Significance
Using the recently introduced Cas9/sgRNA technique, we have developed a method for specifically targeting Drosophila germ-line cells to generate heritable mutant alleles. We have established transgenic lines that stably express Cas9 in the germ line and compared different promoters and scaffolds of sgRNA in terms of their efficiency of mutagenesis. An overall mutagenesis rate of 74.2% was achieved with this optimized system, as determined by the number of mutant progeny out of all progeny screened. We also evaluated the off-targets associated with the method and established a Web-based resource, as well as a searchable, genome-wide database of predicted sgRNAs appropriate for genome engineering in flies. Our results demonstrate that this optimized Cas9/sgRNA system in Drosophila is efficient, specific, and cost-effective and can be readily applied in a semi-high-throughput manner.
Keywords: nanos-Cas9, HRMA
Abstract
The ability to engineer genomes in a specific, systematic, and cost-effective way is critical for functional genomic studies. Recent advances using the CRISPR-associated single-guide RNA system (Cas9/sgRNA) illustrate the potential of this simple system for genome engineering in a number of organisms. Here we report an effective and inexpensive method for genome DNA editing in Drosophila melanogaster whereby plasmid DNAs encoding short sgRNAs under the control of the U6b promoter are injected into transgenic flies in which Cas9 is specifically expressed in the germ line via the nanos promoter. We evaluate the off-targets associated with the method and establish a Web-based resource, along with a searchable, genome-wide database of predicted sgRNAs appropriate for genome engineering in flies. Finally, we discuss the advantages of our method in comparison with other recently published approaches.
Much of our knowledge of the mechanisms underlying biological processes relies on genetic approaches, whereby gene activity is perturbed and the phenotypic consequences of perturbation are analyzed in detail. In recent years, several major advances have been made in the design of methods for specifically and efficiently perturbing genomes. Arguably, the most exciting advances rely on the ability to induce double-strand breaks (DSBs) by targeting a nuclease to a specific genomic sequence. Repair of DSBs by the error-prone nonhomologous end-joining (NHEJ) mechanism allows for the recovery of small deletions; moreover, repair of DSBs by homologous recombination (HR) in the presence of a donor template opens the door to a wide range of specifically engineered changes at the targeted site (1).
Two nuclease-based systems, the zinc-finger nuclease (ZFN) and transcription activator-like effector nuclease (TALEN) systems, work effectively in a number of organisms (2–7). But because these approaches require the production of a construct encoding a unique DNA-binding protein fused to the nuclease domain, they can be both cumbersome and costly. In contrast, the recent approach based on the bacterial CRISPR- associated single-guide RNA (Cas9/sgRNA) system does not require production of specific fusion proteins for each targeted sequence (8–10).
Cas9 was first identified in type II Streptococcus pyogenes as an RNA-guided defense system against invading viruses and plasmids (11–13). This adaptive immune-like system contains three components: CRISPR RNA (crRNA), trans-activating CRISPR RNA (tracrRNA), and Cas9. The tracrRNA triggers Cas9 nuclease activity and the crRNA guides Cas9 to cleave the specific foreign dsDNA sequence via base-pairing between the crRNA and the target DNA. Importantly, a single-guide RNA (sgRNA, also known as chiRNA), comprising the minimal crRNA and tracrRNA, can function similarly to the crRNA and tracrRNA, thereby providing a simplified method for genome editing (8–10, 14–20).
Given the great promise of the Cas9/sgRNA method for genome engineering, we set out to test the system in Drosophila melanogaster. Although many genomic engineering tools are already available for Drosophila (21, 22), including ZFNs and TALENs (2–7), we reasoned that the Cas9/sgRNA system might provide a more cost-effective approach, allowing its use at a genome scale. Similar to other recent reports (14, 16, 20), we found that the Cas9/sgRNA system works in Drosophila. Moreover, we found that injection of sgRNAs into a transgenic line expressing Cas9 in the germ line (nanos-Cas9) provides a straightforward, effective, and inexpensive method that can facilitate the production of large mutant collections. We also evaluated the frequency of off-target events associated with Cas9/sgRNA. In addition, to facilitate prediction and testing of off-target events, we established a searchable database of in silico predicted sgRNAs that includes information about predicted off-target events. Finally, we compared our approach with methodologies recently reported by others (14, 16, 20).
Results
Optimization of the Cas9/sgRNA System.
