Significance
Macromolecules (DNA, proteins, and lipids) in all cells are constantly damaged by reactive oxygen species (ROS). In particular, ROS cause 1,000–7,000 DNA damages per day. Due to its lowest redox potential, the base guanine is mostly affected, resulting in the formation of 8-oxo-7,8-dihydroguanine. This modified base instructs incorporation of adenosine, instead of cytidine, by replicative DNA polymerases, potentially leading to GC→TA transversion mutations. DNA polymerase λ is the most efficient enzyme in performing accurate translesion synthesis over 8-oxo-7,8-dihydroguanine, since it preferentially incorporates the correct cytidine. In this paper we found that the protein called “DNA polymerase δ interacting protein 2” supports DNA polymerase λ in its important task and can protect cells from ROS DNA damage.
Keywords: oxidative damage, auxiliary factor, DNA repair
Abstract
The bypass of DNA lesions by the replication fork requires a switch between the replicative DNA polymerase (Pol) and a more specialized translesion synthesis (TLS) Pol to overcome the obstacle. DNA Pol δ-interacting protein 2 (PolDIP2) has been found to physically interact with Pol η, Pol ζ, and Rev1, suggesting a possible role of PolDIP2 in the TLS reaction. However, the consequences of PolDIP2 interaction on the properties of TLS Pols remain unknown. Here, we analyzed the effects of PolDIP2 on normal and TLS by five different human specialized Pols from three families: Pol δ (family B), Pol η and Pol ι (family Y), and Pol λ and Pol β (family X). Our results show that PolDIP2 also physically interacts with Pol λ, which is involved in the correct bypass of 8-oxo-7,8-dihydroguanine (8-oxo-G) lesions. This interaction increases both the processivity and catalytic efficiency of the error-free bypass of a 8-oxo-G lesion by both Pols η and λ, but not by Pols β or ι. Additionally, we provide evidence that PolDIP2 stimulates Pol δ without affecting its fidelity, facilitating the switch from Pol δ to Pol λ during 8-oxo-G TLS. PolDIP2 stimulates Pols λ and η mediated bypass of other common DNA lesions, such as abasic sites and cyclobutane thymine dimers. Finally, PolDIP2 silencing increases cell sensitivity to oxidative stress and its effect is further potentiated in a Pol λ deficient background, suggesting that PolDIP2 is an important mediator for TLS.
DNA polymerase (Pol) δ-interacting protein 2 (PolDIP2, also known as PDIP38) is a 368-aa protein associated with the p50 subunit of DNA Pol δ both in vitro and in vivo (1), as well as with the Pol δ auxiliary factor proliferating cell nuclear antigen (PCNA) (2). However, the physiological significance of this interaction remains unknown. PolDIP2 has been found in mitochondria (2, 3), as well as shuttling from the cytoplasm to the nucleus and at the plasma membrane (4). Its subcellular localization depends on the proliferative state of the cells and on its physical interaction with a cell-adhesion receptor (4).
PolDIP2 was also found to increase endogenous reactive oxygen species (ROS) production by the NADPH oxidase Nox4, an enzyme with a role in cell migration, growth, and senescence (5). PolDIP2 seemed to be involved in recruiting Nox4 to the nucleus, suggesting a role in regulating the nuclear redox environment.
More recently, PolDIP2 has been shown to physically interact with translesion synthesis (TLS) enzymes such as Pols η and ζ and Rev1 (6). PolDIP2 binds to the ubiquitin-binding zinc finger domain of Pol η, a region involved in the recognition of the ubiquitinated form of PCNA and mediating the switch between Pol η and the replicative Pols during TLS (7, 8). Depletion of PolDIP2 caused an increase of Pol η nuclear foci, even in the absence of DNA damage and a decrease in cell survival after UV irradiation, leading to the proposal that PolDIP2 may be a factor regulating the switch between Pol η and the replicative enzymes in the TLS process (6). However, it is not known if and how PolDIP2 exerts influence on the enzymatic properties of Pol η or any other specialized Pol.
In an attempt to clarify the role of PolDIP2 in the TLS switch, we have analyzed the effects of PolDIP2 on normal and TLS catalyzed by five different human Pols (Pol δ, Pol η, Pol ι, Pol λ, and Pol β) across three major DNA lesions [8-oxo-7,8-dihydroguanine (8-oxo-G), abasic (AP) site, and cyclobutane thymine dimers (T-T)]. Our results suggest that PolDIP2 is a pivotal factor in oxidative damage response both in vitro and in vivo.
Results
PolDIP2 Stimulates the Activity of Pols λ and η.
