely on the functionality of IFI16 rather than whether the protein is expressed in a given cell type. In addition, these results provide a basis for the further investigation of IFI16 activity in the absence of other restrictive phenotypes.
Differences in the ability of different cell lines to express transfected DNA may also be explained at least in part by differences in IFI16. Previous studies have shown that tumor cells have lost the ability to sense immunostimulatory DNA (51), and this often includes loss of IFI16 expression or functionality (as shown in the present study). Furthermore, cell lines, such as 293T cells, that do not express IFI16 (52), or cell lines, such as PC3 cells, that have mutations that prevent the nuclear localization of IFI16 (53), demonstrate enhanced mRNA expression of transfected DNA compared with other cell lines (54). Our results show the possible explanation for this difference in the behavior of these cell lines. In addition, infection with an ICP0-null HSV-1 mutant could provide an assay for the functionality of IFI16 for repression in any cancer or normal cell line.
Potential Normal Cellular Role for the IFI16 Restriction Function.
In addition to its role as a DNA sensor for intrinsic resistance and innate responses to certain forms of foreign DNA, IFI16 has been postulated to have a role in cell growth control and senescence. As discussed earlier, IFI16 is expressed at low levels or is not functional in several transformed or tumor cell lines. Human prostate cancer lines either did not express IFI16 or expressed a variant that was primarily in the cytoplasm, but reintroduction of IFI16 in these cancer cells inhibited colony formation (53). Cancer cells frequently have mutated epigenetic mechanisms and this may involve defects in histone loading, leading to underchromatinized DNA. The ability of IFI16 to recognize underchromatinized DNA may lead to IFN expression, which inhibits cell growth (55). In contrast, senescent cells show elevated levels of IFI16 expression (53). Furthermore, senescent cells show reduced protein synthesis, which leads to decreased levels of histones (56). This may lead to underchromatinized cellular DNA, which would lead to IFI16 binding, activation of IFN and subsequently inhibition of cell growth. Therefore, IFI16 may provide a general chromatin surveillance mechanism for detecting underchromatinized viral and cellular DNA.
Materials and Methods
Cell Culture and Viruses.
HFFs, U2OS, and HEK293 cells were obtained from the American Type Culture Collection. HFFs were cultured as described previously (15). U2OS and HEK293 cells were grown in DMEM supplemented with 5% (vol/vol) heat-inactivated fetal bovine serum (FBS) and 5% (vol/vol) heat-inactivated bovine calf serum (BCS).
To generate HEK293 cells stably expressing FLAG-HA-IFI16, an IFI16 isoform B ORF clone was obtained from the human ORFeome collection and recombined into the Gateway destination vector MSCV-N-Flag-HA-IRES-PURO with LTR-driven expression. A retrovirus carrying the FLAG-HA-IFI16 plasmid was produced by cotransfecting the FLAG-HA-IFI16, GAG/POL, and VSVG plasmids into HEK293 cells (57). The resulting virus was used to infect HEK293 cells and clonal populations were selected by supplementing the growth medium with 1 μg/mL puromycin.
The ICP0-null (7134) and rescued 7134R viruses were grown and titrated in parallel on U2OS cells (58). The d109 virus was propagated and titrated on FO6 cells (22). WT SV40 virus was prepared as described previously (59). Ad5-GFP was from the Gene Transfer Vector Core (University of Iowa).
Virus Infections.
HSV and SV40 were diluted in PBS solution containing 0.1% glucose and 1% heat-inactivated BCS. Cells were infected at the stated MOI for 1 h at 37 °C, washed twice with PBS solution, and overlaid with DMEM containing 1% heat-inactivated BCS. Infected cells were incubated at 37 °C for the indicated length of time.
Ad5GFP stocks were diluted in DMEM containing 1% heat-inactivated BCS (DMEV), and cells were infected for 6 h at 37 °C. Virus inoculum was replaced with fresh DMEV and incubated at 37 °C for 24 h.
siRNA Transfections.
Double-stranded IFI16-specific, STING-specific, and nontarget control siRNAs were purchased from Dharmacon. The pooled siRNAs were transfected into HFF using the DarmaFECT 2 transfection reagent (Dharmacon) at a final siRNA concentration of 50 nM according to the manufacturer’s instructions. The siRNA containing media was replaced at 24 hpt, and cells were assayed for IFI16 or STING levels by qRT-PCR and immunoblot at 72 hpt.
Plasmids and DNA Transfection.