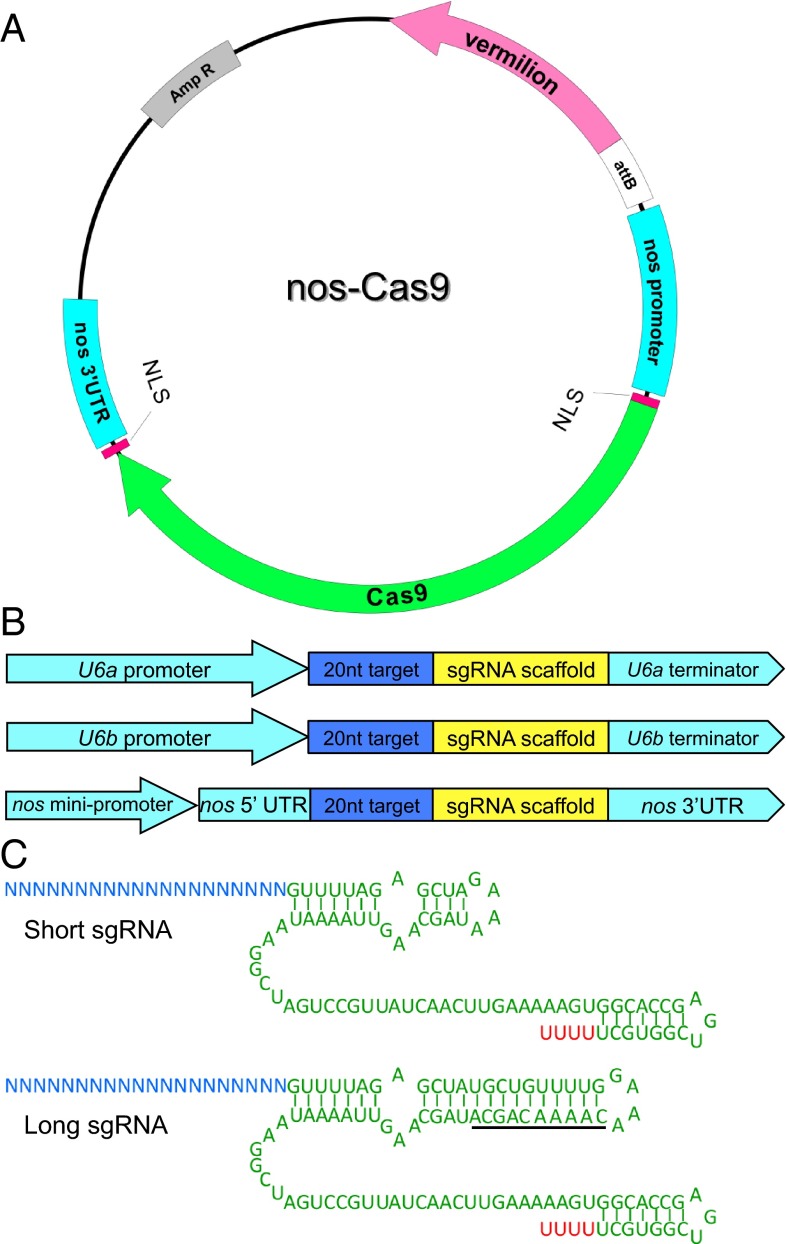
To test the efficacy of the Cas9/sgRNA system in inducing germ-line mutations in D. melanogaster, we generated a vector that expresses the Cas9 gene under the nanos (nos) promoter (Fig. 1A). We reasoned that the nos promoter would induce strong and restricted expression of the transgene in germ cells, which might help avoid viability issues associated with disruption of essential genes in the surrounding somatic cells. The nos-Cas9 DNA was coinjected with U6b-sgRNA DNA targeting the white (w) gene (Fig. 1B). We initially used the U6b core promoter, which has been shown to be an effective promoter in both mammals and Drosophila. With the nos-Cas9 DNA and a single sgRNA plasmid combination, white-eyed mutants were recovered at an overall heritable mutation rate as high as 2.7% (i.e., the percentage of mutant F1s out of all F1s screened) (Table 1).
Fig. 1.
Optimization of the Drosophila Cas9/sgRNA system. (A) The nos-Cas9 plasmid encodes the S. pyogenes Cas9 gene under the regulation of the nos promoter 5′ UTR and 3′ UTR. The attB sequence and a vermillion+ marker are also included in this plasmid. This plasmid was used either directly for mutagenesis in microinjection experiments or for insertion into attP sites to generate Cas9 transgenic flies. (B and C) Schematics of the sgRNA constructs used in this study. (B) Three versions of plasmids expressing sgRNA (U6a, U6b, or minimal nos promoter). (C) Two sgRNA scaffold versions, one similar to that described by Mali et al. (10) and the other with 10 extra base pairs inserted between the crRNA and the tracrRNA modules. The extra base pairs are underscored.
Table 1.
Heritable mutation rates using nos-Cas9 and sgRNA plasmids
| G0 adults |
Overall heritable mutation rate (individual), %, median (range)† | |||||
| sgRNA | Embryos, n | Total n | Survival rate, % | Fertile, n | Germ-line mutants, n (%)* | |
| w1 | 65 | 14 | 21.5 | 13 | 4 (30.8) | 2.7 (3.1–15.1) |
| w2 | 60 | 15 | 25.0 | 13 | 3 (23.1) | 0.6 (1.2–3.2) |
The percentage of germ-line mutants was calculated as the proportion of fertile G0 flies that gave rise to white-eyed progeny.
The overall heritable mutation rate was calculated as the number of white-eye F1s divided by the number of all F1s observed. Mutation rates for each G0 fly that generated white-eyed progeny are shown in parentheses and are calculated as the number of white-eyed F1s divided by the total F1s observed from a single G0 fly.