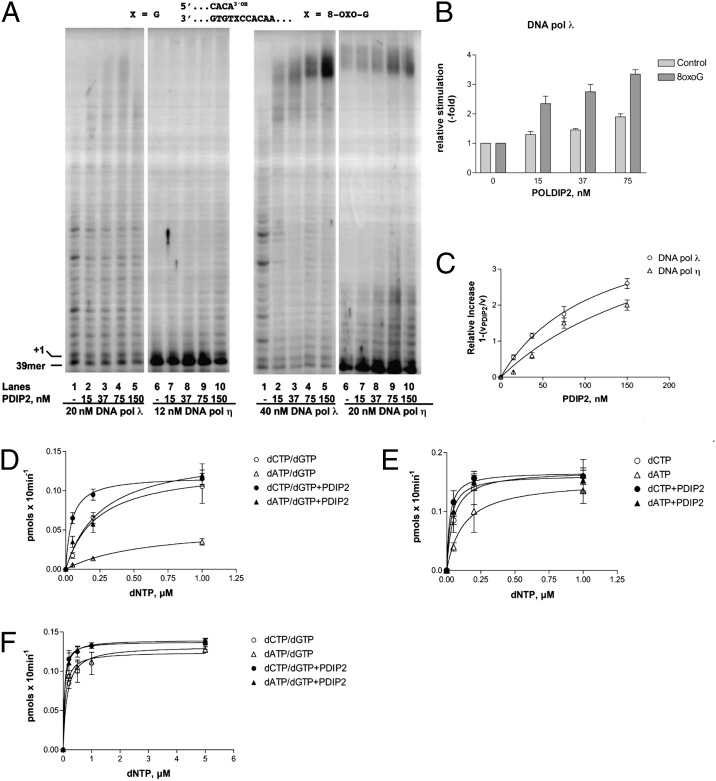
When titrated in the presence of a 5′-labeled 39/100-mer oligonucleotide primer/template (p/t), containing either a normal G or an 8-oxo-G lesion at position +1, GST-tagged PolDIP2 promoted, in a dose-dependent manner, the amount and the length of the DNA products synthesized by Pols λ and η either on the undamaged (Fig. 1A, Left) or on the 5′-labeled 8-oxo-G containing substrate (Fig. 1A, Right). The stimulation was more pronounced for Pol λ than for Pol η (compare lanes 1–5 with lanes 6–10) and for the damaged vs. control template, as shown in Fig. 1B for Pol λ. The apparent affinity (Kd) for PolDIP2 was 113 ± 12 nM for Pol λ and 233 ± 20 nM for Pol η (Fig. 1C). The stimulation was not due to the GST tag, as GST alone did not stimulate either Pol λ or η, (Fig. S1 A and B) and the untagged PolDIP2 was still able to stimulate both Pol λ and η to an extent comparable to the GST-tagged one (Fig. S1 C and D). GST-PolDIP2 alone, in the presence of dNTPs, did not show any nucleotide incorporation (Fig. S1E, lanes 2–5). Because the purification of GST-PolDIP2 by affinity chromatography was easier and more efficient than the purification of the untagged version by conventional chromatography, GST-PolDIP2 was used for the subsequent experiments.
Fig. 1.
PolDIP2 stimulates the bypass of 8-oxo-G by DNA Pols λ and η. (A) Pol λ (lanes 1–5) or Pol η (lanes 6–10) were titrated in the absence (lanes 1 and 6) or presence of increasing amounts of GST-tagged PolDIP2 in the presence of a 5′-labeled undamaged (Left) or 8-oxo-G–containing (Right) 39/100-mer p/t. (B) Stimulation by PolDIP2 of nucleotide incorporation by Pol λ on the normal (light gray bars) or damaged (dark gray bars). Values are the means of three independent determinations. Error bars are ± SD. (C) Relative increase of Pol λ (circles) or Pol η (triangles) nucleotide incorporation on the 5′-labeled 8-oxo-G containing 39/100-mer p/t as a function of GST-tagged PolDIP2 concentrations. Data were fitted to the Michelis–Menten equation. Values are the means of three independent determinations. Error bars are ± SD. (D) Variation of the apparent velocity of the reaction catalyzed by Pol λ on the 5′-labeled 8-oxo-G containing 39/100-mer p/t as a function of increasing equimolar concentrations of dCTP and dGTP (circles) or dATP and dGTP (triangles) in the absence (open symbols) or presence (filled symbols) of 75 nM GST-tagged PolDIP2. Values are the means of three independent determinations. Error bars are ± SD. (E) Variation of the apparent velocity of the reaction catalyzed by Pol λ on the 5′-labeled 8-oxo-G containing 39/100-mer p/t as a function of increasing equimolar concentrations of dCTP (circles) or dATP (triangles) in the absence (open symbols) or presence (filled symbols) of 75 nM GST-tagged PolDIP2. Values are the means of three independent determinations. Error bars are ± SD. (F) Variation of the apparent velocity of the reaction catalyzed by Pol η on the 5′-labeled 8-oxo-G containing 39/100-mer p/t as a function of increasing equimolar concentrations of dCTP and dGTP (circles) or dATP and dGTP (triangles) in the absence (open symbols) or presence (filled symbols) of 75 nM GST-tagged PolDIP2. Values are the means of three independent determinations. Error bars are ± SD.
Pols β and ι Are Not Stimulated by PolDIP2.
PolDIP2 did not stimulate Pol β on the 39/100-mer p/t, whether undamaged or bearing an 8-oxo-G (Fig. S2 A and B). When PolDIP2 was tested with Pol ι in the presence of its preferred metal ion, Mn2+ (Fig. S2C), only very slight stimulation was observed for both dATP and dCTP incorporation, either opposite a normal G (lanes 2–10) or opposite 8-oxo-G (lanes 12–20). Thus, PolDIP2 did not influence the fidelity of TLS by Pol ι across the lesion (9). In summary, these initial experiments suggested that PolDIP2 might be a specific auxiliary factor for Pols λ and η.
PolDIP2 Increases the Efficiency of TLS Past 8-oxo-G by Pols λ and η Without Affecting Their Fidelity.
The 39/100-mer p/t substrate used in this study was designed to have only As and Cs on the template strand downstream of the lesion (10), and incorporation of dATP or dCTP could occur only opposite the lesion. Using combinations of either dCTP and dGTP or dATP and dGTP, the complete TLS reaction (incorporation opposite the lesion and subsequent elongation) could be visualized as the accumulation of a +3 product, resulting in the incorporation of two dGMP opposite the two Cs at template positions +2 and +3. This allowed for the possibility to distinguish between error-free and error-prone TLS. As shown in Fig. S3A, PolDIP2 stimulated both the error-free (compare lanes 2–4 with 8–10) or the error-prone (compare lanes 5–7 with 11–13) bypass of 8-oxo-G by Pol λ. In single nucleotide incorporation reactions with dCTP or dATP only (Fig. S3B), PolDIP2 similarly increased the incorporation of both nucleotides opposite the lesion. The kinetic parameters for the reaction are reported in Table 1. PolDIP2 increased the catalytic efficiency of both dATP and dCTP utilization by Pol λ by 2- to 3-fold (Fig. 1 D and E), but the fidelity of TLS was not affected (Table 1). In single nucleotide incorporation reactions, Pol λ showed a 3-fold preference for dCTP vs. dATP incorporation, which increased to 5.6-fold in the TLS reactions including dGTP (incorporation followed by elongation). Inclusion of PolDIP2 did not significantly affect these values (Table 1). Similarly, PolDIP2 did not affect the fidelity of TLS by Pol λ on a single nucleotide gapped substrate bearing an 8-oxo-G on the template strand (Fig. S3 C and D and Table 1). PolDIP2 stimulated the catalytic efficiency of Pol η by 2-fold (Fig. S3E and Fig. 1F) without affecting its fidelity (∼2-fold preference of dCTP vs. dATP incorporation, Table 1) similarly to Pol λ. PolDIP2 increased the efficiency of nucleotide incorporation (Vmax/Km) of Pol λ and Pol η, by decreasing the apparent Km for dNTP binding, while leaving Vmax unaffected (Table 1). Because for Pols λ (11) and η (12) the rate of dissociation of the enzyme from the p/t (koff) is slower than the rate of incorporation (kpol), under the conditions used (i.e., with incorporation of a limited number of nucleotides), Km = (Ks·koff)/kpol and Vmax = [ES]·koff, where Ks is the true Michelis constant for dNTP binding and [ES] is the concentration of the catalytically competent enzyme–substrate complex. Thus, the decrease in the apparent Km, without affecting the Vmax, can be explained by an increase of the rate of nucleotide incorporation kpol by PolDIP2.
