Significance
Evolution functionally diversifies conserved protein architectures, precluding assignment of function from structure alone. The HD structural domain was first recognized in a group of phosphohydrolases and came to be associated with that activity, but characterization of the archetypal mixed-valent diiron oxygenase (MVDO), myo-inositol oxygenase, attributed a very different activity, O2-mediated C-C bond cleavage, to an HD protein. We demonstrate that the recently discovered C-P bond-cleaving enzyme, PhnZ, is another example of an HD-domain MVDO. Sequence and functional data for the dimetal HD proteins reveal that they segregate into well-defined clades, of which several are more likely to comprise MVDOs than phosphohydrolases. This study provides a basis to assign hydrolase or oxygenase activity to proteins in this largely uncharacterized enzyme superfamily.
Keywords: nonheme diiron enzymes, C-H activation, superoxo intermediate, PhnY, structural genomics
Abstract
The founding members of the HD-domain protein superfamily are phosphohydrolases, and newly discovered members are generally annotated as such. However, myo-inositol oxygenase (MIOX) exemplifies a second, very different function that has evolved within the common scaffold of this superfamily. A recently discovered HD protein, PhnZ, catalyzes conversion of 2-amino-1-hydroxyethylphosphonate to glycine and phosphate, culminating a bacterial pathway for the utilization of environmentally abundant 2-aminoethylphosphonate. Using Mössbauer and EPR spectroscopies, X-ray crystallography, and activity measurements, we show here that, like MIOX, PhnZ employs a mixed-valent FeII/FeIII cofactor for the O2-dependent oxidative cleavage of its substrate. Phylogenetic analysis suggests that many more HD proteins may catalyze yet-unknown oxygenation reactions using this hitherto exceptional FeII/FeIII cofactor. The results demonstrate that the catalytic repertoire of the HD superfamily extends well beyond phosphohydrolysis and suggest that the mechanism used by MIOX and PhnZ may be a common strategy for oxidative C-X bond cleavage.
With the number of known protein sequences rising exponentially, assignment of functions to the ever-expanding proteome is a central, largely unmet challenge in the molecular life sciences. Assignment to a known superfamily based on sequence or structure is usually the first approach to predict the function of an uncharacterized protein and often provides valuable hints for further studies. However, nature uses divergent evolution to create new functions, leading to functional diversification within superfamilies that can be as modest as different substrate specificities or as profound as fundamentally different reaction types (1). Without any biochemical or biological information, functional predictions based solely on superfamily assignment can thus be misleading or incorrect (2).
The HD domain superfamily, recognized by Aravind and Koonin in 1998 on the basis of sequence analysis (3), now contains more than 37,000 members occurring in a broad range of organisms in all three domains of life and in more than 240 distinct domain architectures (4). HD proteins possess a conserved α-helical core containing their characteristic H…HD…D sequence motif that binds a divalent metal ion (often ZnII, MgII, or MnII) (5). HD proteins with a dinuclear metal cluster, in which two additional histidines from within the D…D region of the domain form a second metal binding site, have also been identified (6, 7). Only a limited number of HD proteins have been biochemically characterized; they almost exclusively catalyze phosphoester hydrolysis, which has led to the general assignment of HD proteins as phosphohydrolases (3, 5, 8–12). The wide phylogenetic distribution and diverse domain architectures of the HD proteins suggest, however, that additional activities might have evolved within the superfamily. Indeed, the enzyme myo-inositol oxygenase (MIOX) exemplifies a mechanistically very different catalytic function for an HD protein (13–15). In MIOX, the conserved H…HD…H…H…D motif binds two iron ions, and the enzyme uses an unprecedented mixed-valent FeII/FeIII cofactor form to catalyze the four-electron (4e–) oxidative C-C bond cleavage converting myo-inositol (MI; cyclohexan-1,2,3,5/4,6-hexa-ol) to d-glucuronate (13, 16, 17). The FeIII site coordinates MI, serving as Lewis acid to activate the substrate, and the FeII site activates O2 to produce a superoxo-FeIII/FeIII intermediate that initiates the reaction by abstracting a hydrogen atom from C1 of MI (6, 18).