We next compared the efficiencies of different promoters in controlling the transcription of sgRNA (Fig. 1B). We tested whether a different U6 promoter, the U6a promoter, could lead to higher expression levels of sgRNA. We also tested a modified nos promoter (Materials and Methods), with the idea that this promoter should allow for high expression of sgRNAs specifically in germ-line cells. We also note that unlike the sgRNAs associated with the U6 promoters, those used in conjunction with the nos cassette also include 5′ and 3′ UTR sequences and should be transcribed by RNA polymerase II.
Based on the frequency of mutations recovered (Table 2), U6b was the most effective of the three promoters tested. Given previous in vitro studies suggesting that crRNA-tracrRNA duplexes with extra nucleotides lead to increased efficiency of genome editing (9), we next constructed a version of the sgRNA cassette with additional nucleotides inserted between the crRNA and tracrRNA to extend the sgRNA further than those previously tested (Fig. 1C). A comparison of two sgRNAs targeting w revealed that the shorter version was approximately twice as effective as the longer version (Table 2); thus, we used the short version in all subsequent constructs. Taken together, our results show that Cas9, expressed under the control of nos regulatory sequence, along with a short sgRNA under the control of U6b appears to provide an optimal combination for genome editing in Drosophila.
Table 2.
Heritable mutation rates using nos-Cas9 and different sgRNA plasmid constructs
| SgRNA promoter | sgRNA scaffold | Embryos, n | G0 adults |
Overall heritable mutation rate (individual), % median (range)† | |||
| Total n | Survival, % | Fertile, n | Germ-line mutants, n (%)* | ||||
| U6b | Short | 78 | 36 | 46.2 | 28 | 6 (21.4) | 3.2 (1.1–64.6) |
| U6a | Short | 68 | 17 | 25.0 | 5 | 4 (80.0) | 2.1 (1.2–6.8) |
| nos-mini | Short | 56 | 9 | 16.1 | 7 | 0 | 0 |
| U6b | Long | 64 | 18 | 28.1 | 13 | 4 (30.8) | 1.7 (1.5–10.0) |
The percentage of germ-line mutants was calculated as the proportion of fertile G0 flies that gave rise to white-eyed progeny.
The overall heritable mutation rate was calculated as the number of white-eyed F1s divided by the number of all F1s observed. Mutation rates for each G0 fly that generated white-eyed progeny are shown in parentheses and are calculated as the number of white-eyed F1s divided by the total F1s observed from a single G0 fly.
Transgenic Expression of Cas9 in the Germ Line Results in Dramatically Increased Efficiency of Mutagenesis.
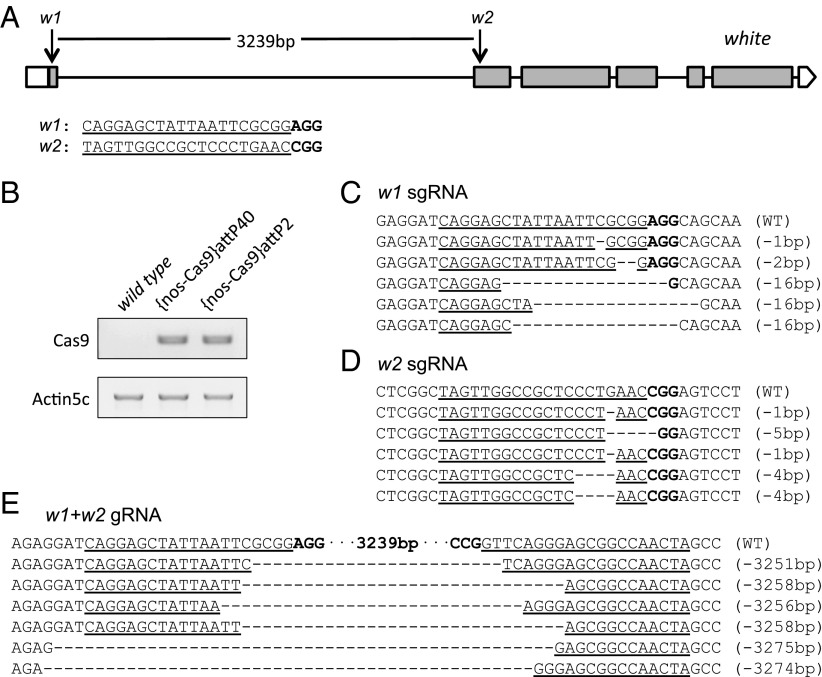
We reasoned that the efficiency of mutagenesis and recovery of injected animals might be compromised by transient expression of Cas9 after DNA injection, variation in the developmental stage of the injected embryos, and/or DNA concentration. With the idea of circumventing these potential issues, we hypothesized that expression of Cas9 in the germ cells via a transgene might improve the efficiency of mutagenesis after injection of sgRNAs into these transgenic flies. We made use of the fact that our nos-Cas9 vector contains an attB sequence and the vermilion reporter gene (Fig. 1A), and generated transgenic fly strains in which nos-Cas9 was integrated into either the attP40 site on chromosome 2 or the attP2 site on chromosome 3. Both transgenic fly stocks are healthy and fertile, and expression of the Cas9 transcript was readily detected by RT-PCR (Fig. 2B).