Table 1.
Effect of PolDIP2 on the kinetics of nucleotide incorporation opposite 8-oxo-G
| Enzyme, substrates, cofactors | Km, µM | Vmax, pmol⋅min−1 | Vmax/Km | Fidelity* |
| Pol λ | ||||
| 39/100mer | ||||
| dCTP | 0.047 ± 0.01 | 0.017 ± 0.001 | 0.36 | 3 |
| dATP | 0.122 ± 0.04 | 0.015 ± 0.002 | 0.12 | |
| dCTP+PolDIP2 | 0.02 ± 0.004 | 0.017 ± 0.001 | 0.85 | 2.9 |
| dATP+PolDIP2 | 0.055 ± 0.01 | 0.016 ± 0.001 | 0.29 | |
| dCTP/dGTP | 0.23 ± 0.07 | 0.013 ± 0.001 | 0.056 | 5.6 |
| dATP/dGTP | 0.6 ± 0.05 | 0.006 ± 0.001 | 0.01 | |
| dCTP/dGTP+PolDIP2 | 0.043 ± 0.003 | 0.011 ± 0.001 | 0.25 | 6.5 |
| dATP/dGTP+PolDIP2 | 0.26 ± 0.03 | 0.01 ± 0.003 | 0.038 | |
| 39/60/100mer | ||||
| dCTP | 0.02 ± 0.003 | 0.016 ± 0.003 | 0.8 | 3.5 |
| dATP | 0.06 ± 0.01 | 0.014 ± 0.002 | 0.23 | |
| dCTP+PolDIP2 | 0.006 ± 0.003 | 0.02 ± 0.003 | 3.3 | 4.1 |
| dATP+PolDIP2 | 0.02 ± 0.01 | 0.016 ± 0.001 | 0.8 | |
| Pol η | ||||
| dCTP/dGTP | 0.07 ± 0.007 | 0.013 ± 0.001 | 0.18 | 1.8 |
| dATP/dGTP | 0.11 ± 0.01 | 0.012 ± 0.001 | 0.1 | |
| dCTP/dGTP+PolDIP2 | 0.035 ± 0.003 | 0.013 ± 0.001 | 0.37 | 1.7 |
| dATP/dGTP+PolDIP2 | 0.05 ± 0.01 | 0.011 ± 0.003 | 0.22 |
Calculated as Vmax/Km(dCTP)/Vmax/Km(dATP).
PolDIP2 Increases the Processivity of Pols λ and η.
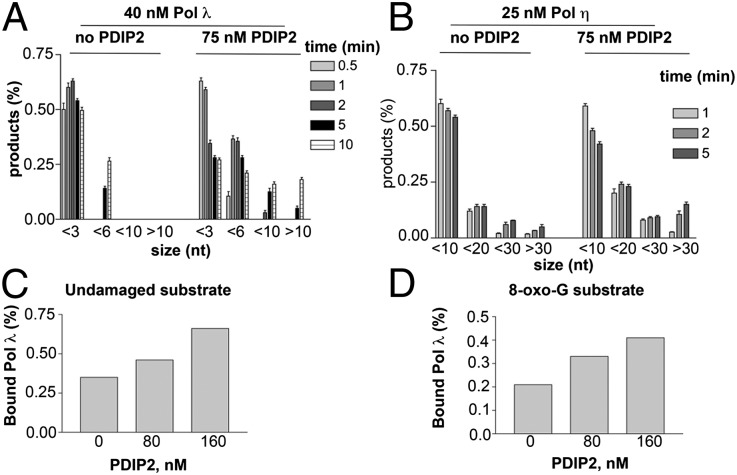
Next, PolDIP2 was tested with Pols λ and η in time-course elongation experiments. The 5′-labeled undamaged 39/100-mer p/t and the enzymes were preincubated to reach equilibrium and the reaction was started by adding nucleotides and an excess of unlabeled poly(dA)/oligod(T) p/t as a trapping agent. Under these conditions, any Pol molecule dissociating from the template should be trapped by the unlabeled competitor DNA, thus not further contributing to the reaction. As shown in Fig. S4A, Pol λ was very distributive, generating only short products (6 nt) in the presence of the trap (lanes 7–11). Addition of PolDIP2 resulted in the earlier accumulation of longer products that reached the length of 17 nt (lanes 18–22). A set of control reactions (lanes 2–6 and 13–17) in the absence of the competitor DNA were also included. In a similar experiment, Pol η proved to be more processive than Pol λ (Fig. S4B), being able to synthesize long products even in the presence of the competitor DNA (lanes 5–7). However, addition of PolDIP2 increased the time-dependent accumulation of long products (lanes 11–13). Plotting the size distribution and relative amounts of the synthesized products over time for Pols λ and η, respectively, as detected in the presence of the competitor DNA (Fig. 2 A and B), shows that the products were longer for both Pols in the presence of PolDIP2. Collectively, these data clearly indicated that PolDIP2 can increase the processivity of Pols λ and η.
Fig. 2.