For construction of the N-Myc IFI16 plasmid the IFI16 isoform B ORF was recombined into the Gateway destination vector pDEST-Myc using LR recombinase (Invitrogen). U2OS cells were plated at a density to ensure 50% confluence on the day of transfection. Cells were transfected with 0.5 μg of empty vector or N-Myc IFI16 plasmid by using the Effectene transfection reagent (Qiagen) and were infected with the indicated viruses at 48 hpt.
HFFs were transfected with 0.5 μg of an empty vector plasmid, pEGFP-C1 (Clontech), pEF1-GFP (provided by C. Cepko, Harvard Medical School, Boston), or SV40 plasmid DNA (60) by using the Lipofectamine LTX reagent (Invitrogen) at 48 h after siRNA treatment. Transfection media was replaced at 6 hpt with 10% DMEM, and whole-cell lysates were harvested and processed for flow cytometry or Western blot at 48 hpt.
Cellular RNA Analysis by qPCR.
Total RNA was extracted using the Qiagen RNeasy Kit and DNase treated using the DNA-free kit (Ambion). Equal amounts of DNase-treated RNAs were reverse-transcribed and quantified by real-time qPCR by using the Power SYBR Green PCR master mix and a Prism 7300 sequence detection system (Applied Biosystems). qPCR reactions were carried out in duplicate, and relative copy numbers were determined by comparison with standard curves. Mock reverse-transcribed samples were included as negative controls. Transcript levels were normalized to 18S rRNA and made relative to mock-infected samples. Experiments were conducted three times, and the values were averaged. A list of primer sequences used is provided in Table S1.
Nuclear DNA Analysis.
Nuclei from d109-infected cells were isolated by using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific), and DNA was harvested from these nuclei by using a Qiagen Generation Capture Column Kit. Viral DNA levels were determined by quantitative real-time PCR using the Power SYBR Green PCR master mix and a Prism 7300 sequence detection system (Applied Biosystems). PCR reactions were carried out in duplicate, and relative copy numbers were determined by comparison with standard curves. Viral DNA was normalized to cellular γ-actin levels. A list of primer sequences used is provided in Table S1.
Western Blots.
Cells were lysed in NuPAGE LDS Sample Buffer, and proteins were resolved on NuPAGE 4% to 12% Bis Tris gels (Invitrogen). Proteins were transferred overnight to PVDF membranes and blocked with 5% milk in PBS solution. Membranes were probed with primary antibody at 4 °C, washed with PBS solution containing 0.05% Tween 20, and incubated in secondary antibody for 1 h at room temperature. Western blots were developed using Luminate Forte Western HRP substrate (Millipore). A list of antibodies and their dilutions is provided in SI Materials and Methods.
Indirect Immunofluorescence.
HSV-1–infected HFFs grown on coverslips were fixed with 2% formaldehyde, permeabilized with 0.5% Nonidet P-40, and blocked in 5% normal goat serum. Fixed cells were incubated with antibodies for 30 min at 37 °C and washed two times with PBS solution containing 0.05% Tween 20 followed by one wash with PBS solution. Alexa Fluor 488- and 594-conjugated secondary antibodies were incubated with cells for 2 h at 25 °C. The coverslips were washed as described earlier and mounted in ProLong Gold antifade reagent (Invitrogen). Images were acquired by using an Axioplan 2 microscope (Zeiss) with a 63× objective and Hamamatsu CCD camera (model C4742-95). Images were arranged in figures by using Adobe Photoshop CS4 (Adobe Systems). A list of antibodies and their dilutions is provided in SI Materials and Methods.
Flow Cytometry.
Transfected or infected HFF were trypsinized, pelleted, and resuspended in 500 μL Accumax cell counting solution (Millipore). Cell suspensions were passed through a 40-μm filter to prevent clumping and stained with a 1:500 dilution of propidium iodide (PI). Fluorescence readings were collected for 20,000 cells. PI-positive cells were gated out during analysis, and GFP+ cells were defined on empty vector-transfected or mock-infected cells. Data analysis was performed using FlowJo (version 9) software, and graphs were constructed by using GraphPad Prism software.
ChIP.