Fig. 2.
Testing the optimized Drosophila nos-Cas9/sgRNA system on the white locus. (A) Schematic of two sgRNAs targeting the white locus, with the 20-nt target sequence underscored and PAM in bold type. Gray boxes represent exons. w1 sgRNA targets the first exon, and w2 targets the second. (B) RT-PCR results confirming the expression of Cas9 in {nos-Cas9}attP40 and {nos-Cas9}attP2 embryos, with actin 5C as an internal control. (C and D) Representative sequencing results showing the indel mutations generated in this study, with w1 sgRNA (C) and w2 sgRNA (D). (E) Schematic showing the defined deletion generated by using w1 and w2 sgRNAs, along with representative sequencing results showing the break points of the deletions generated. The genomic sequence between the sgRNA target sites is not shown. In C–E, the targeted genomic DNA sequence is underscored, and the NGG PAM sequence is in bold type. Dashed lines represent the locations of genomic deletions detected by sequencing.
To test the Cas9/sgRNA system with these transgenic animals, we next injected {nos-Cas9}attP2 fly embryos with w1 sgRNA and/or w2 sgRNA targeting the w gene. Specifically, the sgRNAs were injected individually to generate mutations by NHEJ or injected together to generate a 3.2-kb deletion (Fig. 2A). Strikingly, the overall heritable mutation rates reached 42.9% for w1 and 12.4% for w2, which are significantly higher than the 2.7% and 0.6% obtained after coinjection of nos-Cas9 and w1 or w2 sgRNA DNAs, respectively (Tables 1 and 3). Note that approximately 25% of embryos from either coinjection of Cas9/sgRNA DNA or injection of sgRNA DNA into {nos-Cas9}attP2 embryos developed into fertile adults, indicating no apparent toxicity associated with either method (Tables 1 and 3).
Table 3.
Heritable mutation rates using transgenic {nos-Cas9}attP2 flies and sgRNA plasmid
| sgRNA | Embryos, n | G0 adults |
Overall heritable mutation rate (individual), %, median (range) † | |||
| Total n | Survival, % | Fertile, n | Germ-line mutants, n (%)* | |||
| w1 | 61 | 16 | 26.2 | 13 | 13 (100.0) | 42.9 (3.3–100) |
| w2 | 65 | 17 | 26.2 | 17 | 15 (88.2) | 12.4 (1.1–58.3) |
| w1 + w2 | 71 | 21 | 29.6 | 12 | 11 (91.7) | 74.2 (13.6–100) |
The percentage of germ-line mutants was calculated as the proportion of fertile G0 flies that gave rise to white-eyed progeny.
The overall heritable mutation rate was calculated as the number of white-eyed F1s divided by the number of all F1s observed. Mutation rates for each G0 fly that generated white-eyed progeny are shown in parentheses and were calculated as the number of white-eyed F1s divided by the total number of F1s observed from a single G0 fly.
To examine whether these mutations also could be detected in G0 animals, we amplified the targeted region from whole embryo genomic DNA. Using high-resolution melt analysis (HRMA), we were able to detect mutations, facilitating isolation of effective mutations at an early stage (Materials and Methods). Of 15 G0 animals tested, 5 contained mutations at w1 but none had mutations at w2, likely owing to the lower mutagenesis rate of w2 (Table S1).
Injection of the two w sgRNAs together generated deletions between the targets and improved the overall heritable mutation rate significantly (74.2% with w1 + w2, compared with 42.9% with w1 and 12.4% with w2) (Table 3). Random testing of 30 white-eyed F1 flies from six G0 lines identified defined sets of deletion mutations in 16 progeny derived from five individual G0 flies, for example, 74.2% with w1 + w2, 53% of which were 3.2-kb deletions. Importantly, defined genomic deletions resulting after the introduction of two different sgRNAs could be readily identified in the F1 flies by PCR amplification. Furthermore, PCR identified 5 of 21 G0 adult flies with defined deletions. In conclusion, generation of deletions using two sgRNAs not only increased the efficiency of mutagenesis, but also facilitated detection of deletion events in G0 flies.
Off-Target Analysis.
Previous studies have shown that a single mismatch between the sgRNA and the target DNA outside the 13-nt neighboring protospacer adjacent motif (PAM), the so-called “seed” region, can be tolerated, given that sgRNA with a single mismatch in nucleotides 14–20 can still guide Cas9 to introduce DSB in the genome (8–10, 23). Furthermore, Cas9 can induce a DSB at genomic sequences with as many as five nucleotide mismatches to the sgRNA (18, 24). To evaluate the potential off-target events associated with w1 and w2 sgRNAs, we identified all possible locations that share high sequence similarity with the target site and include a PAM sequence (see Online Resource of sgRNA Designs for Drosophila Genome Editing and Fig. 3). The tool did not identify any off-target sequences using default conditions, but when search criteria were relaxed, 10 potential off-target sites were found (five for each sgRNA), with the closest match containing six mismatches to the on-target sequence (Fig. 3).