PolDIP2 increases the processivity of DNA Pols λ and η. (A) Relative amounts (percent with respect to the total products) of the products synthesized by Pol λ under the conditions shown in Fig. S4A in the presence of the trap as a function of the product size. Values are the means of three independent determinations. Error bars are ± SD. (B) Relative amounts (percent with respect to the total products) of the products synthesized by Pol η under the conditions shown in Fig. S4B in the presence of the trap as a function of the product size. Values are the means of three independent determinations. Error bars are ± SD. (C) Effects of PolDIP2 on the amount of Pol λ bound to the 5′-labeled undamaged 39/100-mer p/t as detected by EMSA. (D) As in C but in the presence of the 5′-labeled 8-oxo-G containing 39/100-mer p/t.
PolDIP2 Increases DNA Binding of Pol λ.
The N-terminal half (amino acids 74–200) of PolDIP2 shares 34% similarity with the YccV Escherichia coli protein (1), which was shown to bind DNA (13). However, electrophoretic mobility shift assays (EMSAs) with the 5′-labeled 39/100-mer p/t, whether undamaged (Fig. S4C, lanes 2–4) or containing the 8-oxo-G lesion (lanes 7–10), showed that PolDIP2 did not affect the mobility of the DNA template even at a 10-fold molar excess over the free probe. These results clearly indicated that, under the conditions used for polymerization assays, PolDIP2 did not bind to the DNA substrate. Next, Pol λ was tested in EMSAs in the presence of either the undamaged (Fig. S4D) or the 8-oxo-G (Fig. S4E) p/t and in the absence or in the presence of increasing amounts of untagged PolDIP2. Addition of PolDIP2 resulted in a 2-fold increase of bound Pol λ to both templates (Fig. 2 C and D). Addition of anti-PolDIP2 IgGs resulted in a supershift of the retarded band, conforming that PolDIP2 was present in the complex (Fig. S4F).
PolDIP2 Allows Efficient Error-Free Bypass of 8-oxoG by Pols λ and η in the Presence of PCNA and RP-A.
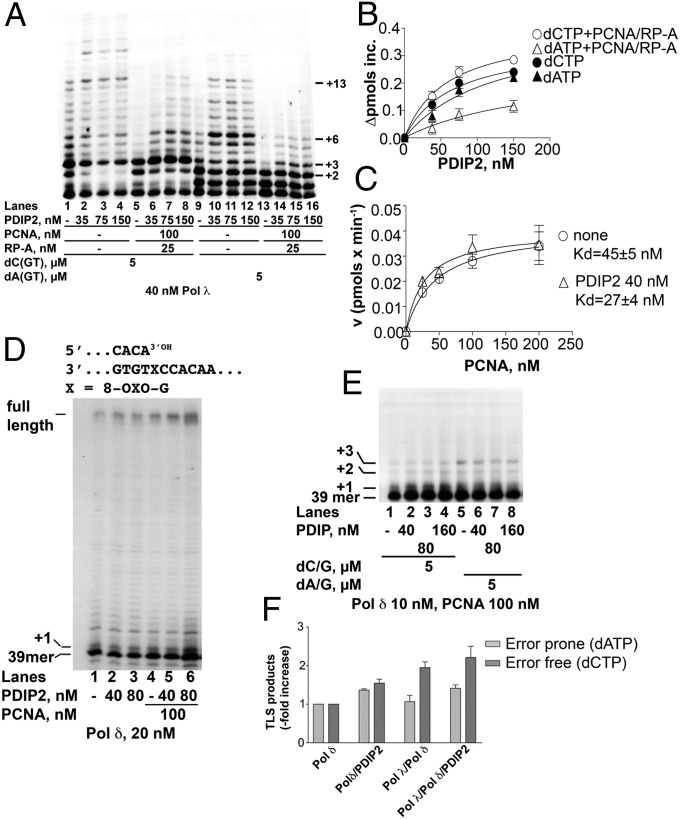
The auxiliary proteins PCNA and RP-A have been shown to suppress the error-prone bypass of 8-oxo-G by Pol λ (14). As shown in Fig. 3A, PCNA and RP-A reduced both error-free (i.e., in the presence of dCTP, dGTP, and dTTP) and error-prone (with dATP, dGTP, and dTTP) TLS by Pol λ (compare lanes 1 and 9 with lanes 5 and 13, respectively). Addition of PolDIP2 to the reactions with dCTP, along with PCNA and RP-A, rescued primer elongation (lanes 6–8), allowing synthesis of products up to +13. Under the same conditions, the addition of PolDIP2 to the reactions in the presence of dATP, PCNA, and RP-A (lanes 14–16) resulted in a lower level of stimulation and in the accumulation of shorter products (+6). In the presence of PCNA/RP-A, a 2.5-fold higher stimulation was observed for dCTP with respect to dATP incorporation (∆max values were 0.53 ± 0.05 pmol⋅min−1 and 0.2 ± 0.01 pmol⋅min−1, respectively) (Fig. 3B). These results indicated that, in the presence of PCNA and RP-A, PolDIP2 specifically stimulated the error-free TLS past an 8-oxo-G lesion by Pol λ. Similar results were obtained with Pol η (Fig. S5A) but the effect was weaker than with Pol λ.
Fig. 3.
PolDIP2 stimulates the error-free bypass of 8-oxo-G by DNA Pol λ in the presence of PCNA and RP-A and increases DNA Pol δ affinity for PCNA. (A) Increasing amounts of GST-tagged PolDIP2 were titrated in reactions containing Pol λ with the 8-oxo-G 5′-labeled 39/100-mer p/t in the presence of dGTP and dTTP and either dCTP, (lanes 1–8) or dATP (lanes 9–16) and in the absence (lanes 1–4 and 9–12) or presence (lanes 5–8 and 13–16) of PCNA and RP-A. (B) Increase in the apparent velocity of the reaction catalyzed by Pol λ on the 8-oxo-G 5′-labeled 39/100-mer p/t as a function of PolDIP2 concentrations in the presence of dGTP and dTTP and either dCTP (circles) or dATP (triangles) either alone (filled symbols) or in combination with PCNA and RP-A (open symbols). Values are the means of three independent determinations. Error bars are ± SD. (C) Variation of the velocity of the reaction catalyzed by Pol δ on the undamaged 39/100-mer p/t as a function of PCNA concentrations in the absence (circles) or presence (triangles) of PolDIP2. Values are the means of three independent determinations. Error bars are ± SD. (D) Pol δ was incubated with the 5′-labeled 8-oxo-G 39/100-mer p/t alone (lane 1) or in the presence of PolDIP2 (lanes 2 and 3), PCNA (lane 4), or both (lanes 5 and 6). (E) Pol δ was incubated with the 5′-labeled 8-oxo-G 39/100-mer p/t in the presence of dCTP and dGTP (lanes 1–4) or dATP and dGTP (lanes 5–8) and in the absence (lanes 1 and 5) or presence (lanes 2–4 and 6–8) of PolDIP2. (F) Stimulation of the TLS past the 8-oxo-G lesion by PolDIP2 under error-prone (light gray bars) or error-free (dark gray bars) conditions. Values are the means of three independent determinations. Error bars are ± SD.