The recently discovered HD protein, PhnZ, catalyzes the 4e– oxidative C-P bond cleavage of 2-amino-1-hydroxyethylphosphonate (OH-AEP) to glycine and phosphate (19) (Fig. 1). It is the second of two enzymes in a recently discovered pathway that degrades the most abundant environmental phosphonate compound, 2-aminoethylphosphonate (2-AEP) (20–23), and its reaction represents the third type of enzymatic C-P bond cleavage to be recognized (23–26). PhnZ was shown to require iron for activity, but the nature of the iron cofactor and its mechanism are unknown (19). The first enzyme in the pathway, the FeII- and α-ketoglutarate(αKG)-dependent oxygenase, PhnY, introduces the hydroxyl group on C1 of 2-AEP, activating the compound for subsequent C-P bond cleavage by PhnZ (19). The assignment of PhnZ to the HD domain superfamily, the conservation of the six residues that coordinate the cofactor in MIOX, the dependence of its catalytic activity on the presence of iron, and the analogy between the PhnZ-catalyzed oxidation and that catalyzed by MIOX all suggest that PhnZ might provide the second example of an HD protein using a mixed-valent diiron cofactor to catalyze an oxygenation (rather than a hydrolysis) reaction.
Fig. 1.
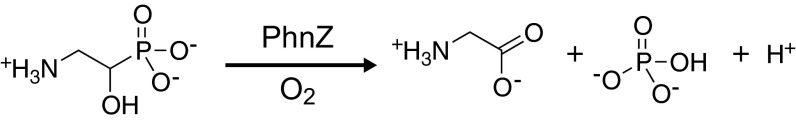
Reaction catalyzed by PhnZ.
Here, we demonstrate by Mössbauer and EPR spectroscopy that PhnZ harbors a diiron cluster and, like MIOX, markedly stabilizes the mixed-valent FeII/FeIII state. The X-ray crystal structure of PhnZ confirms the presence of a diiron cofactor and shows that the substrate is bound to the site 2 Fe ion in a manner analogous to the binding mode of MI in MIOX. PhnZ is an oxygenase requiring O2 for its reaction, as determined by activity assays and isotope-tracer experiments. Freeze-quench (FQ) EPR experiments demonstrate that the FeII/FeIII state is competent to react rapidly with O2 to effect the 4e− oxidation of OH-AEP. Our results thus confirm that PhnZ is the second example of an HD protein using a FeII/FeIII diiron cofactor for oxidative bond cleavage and imply that the mechanism used by MIOX and PhnZ might be a more widespread strategy for the oxidative cleavage of C-X bonds in yet unknown reactions. Indeed, phylogenetic analysis of the HD superfamily