Fig. 3.
No off-targets were detected using the optimized Drosophila Cas9/sgRNA system to generate heritable mutations. (A and B) Potential off-target sites for w1 sgRNA (A) and w2 sgRNA (B). Genomic regions that have at least 11-nt homology to the sgRNA seed region and a neighboring PAM sequence were sequenced from F1 white-eyed flies generated by the optimized system. Mismatches between the potential off-targets and the targeted region are underscored. The PAM sequences are in bold type.
We next PCR-amplified the potential off-target regions from genomic DNA of white-eyed F1 flies with defined deletions generated by injection of both w1 and w2 sgRNAs and assayed for the presence of mutations using HRMA or sequencing. Using HRMA, we analyzed eight independent F1 adult flies for each sgRNA and failed to detect any mutations at any of these sites (Table S1). Finally, we sequenced 20 cloned PCR products from each of eight F1 adults per off-target site and did not detect any mutations, further confirming the HRMA results (Fig. S1).
Online Resource of sgRNA Designs for Drosophila Genome Editing.
To facilitate the use of sgRNAs in Drosophila, we established a freely available online resource of precomputed sgRNAs (www.flyrnai.org/crispr). We developed the resource in two steps. We first extracted all possible 23mers from both the forward and reverse strands of the genomic sequence of D. melanogaster based on FlyBase release 5.52 (July 15, 2013). Some 7.6 million of these 23mers end with a PAM sequence (NGG) and have a unique 15-bp sequence, which includes the seed region sequence and PAM sequence, in the genome. We selected this subset of 23mers as potential CRISPR sgRNA designs and aligned them to the genome sequence. We found that 28% of these sgRNA sequences aligned to intergenic regions, 37% aligned to introns, and 35% targeted the exon region of annotated genes [coding sequence (CDS) or noncoding regions; Table 4].
Table 4.
Statistics for the sgRNA resource
| CRISPR category | All CRISPRs | No off-target | No off-target in CDS regions | Off-target in CDS regions |
| All CRISPRs | 7,566,724 | 300,034 | 2,894,160 | 4,372,530 |
| CRISPRs target intergenic region | 2,110,255 | 76,182 | 907,314 | 1,126,759 |
| CRISPRs target gene region | 5,456,469 | 223,852 | 1,985,158 | 3,247,459 |
| CRISPRs target intron region | 2,786,650 | 95,393 | 1,231,172 | 1,460,085 |
| CRISPRs target exon region | 2,669,819 | 128,459 | 753,986 | 1,787,374 |
| CRISPRs target exon-CDS region | 2,107,960 | 104,892 | 506,998 | 1,496,070 |
| CRISPRs target exon-noncoding region | 561,859 | 23,567 | 246,988 | 291,304 |
We next systematically analyzed potential off-target sites, which share a certain level of sequence similarity (up to five mismatches) and end with a PAM sequence (NGG), for each CRISPR sgRNA design. Although the presence of the NGG PAM sequence on the genomic DNA is required for Cas9 recognition, variations in the PAM (e.g., NAG) can be tolerated in bacteria (18, 24); thus, to be cautious, in the off-target analysis we also included the sites ending with NAG. We found that for CRISPR sgRNA designs targeting gene CDS regions within exons, 104,892 (5% of all designs targeting CDS) have no predicted off-targets, whereas 506,998 (24%) have no predicted off-targets in CDS regions of other genes (Table 4). The designs that target a CDS and have no predicted off-targets correspond to 11,950 Drosophila genes (i.e., 86% coverage of protein-coding genes). Together, the 611,890 designs that target a CDS and have no predicted off-targets in another CDS correspond to 12,996 protein-coding genes (93% coverage of protein-coding genes).
To make the designs available, we developed a user interface that allows the community to query all relevant CRISPR designs by gene identifier (gene symbol, CG number, or FBgn) or genome coordinates and to view all corresponding sgRNA designs on a genome browser (Fig. S2). To identify CRISPR sgRNAs targeting intergenic regions, such as promoters, users can enter an identifier corresponding to a nearby gene, then use the navigator to find the intergenic region of interest, or can enter genome coordinates directly. To help users select relevant designs, CRISPR sgRNAs with different predicted properties regarding the target region and potential off-targets are separated into different tracks, for example “CRISPRs target CDS region/without any off-target” and “CRISPRs target intron/without any off-target.” By clicking on the sgRNA of interest at the genome browser, the user can access detailed information, such as sequence and potential off-target genes (Fig. S2).
Discussion
We report a simple and effective method for efficiently generating heritable loss-of-function alleles in D. melanogaster using the Cas9/sgRNA system. Specifically, we show that injection of sgRNAs into a transgenic line that expresses Cas9 in the germ line (nos-Cas9) is an effective and relatively inexpensive technique for genome engineering.