PolDIP2 Increases the Affinity for PCNA of Pol δ Without Affecting Its Fidelity for 8-oxo-G Bypass.
When PolDIP2 was tested in the presence of Pol δ on the undamaged 39/100-mer p/t, a slight stimulation was observed (Fig. S5B, compare lane 2 with lanes 7 and 12). However, stimulation was more evident in the presence of PCNA (compare lanes 3–6 with lanes 8–11 and 13–16). Titration experiments revealed that the affinity of Pol δ for PCNA was increased about 2-fold by PolDIP2 (Fig. 3C). Similarly, PolDIP2 showed maximal stimulation of Pol δ on the 8-oxo-G–containing template in the presence of PCNA (Fig. 3D, compare lanes 2 and 3 with lanes 5 and 6). When tested on the 8-oxo-G template in the presence of dCTP and dGTP or dATP, and dGTP only (so that synthesis was limited to +3 products), such a stimulatory effect, however, was lost (Fig. 3E). These data suggested that PolDIP2 stimulates the processivity of Pol δ by increasing its affinity for PCNA but does not affect the efficiency of 8-oxo-G bypass per se.
PolDIP2 Increases Error-Free 8-oxo-G Bypass During the Switch from Pol δ to Pol λ.
We have previously shown that the efficient error-free bypass of an 8-oxo-G lesion can be achieved via a switch from Pol δ to the more faithful Pol λ (10). To investigate whether PolDIP2 plays any role in this reaction, bypass of an 8-oxo-G lesion was tested under running start conditions, on a 39/100-mer p/t with the lesion at position +26 on the template strand and bearing only As and Cs on the template strand downstream of the lesion as previously described (10). Reactions were run in parallel under error-prone (lanes 2–7) or error-free (lanes 8–13) conditions in the presence of various combinations of Pol δ, Pol λ, and PolDIP2 (Fig. S5C). The intensities of the bands corresponding to the 8-oxo-G lesion and the corresponding TLS products were normalized in each lane to the amount of a 39-mer primer and the percentage of TLS was calculated for each set of samples. These values were used to calculate the increase in TLS products with respect to the control without PolDIP2. As shown in Fig. 3F, PolDIP2 increased the efficiency of error-free bypass by the concerted action of Pols δ and λ to a larger extent than the error-prone one.
Pol λ Forms a Complex with PolDIP2 and Its Catalytic Domain Is Sufficient for PolDIP2 Stimulation.
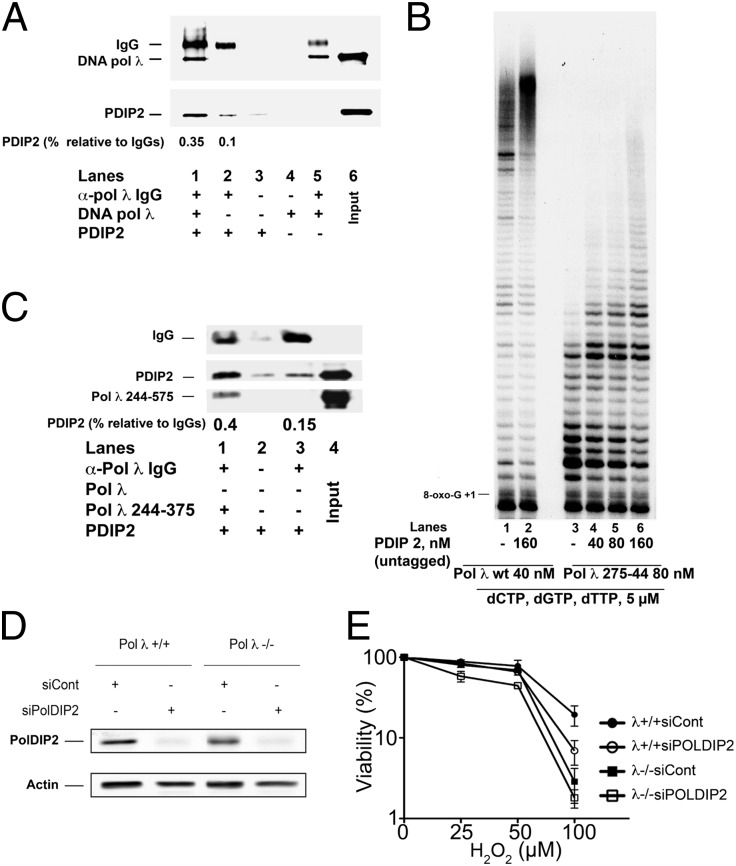
Polyclonal antibodies against Pol λ coupled to Protein A agarose beads were able to immunoprecipitate both recombinant Pol λ and PolDIP2 (Fig. 4A, lane 1). As expected, omission of Pol λ (lane 2), IgGs (lane 4), or both (lane 3) in Fig. 4A did not result in significant recovery of either Pol λ or PolDIP2. These results indicated that Pol λ directly forms a complex with PolDIP2. Pol λ contains in its N-terminal half a breast cancer susceptibility protein C-terminal domain and a proline-rich domain, which have been shown to mediate protein–protein interactions (15). We have previously shown (16) that these two domains are fully dispensable for the catalytic activity. The deletion mutant Pol λ (aa 255–574) lacking both domains but fully competent for catalysis (16), was tested on the p/t containing the 8-oxo-G lesion (Fig. 4B). The mutant was capable of efficient TLS even though with lower processivity than the wild type, resulting in shorter products (compare lane 1 with lane 3 in Fig. 4B). Addition of PolDIP2 resulted in increased TLS and generation of longer products (lanes 4–6). Immunoprecipitation experiments with anti-Pol λ IgGs (Fig. 4C), confirmed that PolDIP2 physically interacted with the recombinant Pol λ (aa 255–574) (lane 1), but not with IgGs alone (lane 3) or with the Sepharose beads (lane 2). Thus, the catalytic domain alone was sufficient for PolDIP2 binding and stimulation of Pol λ.