HFFs (5.5 × 105) were plated in 60-mm dishes and transfected with siRNA as described earlier. Cells were infected at 72 hpt and fixed with 1% formaldehyde for 15 min. The formaldehyde was quenched by the addition of cold glycine at final concentration of 125 mM for 3 min. Cells were washed twice with PBS solution and scraped into PBS solution supplemented with Complete Protease Inhibitor tablets (Roche Diagnostics). Cells were resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) containing PMSF and incubated on ice for 30 min. Lysates were sonicated in 30 s pulses (Biorupter; Diagnode) for a total of 25 min to produce DNA fragments ∼500 bp in length. Samples were clarified by microcentrifugation (5415D, eppendorf) at 14,000 rpm for 10 min. Equal amounts of chromatin (15 μg per antibody) were diluted 10-fold in ChiP dilution buffer (150 mM NaCL, 10 mM Na2PO4, 2 mM EDTA, 1.1% Triton, 0.1% SDS, protease inhibitor tablet), and 1% of the diluted sample was removed for input calculation. Immunocomplexes were immunoprecipitated overnight at 4 °C with 2.5 μg of anti-histone H3 IgG (Abcam), anti-histone H3K9me3 (Abcam), or anti-histone H3K4me3 (Abcam).
Antibody was captured by addition of 20 μL of Magna ChIP protein A magnetic beads (Millipore) for 1 h at 4 °C with rotation. Beads were washed three times with ChIP dilution buffer, three times with LiCl wash buffer (50 mM Hepes, 250 mM LiCl, 1 mM EDTA, 1.0% Nonidet P-40, 0.7% sodium deoxycholate, 1 mM PMSF), and two times with 1× Tris EDTA buffer (10 mM Tris⋅HCl, pH 8.1, 1mM EDTA). The DNA–protein complexes were eluted from antibody by the addition of 200 μL 65 °C elution buffer (1.0% SDS, 100 mM NaHCO3) with rotation for 10 min at room temperature, followed by incubation at 65 °C for 10 min. Immunoprecipitate and input samples were reverse cross-linked overnight at 65 °C by the addition of NaCl to a final concentration of 200 mM and 1 μg RNase A (Ambion). The samples were then treated with proteinase K for 1 h, and DNA was purified by using a QIAquick PCR purification kit (Qiagen). A list of primers is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Connie Cepko (Harvard Medical School) for providing the pEF1α-GFP construct. This research was supported by National Institutes of Health (NIH) Grants AI063106, AI099081 (to D.M.K.), and F31CA177274 (to C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316194110/-/DCSupplemental.
References
- 1.Youell J, Firman K. Mechanistic insight into Type I restriction endonucleases. Front Biosci (Landmark Ed) 2012;17:2122–2139. doi: 10.2741/4041. [DOI] [PubMed] [Google Scholar]
- 2.Cereghini S, Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 1984;3(6):1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wigler M, et al. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 4.Oh J, Fraser NW. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J Virol. 2008;82(7):3530–3537. doi: 10.1128/JVI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cliffe AR, Knipe DM. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J Virol. 2008;82(24):12030–12038. doi: 10.1128/JVI.01575-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knipe DM, Cliffe AR. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6(3):211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 7.Sacks WR, Schaffer PA. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61(3):829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stow ND, Stow EC. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67(pt 12):2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 9.Everett RD, Boutell C, Orr A. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J Virol. 2004;78(4):1763–1774. doi: 10.1128/JVI.78.4.1763-1774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett RD, Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol. 2005;79(8):5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass M, Everett RD. Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J Virol. 2013;87(4):2174–2185. doi: 10.1128/JVI.02950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gariano GR, et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog. 2012;8(1):e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: More than interferon-inducible genes? Exp Cell Res. 2005;308(1):1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 14.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci USA. 2012;109(26):10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA. 2012;109(44):E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013;87(9):5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paladino P, Collins SE, Mossman KL. Cellular localization of the herpes simplex virus ICP0 protein dictates its ability to block IRF3-mediated innate immune responses. PLoS ONE. 2010;5(4):e10428. doi: 10.1371/journal.pone.0010428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samaniego LA, Neiderhiser L, DeLuca NA. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72(4):3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217(1):67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 24.Uprichard SL, Knipe DM. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229(1):113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 25.Boutell C, et al. Herpes simplex virus type 1 ICP0 phosphorylation mutants impair the E3 ubiquitin ligase activity of ICP0 in a cell type-dependent manner. J Virol. 2008;82(21):10647–10656. doi: 10.1128/JVI.01063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2013;94(pt 3):465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 27.Cristea IM, et al. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84(15):7803–7814. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva L, Cliffe A, Chang L, Knipe DM. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 2008;4(5):e1000071. doi: 10.1371/journal.ppat.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taylor TJ (2002) Intranuclear localization of the herpes simplex virus ICP8 protein. PhD dissertation (Harvard Univ, Cambridge, MA)