Several recent studies also have reported success in using the Cas9/sgRNA system to edit genomic DNA in Drosophila (14, 16, 20). Gratz et al. (16) coinjected two DNA plasmids, one encoding Cas9 and the other encoding a single sgRNA targeting the X-linked yellow (y) gene. In that study, Cas9 was expressed under the control of the hsp70 promoter and 3′ UTR, and the sgRNA was under the control of the snRNA U6b promoter. The system was effective in generating somatic patches of yellow tissues in 62% of injected G0 males. Furthermore, germ-line transmission, as detected by the presence of at least one yellow offspring, was observed in 6.4% of the G0 males. The overall germ-line transmission rate (i.e., the number of yellow offspring as a percentage of all progeny) was 0.25%. Gratz et al. (16) also showed that when Cas9 was coinjected with a pair of sgRNAs designed to work together to precisely delete y, 25% of the injected males produced yellow progeny, with an overall germ-line transmission rate of 1.4%.
A different approach was taken by Bassett et al. (14) and Yu et al. (20), who injected fly embryos with in vitro transcripts encoding Cas9 and sgRNA. Basset et al. (14) injected a mix of Cas9 mRNA and sgRNA transcripts targeting either y or w. In their system, both Cas9 and sgRNA were under the control of a bacterial T7 promoter, and a poly-A tail was added posttranscriptionally to the in vitro-transcribed Cas9 mRNA. This system was more effective than the system reported by Gratz et al. (16) at generating somatic mosaics, with 86% of the injected G0 males exhibiting patches of yellow tissues; however, the adult survival rate was only 3%. For the w gene, 25% of injected G0 males had visible phenotypes, and the survival rate was <3%. Although the authors reported that reducing the concentration of mRNA increased the survival rate from 3% to 11%, the proportion of mosaic adults decreased dramatically, from 86% to 10%. Furthermore, germ-line transmission, as detected by the presence of at least one yellow offspring, was observed in 58% of the G0 males, and 34.5% of all offspring contained a mutation in the y gene.
Similarly, Yu et al. (20) injected the in vitro synthesized Cas9 mRNA and sgRNA, and reported a similar efficiency of mutagenesis. In that study, the Cas9 mRNA was transcribed under the control of the Sp6 promoter, and transcription of the sgRNA was under the control of the T7 promoter.
Compared with the foregoing methods, the approach that we describe herein has three key advantages. First, we did not observe any visible phenotypes in G0-injected flies, most likely because control by the nos promoter limits Cas9 expression to the germ cells. Importantly, the absence of somatic events will circumvent the potentially deleterious effects of inducing mutations into the somatic tissues of G0 animals when genes required for cell viability or developmental processes are targeted. Second, the frequency of germ-line transmission that we observed is much higher than that reported for other methods. Injection of a single sgRNA plasmid into Cas9 transgenic flies produced a G0 rate of 93.3% (28 of 30) and an overall heritable mutation rate as high as 42.9%. In addition, we found that injection of a pair of sgRNAs can lead to the generation of defined genome deletion of the region between the targets, with a further improved overall heritable mutation rate (74.2%; Table 3). Indeed, after injection of two sgRNAs, 91.7% (11 of 12) of fertile G0 flies produced F1 mutants. Among all mutant-producing fertile G0 flies, 18.2% (2 of 11) produced mutant progeny at a rate of 50–80%, 18.2% (2 of 11) did so at a rate of 80–100%, and 54.5% (6 of 11) generated only mutant progeny (100%). We found a much higher survival rate of G0 flies (26.2%) after injection of only w1 sgRNA plasmid. This rate compares favorably with a previously reported 3% survival obtained after direct injection of Cas9 mRNA and sgRNA (14). At 81%, the G0 fertility rate of the optimized system was also higher than the 72% rate found with the Cas9 mRNA method. Third, the direct material costs associated with our method are roughly 10–20% of those associated with direct injection of Cas9 mRNA, because our approach avoids the need for in vitro transcription. In addition, because of its high efficiency and low lethality, our approach requires microinjection of only approximately 20 embryos (roughly 1/15th of that in previously reported methods) to achieve the same level of mutagenesis, further contributing to the cost-effectiveness of our approach.
To be useful, a genome engineering technique must be specific. Experience with other mutagenesis methods and previous reports with CRISPRs have taught us that off-targets are a major concern. Previous in vitro studies have indicated that sgRNA/Cas9 specificity extends past a 7- to 12-bp seed sequence and that up to five mismatches are tolerated depending on their positions along the sgRNA (23, 24). Using various methods, we detected no off-targets in either CDS or intergenic regions (Fig. S1), suggesting that the Cas9 protein is expressed at appropriate levels in the transgenic animals, and that specificity can be improved by designing sgRNAs to avoid highly similar off-targets.
Several CRISPR Web tools are available online; however, these do not provide much information about potential off-targets. We have developed a user interface that allows the community to query all relevant sgRNA designs by gene identifier (gene symbol, CG number, or FBgn) or genome coordinates, and also to view all corresponding sgRNA designs on a genome browser (Fig. S2). To facilitate the use of sgRNAs in Drosophila, we have established a freely available online resource of precomputed sgRNAs (www.flyrnai.org/crispr) (Table 4). The designs are organized into subset “tracks” based on off-target potential. Moreover, a detail page that includes a summary list of potential off-targets can be viewed for each design.