Fig. 4.
PolDIP2 interacts with the catalytic domain of DNA Pol λ and its silencing increases cell sensitivity to oxidative damage. (A) Western blot analysis of the Protein A Sepharose immunoprecipitated material with anti-Pol λ IgGs in the presence of Pol λ and untagged PolDIP2 (lane 1), untagged PolDIP2 alone (lane 2), or Pol λ alone (lane 5). Lanes 3 and 4 show control mock immunoprecipitations in the absence of IgGs and in the presence of PolDIP2 or Pol λ, respectively. The relative amounts of PolDIP2 present in lanes 1 and 2 were expressed as the ratio of the PolDIP2 signal intensity toward the IgG signal intensity. (B) Pol λ wt (lanes 1 and 2) or the truncated mutant 275–544 (lanes 3–6) were incubated with the 8-oxo-G 5′-labeled 39/100-mer p/t in the absence (lanes 1 and 3) or presence of increasing amounts of PolDIP2. (C) Western blot analysis of the Protein A Sepharose immunoprecipitated material with anti-Pol λ IgGs in the presence of Pol λ truncated mutant 275–544 and untagged PolDIP2 (lane 1) or untagged PolDIP2 alone (lane 3). Lane 2 shows control mock immunoprecipitation in the absence of IgGs and presence of PolDIP2. The relative amounts of PolDIP2 present in lanes 1 and 3 were expressed as the ratio of the PolDIP2 signal intensity toward the IgGs signal intensity. (D) Silencing of PolDIP2 by siRNA in wild-type (first and second lane) or Pol λ−/− (third and fourth lane) MEFs. The first and third lanes show scrambled siRNA negative controls. (E) Cell viability assays on wild type (circles) or Pol λ−/− (squares) MEFs after treatment with scrambled (filled symbols) or PolDIP2 specific (open symbols) siRNA in the presence of increasing doses of H2O2.
The Presence of PolDIP2 Stimulates the Bypass of Different Lesions by Pols λ and η.
When tested on a p/t containing an AP site at position +1 (Fig. S5D), PolDIP2 was able to stimulate TLS by Pol λ (lanes 2–5) and Pol η (lanes 6–9), but the stimulation was lower for Pol λ than for Pol η (1.5-fold and 2.2-fold, respectively; Fig. S5F). When challenged with a p/t containing a T-T lesion at positions +1 and +2 (Fig. S5E), Pol λ was unable to synthesize across this lesion even with PolDIP2 (lanes 2–5). On the other hand, Pol η was able to bypass the T-T lesion (Fig. S5E, lane 6) and addition of PolDIP2 (lanes 7–9) increased TLS (1.7-fold stimulation, Fig. S5F). These data indicate that PolDIP2 is capable of increasing TLS by Pols λ and η over different lesions. However, the nature of the lesions may affect the extent of stimulation displayed by PolDIP2 on different Pols.
Increased Sensitivity of Mammalian Cells to Oxidative DNA Damage in Response to Silencing Expression of PolDIP2 Is Further Enhanced by Lack of Pol λ.
PolDIP2 silencing in mammalian cells has been already shown to confer UV sensitivity similar to the one displayed by xeroderma pigmentosum variant cells lacking Pol η (6). Here, PolDIP2 expression was silenced by siRNA in mouse embryonic fibroblast (MEF) cells either wild type (λ+/+) or lacking the Pol λ gene (λ−/−). Silencing was very efficient in both cell lines (Fig. 4D). Cell viability was measured upon exposure to increasing doses of H2O2. As expected (17), λ−/−cells were more sensitive to H2O2 treatment than the wild type (Fig. 4E, filled squares). Interestingly, PolDIP2 silencing conferred a sensitivity similar to the one displayed by λ−/− cells (Fig. 4E, open circles). In addition, silencing of PolDIP2 in a λ−/− genetic background, further significantly increased H2O2 sensitivity (Fig. 4E, open squares). Collectively, these results indicate that PolDIP2 is important in protecting cells from oxidative damage and its action is not epistatic with Pol λ.
Discussion
In response to the block of replicative Pols δ and ε when certain DNA lesions on the template strand are encountered by the advancing replication fork, PCNA is monoubiquitinated by the Rad6–Rad18 protein complex and promotes the switch to TLS Pols at the site of damage (7). TLS is mainly performed by the Y family Pols η, ι, and κ (15). We have shown that the X family enzyme Pol λ is also essential for the TLS across the 8-oxo-G lesion (14) both for the postreplicative MutYH-dependent base excision repair of 8-oxo-G:A mismatches (18, 19) and for the bypass of 8-oxo-G lesions that can stall Pol δ during DNA replication (10). PolDIP2 has been shown to interact with Pol δ, PCNA, Rev 1, Pol ζ, and Pol η and has been proposed to act as a switch factor for Pol η (6). In this work, we have shown that PolDIP2 physically interacts also with Pol λ and can specifically increase the efficiency of elongation past three major DNA lesions by Pols λ or η, whereas it has no effect on Pols β or ι. Whereas PolDIP2 did not alter the intrinsic fidelity of Pols, it favored error-free TLS (i.e., incorporation of C) opposite 8-oxo-G by both Pols λ and η in the presence of PCNA and RP-A. Stimulation by PolDIP2 was stronger in the presence of an 8-oxo-G lesion than with an AP site or a T-T lesion and our results suggest that PolDIP2 stimulates Pols λ and η by increasing the polymerization rate and providing further stability to the enzyme–DNA complex.