suggests that there are potentially many other HD domain mixed-valent diiron oxygenases (HD-MVDOs). These findings demonstrate that the catalytic repertoire of the HD superfamily extends well beyond phosphohydrolysis and illustrate once more how nature creates distinct catalytic functions within a common protein scaffold by divergent evolution.
Results
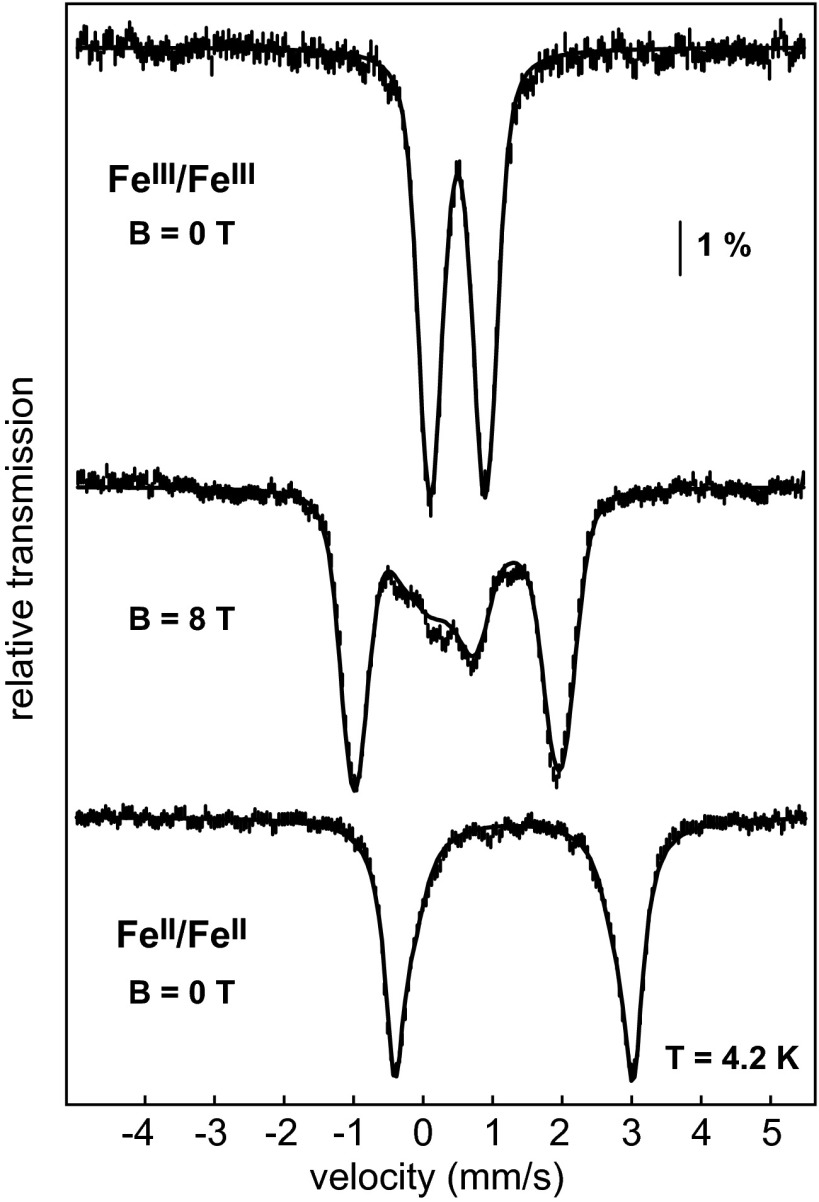
To address the nature of the iron cofactor in PhnZ, we turned first to Mössbauer spectroscopy. Overexpression of N-terminally His6-tagged PhnZ from the uncultured bacterium HF130_AEPn_1 in Escherichia coli and purification by NiII-affinity chromatography yielded protein containing 1.2 ± 0.2 Fe per monomer. Production of PhnZ in minimal medium supplemented with 57FeII afforded protein suitable for Mössbauer spectroscopy. The spectrum of the aerobically isolated PhnZ recorded at 4.2 K without an applied magnetic field (4.2-K/0-T) consists of a single broad quadrupole doublet with isomer shift (δ) of 0.50 mm/s and quadrupole splitting constant (ΔEQ) of 0.79 mm/s, which are characteristic of N/O-coordinated high-spin (S = 5/2) FeIII ions (Fig. 2). The spectrum recorded in an external field of 8.0 T applied parallel to the γ-beam can be simulated with the above parameters, an asymmetry parameter (η) of 0.6, and the assumption that the FeIII species in PhnZ has a diamagnetic electronic ground state (STotal = 0), consistent with the presence of an antiferromagnetically (AF) coupled pair of high-spin (S = 5/2) FeIII ions (Fig. 2). The spectra thus provide definitive evidence that PhnZ contains a dinuclear iron cluster. An 8-T spectrum recorded over a wider range of Doppler velocities shows that ∼90% of the Fe in the sample is in the form of FeIII/FeIII clusters (SI Appendix, Fig. S1). Following treatment of PhnZ with the strong reductant, dithionite, the 4.2-K/0-T Mössbauer spectrum comprises a broad quadrupole doublet with average parameters δ = 1.3 mm/s and ΔEQ = 3.1 mm/s, which are characteristic of high-spin (S = 2) FeII ions with octahedral N/O coordination (Fig. 2). The signals are broad and suggest the presence of two or more FeII complexes (SI Appendix, Fig. S2), as has been seen in MIOX and other nonheme diiron proteins (16, 27–29).
Fig. 2.