Our simplified Cas9/sgRNA system will facilitate the generation of large mutant collections, including both loss-of-function indel mutations and defined deletions. Fig. S3 shows a flowchart designed to expedite the production of such collections in a semi-high-throughput manner. In addition to generation of deletions via NHEJ after production of DSB, based on a previous report (16), we expect that using our system with a donor DNA will facilitate HR-mediated knock-in approaches, such as the introduction of visible markers. This would expand the applications of our approach, including easier mutation identification (e.g., after knock-in of a marker), introduction of site-directed mutagenesis in endogenous loci, and in-frame insertion of ectopic tags into genomic sequence to detect gene expression patterns.
Materials and Methods
Cas9 and sgRNA Plasmid Design.
To express Cas9 in Drosophila germ cells, we constructed a nos cassette plasmid containing approximately 700 base pairs of the nos promoter, the nos 5′ UTR, and the nos 3′ UTR. The attB donor sequence and a vermillion+ marker were also included in the nos cassette plasmid. The Cas9 coding sequence was then excised from pcDNA3.1-Cas9 (25) and cloned into the XbaI and AvrII restriction sites of the nos cassette plasmid.
Two sgRNA scaffolds were designed: a short scaffold, as described by Mali et al. (10), and a long scaffold with 10 extra base pairs between the crRNA and the tracrRNA modules (Fig. 1C). To express sgRNA in vivo, we selected three different regulatory sequences: U6a (CR31379) and U6b (CR32867) (26, 27), both of which direct transcription of Drosophila U6 snRNAs, and nos-mini, which contains the nos promoter and a minimal 5′ UTR (28, 29). We used the 404-bp upstream and 93-bp downstream regulatory sequences of U6a to construct U6a-sgRNA-short and U6a-sgRNA-long. Similarly, we used the 400-bp upstream and 95-bp downstream regulatory sequences of U6b to construct U6b-sgRNA-short and U6b-sgRNA-long. The nos 700-bp promoter, 80-bp minimal 5′ UTR, and 900-bp 3′ UTR were used to construct nos-minisgRNA-short and nos-minisgRNA-long. In all six versions of the sgRNA plasmid, the 20-bp target sequence is inserted into two BbsI sites at the beginning of the sgRNA scaffold. A 680-bp fragment was placed between the BbsI sites as a marker for detecting incomplete enzyme digestion. The sequence of the nos-Cas9 plasmid is shown in Fig. S4, and partial sequences of the six different versions of the sgRNA constructs used are shown in Fig. S5.
Fly Stocks.
All flies were cultured on standard cornmeal food at 25 °C. The {nos-Cas9}attP40 and {nos-Cas9}attP2 fly stocks were established according to a previously described protocol (30). In brief, nos-Cas9 was inserted into attP40 on the second chromosome or attP2 on the third chromosome by microinjection into y[1] sc[1] v[1] P{y[+t7.7]=nos-phiC31\int.NLS}X; P{y[+t7.7]=CaryP}attP40 or y[1] sc[1] v[1] P{y[+t7.7]=nos-phiC31\int.NLS}X; P{y[+t7.7]=CaryP}attP2 fly embryos, respectively, followed by screening for the vermillion+ marker present in the nos-Cas9 plasmid. Canton-S and y[1]w[67c23] flies were used.
Genomic DNA Extraction.
Fly genomic DNA was purified via phenol-chloroform extraction. Single flies were homogenized in 400 µL of lysis buffer (1X PBS, 0.2% SDS, and 200 µg/mL proteinase K; Roche) and incubated at 50 °C for 1 h, followed by extraction in 400 µL of phenol-chloroform. The mixture was then centrifuged at 21,000 X g for 20 min at 4 °C, after which the supernatant was transferred to a new tube. An equal volume of isopropanol was added, and the tube was vortexed thoroughly. The mixture was then kept at −20 °C for at least 1 h, followed by centrifugation at 21,000 X g for 20 min at 4 °C. The supernatant was removed, and the pellet was washed with 500 µL of 75% ethanol, followed by centrifugation at 21,000 X g for 5 min at 4 °C. Finally, the pellet was dried for 10 min and resuspended in 30 µL of DNase-free water.
Embryo Injection.
Plasmid DNA was microinjected into Drosophila embryos following standard protocols. To mutagenize flies with Cas9 and sgRNA, the nos-Cas9 plasmid (250 ng/µL) and sgRNA plasmid (250 ng/µL) were coinjected as a mixture. For coinjection of the plasmids, the injection solution consisted of 250 ng/µL of the nos-Cas9 plasmid and 250 ng/µL of each sgRNA plasmid. When using transgenic Cas9 flies, a single sgRNA plasmid at 250 ng/µL was injected or two sgRNA plasmids at 250 ng/µL each were coinjected.
Screening of Mutations.