In current models for TLS, the specialized Pol needs to synthesize a certain stretch of DNA, either to fill the gap or to provide the replicative Pol with a suitable primer to reinitiate DNA synthesis (8). Thus, the ability of PolDIP2 to increase the elongation efficiency and the processivity during TLS will allow faster completion of the bypass process. Thus, our results suggest that PolDIP2 is important in the Pol switch and elongation steps during TLS. Previous observations reported that PolDIP2 inhibited Pol δ. However, such an inhibitory effect was apparent only at very high doses of PolDIP2 (high micromolar range). Here, we showed that at more physiological concentrations (low nanomolar range), PolDIP2 stimulated Pol δ by increasing its affinity for PCNA binding. Because PolDIP2 interacts with both PCNA and the p50 subunit of Pol δ, which is also involved in the interaction with PCNA, our data suggest that PolDIP2 may act as a chaperone coordinating the interaction of different Pols with PCNA.
We found that PolDIP2 is required for maximal efficiency of 8-oxo-G bypass during the switch from Pol δ to Pol λ. In addition, we have shown that silencing of PolDIP2 caused cell sensitivity to oxidative agents to a similar extent as Pol λ silencing and that this effect was further increased when PolDIP2 was silenced in a Pol λ null genetic background. These data clearly suggest that both Pol λ and PolDIP2 are required by cells for maximal efficiency in oxidative damage response and that lacking both proteins severely compromises the ability of the cell to tolerate oxidation. Under this respect, it is worth noting that PolDIP2 has been identified as a positive regulator of the activity of the cellular oxidase Nox 4 in the nucleus (5). Endogenous ROS generation by Nox proteins mediates various growth-related responses, so their presence is essential for cellular metabolism (20). Thus, having a protein such as PolDIP2 that plays a role in increasing both the scheduled ROS production by cellular oxidases and the efficiency of oxidative DNA damage tolerance, may provide a unique and efficient defense mechanism to counteract the mutagenic effects of endogenous ROS.
Materials and Methods
Chemicals.
Deoxynucleotides were purchased from Gene Spin. Labeled [γ-32P]ATP was purchased from Hartmann Analytic GmbH. All of the other reagents were of analytical grade and purchased from Fluka or Merck.
Oligonucleotides.
DNA oligonucleotides were synthesized by Purimex and purified from polyacrylamide denaturing gels (PAGE). The oligonucleotide containing the T-T lesion was a kind gift from Korbinian Heil and Thomas Carell (Ludwig Maximilians University of Munich). See SI Materials and Methods for complete sequences. When indicated, oligonucleotides were 5′-labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP according to the manufacturer’s protocol. Each labeled primer was mixed to the complementary template oligonucleotide at 1:1 (M/M) ratio in the presence of 25 mM Tris⋅HCl pH 8 and 50 mM KCl, heated at 75 °C for 10 min, and then slowly cooled down at room temperature.
Enzymes and Proteins.
Bacterial clones for the expression of human Pols η and ι were a kind gift from R. Woodgate [National Institutes of Health (NIH)]. Human recombinant Pols η and ι were expressed and purified as described previously (21). Human recombinant Pols λ and β and human recombinant RP-A and PCNA were expressed as described (14). Human recombinant Pol δ was expressed and purified as described in ref. 10. The full length ORF of PolDIP2 was obtained from a testis cDNA library by PCR and cloned into a modified pETM33 expression vector encoding an N-terminal GST tag. The recombinant protein was expressed in E. coli and purified through affinity and ion exchange chromatography. See SI Materials and Methods for detailed protocols.
Pol Assays.
All reactions were done in a final volume of 10 µL and incubated for 10 min at 37 °C in the presence of 20 nM 5′-labeled p/t, unless otherwise stated in the figure legends, under optimal buffer conditions for each Pol. See SI Materials and Methods for details. Auxiliary proteins and nucleotides were in the concentration specified in the figures and figure legends. For denaturing gel analysis of the DNA products, the reaction mixtures were stopped by addition of standard denaturing gel loading buffer [95% (vol/vol) formamide, 10 mM EDTA, and xylene cyanol and bromophenol blue], heated at 95 °C for 5 min, and loaded on a 7-M urea 12% (vol/vol) polyacrylamide gel. The reaction products were analyzed by using Molecular Dynamics Phosphoimager (Typhoo Trio, GE Healthcare) and quantified by Image Quant.
EMSAs.
Pol λ and PolDIP2 were incubated with 20 nM 5′-labeled p/t for 10 min at 37 °C in the concentrations specified in the figures and figure legends in a 10-µL final volume of 25 mM Tris⋅HCl pH 7.5, 0.25 mg/mL BSA, 1.6% (vol/vol) Ficoll, 0.4 mM MgCl2, and 0.5 mM DTT. Protein-DNA complexes were separated from the free probe by nondenaturing 7.5% PAGE (19:1). The gel was run at 10 V/cm at room temperature in Tris-borate EDTA buffer 0.5X.
Immunoprecipitation.
One microgram of polyclonal IgGs against Pol λ (Bethyl) was incubated with 1 µg of purified proteins as specified in the figure legends in 10 µL of 20 mM Hepes⋅potassium hydroxide pH 7.9, 2 mM MgCL2, 0.2 mM EGTA, 10% (vol/vol) glycerol, 0.1 mM PMSF, 2 mM DTT, 140 mM NaCl, 0.05% Nonidet P-40, and protease inhibitors. Samples were rotated for 2 h at 4 °C. Protein A Sepharose beads (Invitrogen), equilibrated in the same buffer were added and kept rotating at 4 °C overnight. Beads were washed 3× with the binding buffer and centrifuged for 30 s at 700 × g and the pelleted beads were resuspended in Laemmli sample buffer 1X. Samples were boiled for 5 min at 100 °C and centrifuged at 10,000 × g and the supernatant loaded on a 10% (vol/vol) SDS/PAGE. Immunoprecipitated proteins were revealed by Western blot analysis with anti-Pol λ or anti-PolDIP2 (Proteintech) antibodies.