Mössbauer spectra of PhnZ. Spectra of the aerobically isolated enzyme (vertical bars) recorded at 4.2 K with a magnetic field, B, of 0 T (Top) or 8 T (Middle) applied parallel to γ beam. The solid lines correspond to simulations with parameters quoted in the text. The high-field spectrum shows a complex with a diamagnetic ground state (STotal = 0), in accordance with an AF-coupled Fe2III/III species. The bottom spectrum is of the dithionite-reduced PhnZ (vertical bars) recorded at 4.2 K and B = 0 T with the corresponding simulation (solid line, parameters quoted in Results and in SI Appendix, Fig. S2).
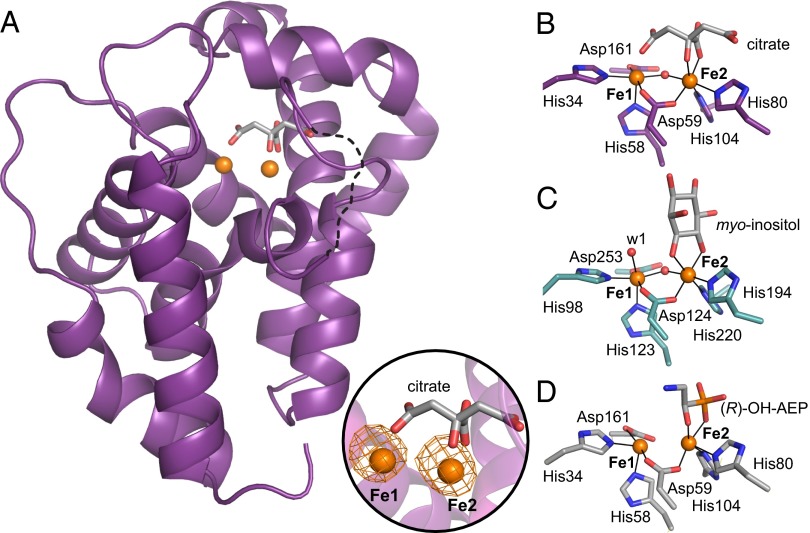
The 3D structure of the aerobically purified PhnZ (citrate-bound; Fig. 3 A and B), solved at a resolution of 1.8 Å with phasing by single-wavelength (Fe K-edge) anomalous diffraction (SI Appendix, Table S1), confirms that the diiron cluster detected in the Mössbauer spectra is coordinated by the residues in the H…HD…H…H…D sequence motif, as in other structurally characterized HD proteins with dinuclear metal clusters [see, for example, Protein Data Bank (PDB) ID codes 3TM8, 2IBN, 3CCG, 2O08, 2OGI, 3GW7, and 2PQ7 discussed below]. The coordination geometry of the PhnZ diiron cluster is nearly identical to the cluster geometries observed in these other proteins (Fig. 3). The enzyme is a monomer, and the core helices near the active site superimpose well with those of MIOX (2.05 Å rmsd for 125 Cα atoms; ref. 30). However, PhnZ is smaller (190 aa) than MIOX (285 aa), and the topology of peripheral structural elements differs significantly between the two proteins (SI Appendix, Fig. S3) (6). The five α-helices characteristic of the HD scaffold contribute the six conserved iron ligands (His34, His58, Asp59, His80, His104, and Asp161). The iron ions, which are modeled at almost full occupancy (0.92), are most likely both in the FeIII oxidation state and are separated by 3.7 Å, similar to the distance seen in the structures of MIOX from Mus musculus and Homo sapiens (3.65 Å and 3.8 Å; PDB ID codes 2HUO and 2IBN) (6, 31). The two Fe sites are bridged by the aspartate of the HD motif and one µ-oxo/hydroxo bridge, as also seen in the MIOX structures (Fig. 3 B and C). Citrate from the crystallization solution binds to the active site with well-defined electron density (SI Appendix, Fig. S5). Its hydroxyl group and Oβ1 