To score for germ-line mutations, all G0 adult flies that developed from injected embryos were crossed to y[1]w[67c23] flies. The F1 progeny were then screened for white eye color over a 6-d period. If the G0 fly was a female, then all progeny were screened. If the G0 fly was a male, then only female progeny were screened. Individual heritable mutation rates were calculated for each single G0 as the number of mutant F1s out of the number of screened progeny. The overall heritable mutation rate was calculated as the number of all mutant F1s out of the number of all screened progeny for a given sgRNA target. Mutagenesis events were confirmed by sequence analysis of F1 adults; the detection primers are listed in Table S2.
Off-Target Analysis.
To investigate the possibility of off-target cleavage by Cas9/sgRNA, we searched the fly genome for potential off-targets containing a match to the sgRNA sequence of at least 11 nt followed by the PAM sequence, as well as a match of 13 or 14 nt but without a neighboring PAM sequence. Primers flanking the potential off-targets were used to PCR-amplify these regions for analysis by HRMA and sequencing. For sequencing analysis, genomic DNA from a single fly was used as template, and the defined DNA fragment was amplified by specific primers carrying EcoRI and XbaI (Table S2). The PCR products can be directly sequenced by one of the primers or cloned into the linearized VALIUM1 vector (30) by the same set of restriction enzymes, and then sequenced by specific primers (Table S2).
HRMA.
For both on-target and off-target HRMA assays, PCR was performed using specific primers (Table S2, off-target sequencing primers) to amplify regions surrounding the target site from genomic DNA isolated from either injected embryos (for on-target analysis) or F1 adults (for off-target analysis). PCR products were diluted 1:500,000 before a second round of PCR using nested primers (Table S2, off-target HRM primers) to generate short amplicons (70–150 bp) including the target sites. Melting curves were generated by heating from 55 °C to 95 °C and measuring fluorescence in the presence of Evagreen DNA dye at increments of 0.1 °C. Both amplification and melt curve steps were performed using a BioRad CFX Real-Time System qPCR machine and BioRad Precision Melt Supermix. Genomic DNA isolated from the uninjected parent strain (y[1] sc[1] v[1] P{y[+t7.7]=nos-phiC31\int.NLS}X; P{y[+t7.7]=CaryP}attP2) was used to generate control samples. HRMA data were analyzed to identify melt curves that differed significantly from controls using custom scripts that will be reported in detail separately. The HRMA analysis data are provided in Table S1.
RT-PCR.
Total RNA was isolated from 0- to 1-h Drosophila embryos using the AxyPrep Multisource Total RNA MiniPrep Kit (Axygen). A total of 3 µg RNA was used to create cDNA, using the GoldScript cDNA Kit (Invitrogen) according to the manufacturer’s protocol, followed by PCR to amplify the target sequence. The Actin 5C gene served as an internal control. Primer sequences are listed in Table S2.
Assembly of Online Resource.
A Perl module was developed in-house to identify all possible CRISPR sequences from both strands of genome sequences downloaded from FlyBase release 5.52 (July 16, 2013). The BLAST program was used to identify all possible off-target sites, and a second Perl module was developed in-house to annotate on-target and off-target sites, as well as to format GFF3 files to upload genome browse. JBrowse was used and configured to display all CRISPR designs.
Supplementary Material
Acknowledgments
We thank Son Nguyen and Ting Wu for helpful discussion. This work was supported by the Tsinghua-Peking Center for Life Sciences, the Ministry of Science and Technology of China 973 Program (Grant 2013CB835100), the PhD Programs Foundation of the Chinese Ministry of Education (Grant 20121018577 and 20120002110056), and the National Institutes of Health, National Institute of General Medical Sciences (Grants R01 GM084947 and R01 GM067761, to N.P.). N.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318481110/-/DCSupplemental.
References
- 1.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161(3):1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao G, McMahon C, Chen J, Rong YS. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc Natl Acad Sci USA. 2008;105(37):13999–14004. doi: 10.1073/pnas.0805843105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100(5):2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J, Zhou W, Dong W, Watson AM, Hong Y. Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci USA. 2009;106(20):8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, et al. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics. 2012;39(5):209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288(5473):2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 8.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: Versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 12.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14(3):321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 14.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4(1):220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013;195(1):289–291. doi: 10.1534/genetics.113.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venken KJ, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat Rev Genet. 2005;6(3):167–178. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- 22.Venken KJ, Bellen HJ. Transgenesis upgrades for Drosophila melanogaster. Development. 2007;134(20):3571–3584. doi: 10.1242/dev.005686. [DOI] [PubMed] [Google Scholar]
- 23.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23(4):465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakiyama M, Matsumoto T, Yokoyama S. Drosophila U6 promoter-driven short hairpin RNAs effectively induce RNA interference in Schneider 2 cells. Biochem Biophys Res Commun. 2005;331(4):1163–1170. doi: 10.1016/j.bbrc.2005.03.240. [DOI] [PubMed] [Google Scholar]
- 27.Das G, Henning D, Reddy R. Structure, organization, and transcription of Drosophila U6 small nuclear RNA genes. J Biol Chem. 1987;262(3):1187–1193. [PubMed] [Google Scholar]
- 28.Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8(4):243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci USA. 2009;106(23):9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni JQ, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5(1):49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.