Kinetic Analysis.
To account for the excess of enzyme over the DNA substrate used, due to the highly distributive nature of the reaction, the variation of the nucleotide incorporation rates (v) as a function of the DNA substrate concentration was fitted to the modified Briggs-Haldane equation
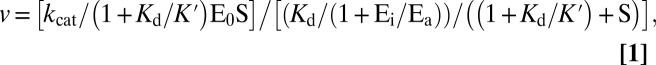 |
where kcat is the apparent catalytic rate, E0 is the input enzyme concentration, S is the variable substrate concentration, Kd is the apparent affinity of the enzyme for the substrate, K′ is the equilibrium dissociation constant for the nonproductive binding of the enzyme to the substrate, Ei is the fraction of enzyme not involved in catalysis, and Ea is the fraction of enzyme involved in catalysis. Fitting was obtained with the GraphPad Prism 3.0.
Cell Culture and siRNA Treatment.
MEFs (14), were grown in DMEM containing GlutaMAX-I supplemented with 10% (vol/vol) FBS and 100 U/mL penicillin–streptomycin (all obtained from Gibco, Invitrogen) at 37 °C in a 5% (vol/vol) CO2 incubator. Cells were transfected with control or PolDIP2-specific siRNA by using Lipofectamine RNAi max (Invitrogen) according to the manufacturer’s protocol and analyzed 72 h after transfection by Western blot analysis using antibodies against Pol λ and PolDIP2 (Proteintech). Control and PolDIP2-specific ON-TARGET plus SMART siRNAs were obtained from Thermo Scientific Dharmacon. A pool of four different siRNAs was used to target PolDIP2.
Cell Viability Assay.
MEFs were seeded 24 h after the siRNA transfection in a 96-well plate and incubated for additional 24 h. Indicated amounts of H2O2 (Sigma-Aldrich) were added and cells exposed to continuous treatment for 24 h. Next, PrestoBlue Cell Viability Reagent (Invitrogen) was added to cells, the fluorescence determined at 560 nm, and the mean from at least four wells used to calculate cell viability. Data presented in Fig. 4E show growth from one representative experiment that has been reproduced four times independently.
Supplementary Material
Acknowledgments
We thank R. Woodgate (NIH) for providing Pol η and ι bacterial expression clone, Korbinian Heil and Thomas Carell for the T-T–containing oligonucleotide, and O. Georgiev for scientific support. This work was supported by the Swiss National Science Foundation (31003A_133100/1 to G.M., E.C., E.M., and U.H.) and by the University of Zürich (R.I., B.v.L., A.F., and U.H.). G.M. was also partially supported by Italian Cancer Research Association (AIRC) Grant IG12084 and by Regione Lombardia, Italy, Project Aree Tematiche Prioritarie Grant 13810040. G.V. was supported by Grant Electricite de France NRB62013-07.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.D.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308760110/-/DCSupplemental.
References
- 1.Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MY. Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen. J Biol Chem. 2003;278(12):10041–10047. doi: 10.1074/jbc.M208694200. [DOI] [PubMed] [Google Scholar]
- 2.Xie B, et al. Further characterization of human DNA polymerase delta interacting protein 38. J Biol Chem. 2005;280(23):22375–22384. doi: 10.1074/jbc.M414597200. [DOI] [PubMed] [Google Scholar]
- 3.Cheng X, et al. PDIP38 associates with proteins constituting the mitochondrial DNA nucleoid. J Biochem. 2005;138(6):673–678. doi: 10.1093/jb/mvi169. [DOI] [PubMed] [Google Scholar]
- 4.Klaile E, et al. The cell adhesion receptor carcinoembryonic antigen-related cell adhesion molecule 1 regulates nucleocytoplasmic trafficking of DNA polymerase delta-interacting protein 38. J Biol Chem. 2007;282(36):26629–26640. doi: 10.1074/jbc.M701807200. [DOI] [PubMed] [Google Scholar]
- 5.Lyle AN, et al. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ Res. 2009;105(3):249–259. doi: 10.1161/CIRCRESAHA.109.193722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tissier A, et al. Crosstalk between replicative and translesional DNA polymerases: PDIP38 interacts directly with Poleta. DNA Repair (Amst) 2010;9(8):922–928. doi: 10.1016/j.dnarep.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Zhuang Z, et al. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc Natl Acad Sci USA. 2008;105(14):5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann AR, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6(7):891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res. 2001;29(4):928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markkanen E, Castrec B, Villani G, Hübscher U. A switch between DNA polymerases δ and λ promotes error-free bypass of 8-oxo-G lesions. Proc Natl Acad Sci USA. 2012;109(50):20401–20406. doi: 10.1073/pnas.1211532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiala KA, Abdel-Gawad W, Suo Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry. 2004;43(21):6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 12.Washington MT, Prakash L, Prakash S. Yeast DNA polymerase eta utilizes an induced-fit mechanism of nucleotide incorporation. Cell. 2001;107(7):917–927. doi: 10.1016/s0092-8674(01)00613-4. [DOI] [PubMed] [Google Scholar]
- 13.d’Alençon E, et al. Isolation of a new hemimethylated DNA binding protein which regulates dnaA gene expression. J Bacteriol. 2003;185(9):2967–2971. doi: 10.1128/JB.185.9.2967-2971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maga G, et al. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447(7144):606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 15.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 16.Blanca G, et al. Human DNA polymerase lambda diverged in evolution from DNA polymerase beta toward specific Mn(++) dependence: A kinetic and thermodynamic study. Biochemistry. 2003;42(24):7467–7476. doi: 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- 17.Braithwaite EK, et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem. 2005;280(36):31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 18.Maga G, et al. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc Natl Acad Sci USA. 2008;105(52):20689–20694. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Loon B, Hübscher U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc Natl Acad Sci USA. 2009;106(43):18201–18206. doi: 10.1073/pnas.0907280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 21.Frank EG, McDonald JP, Karata K, Huston D, Woodgate R. A strategy for the expression of recombinant proteins traditionally hard to purify. Anal Biochem. 2012;429(2):132–139. doi: 10.1016/j.ab.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.