of the central carboxyl group coordinate to the Fe2 ion, forming a five-membered ring. The analogous Fe2 (FeIII) site in MIOX is chelated by the C1 and C6 hydroxyl groups of the MI substrate in a very similar five-membered ring that includes the scissile C1-C6 bond. A structure of PhnZ with its natural substrate OH-AEP bound was obtained at a lower resolution (3.0 Å). PhnZ was previously shown to accept only one enantiomer of the chiral OH-AEP, but the active enantiomer was not identified (19). The PhnZ substrate was prepared in stereochemically pure form by enzymatic synthesis using PhnY to hydroxylate 2-AEP. Polarimetry on the purified product established that PhnY produces (R)-OH-AEP. The crystal structure of the substrate-bound PhnZ shows that the modeled (R)-OH-AEP coordinates to Fe2 with its vicinal hydroxyl and phosphonate O-atoms bracketing the scissile C-P bond to form a five-membered ring, as in the MIOX and citrate-bound PhnZ structures (Fig. 3D and SI Appendix, Fig. S6). In this binding mode, the C1 hydrogen that must be abstracted during the reaction projects toward Fe1. By analogy to the mechanism of MIOX, O2 should add to Fe1 (the FeII ion) at its open coordination site (the position occupied by the Oγ1 of the citrate; Fig. 3B), leading to formation of a superoxo-FeIII/FeIII complex that should abstract the H atom from C1 of (R)-OH-AEP. Residues within hydrogen-bonding distance to the substrate and most likely crucial for stabilizing the complex include the largely conserved Lys108, Ser126, Thr129, Gln133, and Arg158 (SI Appendix, Fig. S4), which bind to the phosphonate oxygens, and His62, which interacts with the hydroxyl group of (R)-OH-AEP. His62, in particular, seems important for proper orientation of the substrate, analogous to Lys127 in MIOX, which is essential for activity (6).
Fig. 3.
Three-dimensional structures of PhnZ shown in comparison with MIOX. (A) Ribbon diagram illustrating the α-helical fold of PhnZ, the location of the diiron site, and coordination by a citrate molecule. A loop region (residues 66–72) that could not be modeled is represented as a dotted line. An Fe anomalous difference Fourier map (orange mesh, contoured at 7.0 σ) is shown in Inset. Also shown are views of the active site in PhnZ with the citrate ligand (B), MIOX with the substrate myo-inositol (C), and PhnZ with the substrate (R)-OH-AEP (D). Amino