- 30.Everett RD, Young DF, Randall RE, Orr A. STAT-1- and IRF-3-dependent pathways are not essential for repression of ICP0-null mutant herpes simplex virus type 1 in human fibroblasts. J Virol. 2008;82(17):8871–8881. doi: 10.1128/JVI.00613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Bruyn Kops A, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55(5):857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 32.Callé A, et al. Nucleolin is required for an efficient herpes simplex virus type 1 infection. J Virol. 2008;82(10):4762–4773. doi: 10.1128/JVI.00077-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greco A, et al. Nucleolin interacts with US11 protein of herpes simplex virus 1 and is involved in its trafficking. J Virol. 2012;86(3):1449–1457. doi: 10.1128/JVI.06194-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferenczy MW, DeLuca NA. Epigenetic modulation of gene expression from quiescent herpes simplex virus genomes. J Virol. 2009;83(17):8514–8524. doi: 10.1128/JVI.00785-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferenczy MW, DeLuca NA. Reversal of heterochromatic silencing of quiescent herpes simplex virus type 1 by ICP0. J Virol. 2011;85(7):3424–3435. doi: 10.1128/JVI.02263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104(43):17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vayda ME, Rogers AE, Flint SJ. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983;11(2):441–460. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roizman B, Knipe DM, Whitley RJ. Herpes Simplex Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6th Ed. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1823–1897. [Google Scholar]
- 39.Paulus C, Nevels M. The human cytomegalovirus major immediate-early proteins as antagonists of intrinsic and innate antiviral host responses. Viruses. 2009;1(3):760–779. doi: 10.3390/v1030760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brázda V, Coufal J, Liao JC, Arrowsmith CH. Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochem Biophys Res Commun. 2012;422(4):716–720. doi: 10.1016/j.bbrc.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 42.Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci USA. 1975;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen CK, Hu WS. DNA supercoiling of recombinant plasmids in mammalian cells. Proc Natl Acad Sci USA. 1986;83(6):1641–1645. doi: 10.1073/pnas.83.6.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karen KA, Hearing P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J Virol. 2011;85(9):4135–4142. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Dosari MS, Gao X. Nonviral gene delivery: Principle, limitations, and recent progress. AAPS J. 2009;11(4):671–681. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson DA, Juranek S, Lipps HJ. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Mol Ther. 2006;14(5):613–626. doi: 10.1016/j.ymthe.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 47.Hancock MH, Corcoran JA, Smiley JR. Herpes simplex virus regulatory proteins VP16 and ICP0 counteract an innate intranuclear barrier to viral gene expression. Virology. 2006;352(1):237–252. doi: 10.1016/j.virol.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 48.Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol. 2010;84(8):4026–4040. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McFarlane S, Preston CM. Human cytomegalovirus immediate early gene expression in the osteosarcoma line U2OS is repressed by the cell protein ATRX. Virus Res. 2011;157(1):47–53. doi: 10.1016/j.virusres.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 50.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Kwak JC, Ongusaha PP, Ouchi T, Lee SW. IFI16 as a negative regulator in the regulation of p53 and p21(Waf1) J Biol Chem. 2003;278(42):40899–40904. doi: 10.1074/jbc.M308012200. [DOI] [PubMed] [Google Scholar]
- 53.Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22(31):4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 54.Karyala P, Namsa ND, Chilakalapudi DR. Translational up-regulation and high-level protein expression from plasmid vectors by mTOR activation via different pathways in PC3 and 293T cells. PLoS ONE. 2010;5(12):e14408. doi: 10.1371/journal.pone.0014408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutterman JU. Cytokine therapeutics: Lessons from interferon alpha. Proc Natl Acad Sci USA. 1994;91(4):1198–1205. doi: 10.1073/pnas.91.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feser J, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39(5):724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai WZ, Schaffer PA. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol. 1989;63(11):4579–4589. doi: 10.1128/jvi.63.11.4579-4589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poulin DL, DeCaprio JA. The carboxyl-terminal domain of large T antigen rescues SV40 host range activity in trans independent of acetylation. Virology. 2006;349(1):212–221. doi: 10.1016/j.virol.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 60.Fine DA, et al. Identification of FAM111A as an SV40 host range restriction and adenovirus helper factor. PLoS Pathog. 2012;8(10):e1002949. doi: 10.1371/journal.ppat.1002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.