acid side chains in the first coordination sphere and substrate molecules are represented in stick format, FeIII ions are shown as orange spheres, and nonprotein oxygen-based ligands are shown as red spheres. Because of the modest resolution (3.0 Å) of the (R)-OH-AEP–bound PhnZ structure, the oxo/hydoxo bridge cannot be resolved and, therefore, was not modeled.
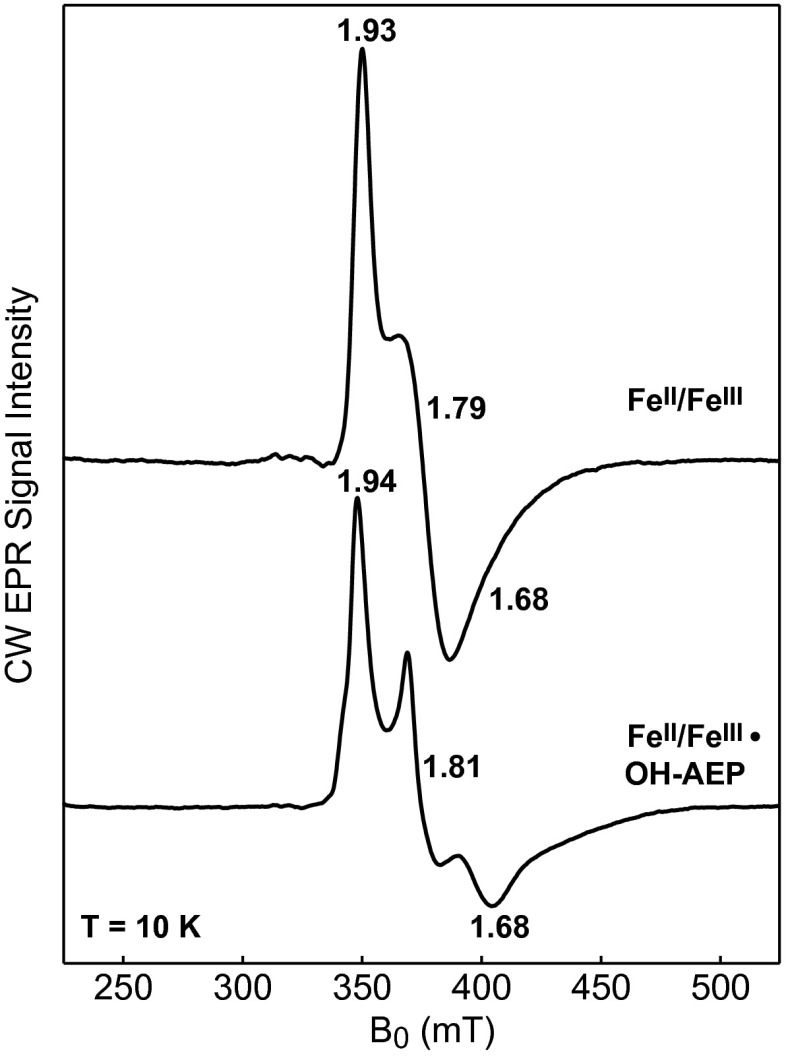
With the exception of MIOX, all dinuclear nonheme-iron oxygenases and oxidases studied to date use the fully reduced forms of their cofactors to activate O2 (32–38), and the mixed-valent FeII/FeIII forms are usually only marginally stable in these enzymes (28, 39, 40). By contrast, MIOX employs the mixed-valent form of its diiron cluster to promote substrate and O2 activation and accordingly stabilizes this state, allowing its accumulation in 60–70% yield (17). For PhnZ to also use an FeII/FeIII cofactor for catalysis, it would be expected to stabilize the mixed-valent state. Indeed, either upon incubation of the fully reduced FeII/FeII PhnZ with a limiting quantity of O2 (in the presence or absence of l-ascorbate) or upon treatment of the fully oxidized FeIII/FeIII protein with l-ascorbate, an EPR signal with apparent principal g values of 1.93, 1.79, and 1.68, characteristic of an AF-coupled mixed-valent FeII/FeIII cluster with an Stotal = 1/2 ground state, develops (Fig. 4). The mixed-valent form in PhnZ can accumulate to 40–70% of the diiron species, consistent with its catalytic relevance. Anaerobic addition of substrate to the FeII/FeIII form of PhnZ alters its EPR spectrum markedly (Fig. 4) and affords changes similar to those observed upon MI binding to MIOX (16), in agreement with the crystallographic observation that (R)-OH-AEP coordinates directly to the diiron cluster to form a five-membered ring, as MI does in MIOX.
Fig. 4.
CW X-Band EPR spectra of PhnZ recorded at 10 K. The spectrum of the l-ascorbate–reduced PhnZ (upper spectrum) shows signals characteristic of an AF-coupled Fe2II/III center. Addition of 3 mM (R)-OH-AEP to the l-ascorbate–reduced PhnZ (lower spectrum) perturbs the spectral features, leading to an increase in anisotropy and decrease in linewidth, suggesting that the substrate binds to the diiron site. Experimental conditions: microwave frequency = 9.48 GHz, microwave power = 2 mW, and modulation amplitude = 1 mT.
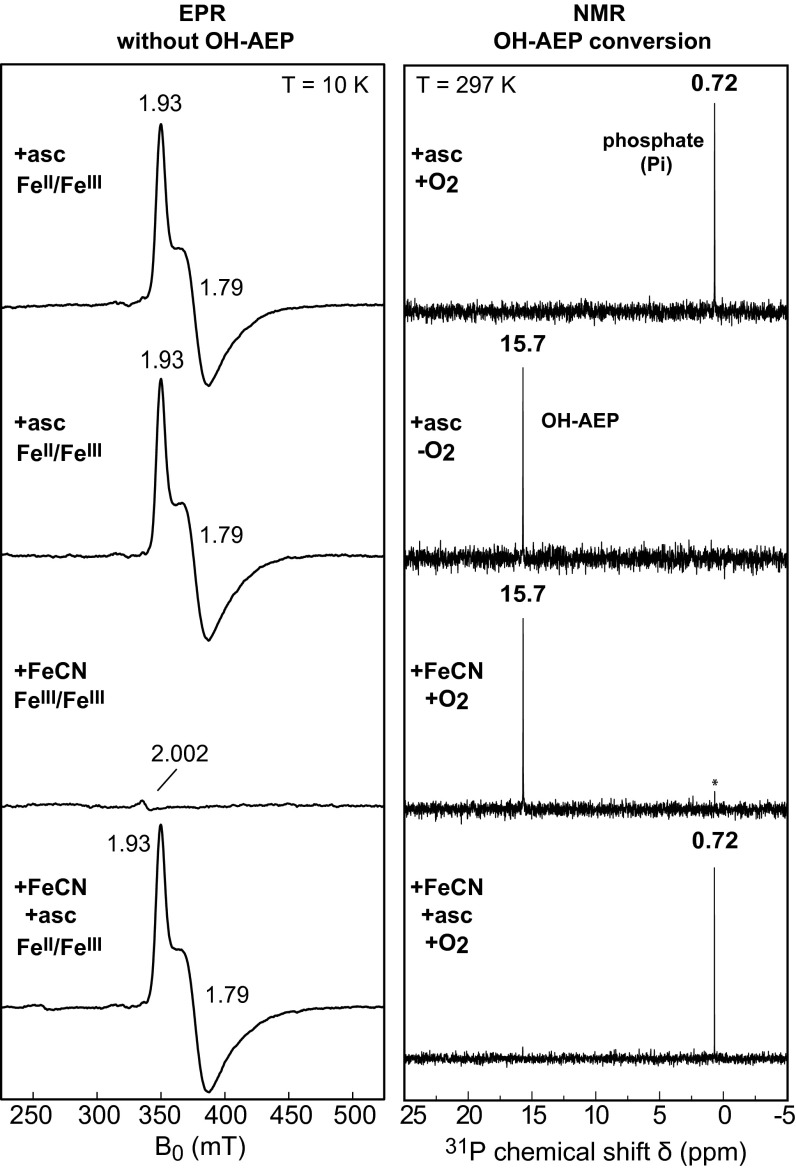
The active redox state(s) of the cofactor in PhnZ and the dependence of its activity on O2 were assessed by monitoring conversion of (R)-OH-AEP to phosphate (Pi) by 31P-NMR spectroscopy. For the activity measurements, the FeII/FeIII cofactor was generated by incubation of the aerobically isolated protein with 5 equivalents (per diiron cluster) of l-ascorbate. Formation of the mixed-valent cofactor to ∼40% of the total diiron centers was confirmed by EPR spectroscopy (Fig. 5). Incubation of 10 µM PhnZ containing 2.7 µM FeII/FeIII cofactor with 2 mM (R)-OH-AEP at ambient temperature while continuously flushing with air resulted in complete conversion of the substrate to Pi within 30 min (Fig. 5). Without exposure to air, no product was detected, even after 6 h (Fig. 5 and SI Appendix, Fig. S7), showing the requirement of the PhnZ reaction for O2. Mass-spectrometric detection of the glycine generated by PhnZ under an atmosphere of either natural-abundance O2 (99.8% 16O) or 18O2 showed the expected two-unit difference in m/z for incorporation of an O atom from O2 (SI Appendix, Fig. S8). This result demonstrates that PhnZ is an oxygenase rather than an oxidase.
Fig. 5.
Dependence of the catalytic activity of PhnZ on O2 and the oxidation state of the diiron cofactor. Conversion of (R)-OH-AEP to phosphate (Pi) by PhnZ was detected by 31P-NMR; the FeII/FeIII cofactor was detected by EPR. EPR samples contained 200 µM PhnZ and spectra were acquired at a microwave frequency of 9.48 GHz, a microwave power of 2 mW, and a modulation amplitude of 1 mT. Enzyme reactions contained 10 μM PhnZ, and 2 mM (R)-OH-AEP, and were carried out for 30 min at room temperature. In the experiments requiring O2, the protein solution was flushed continuously with hydrated air.
To assess whether the FeIII/FeIII cofactor form of PhnZ is catalytically active, the enzyme was first treated with a substoichiometric quantity (0.2 equivalents) of ferricyanide to oxidize the residual FeII/FeIII enzyme form that is generally present along with the predominant FeIII/FeIII complex in the aerobically isolated protein. This treatment diminished the quantity of the FeII/FeIII enzyme form to a level not detectable by EPR, and the resultant protein converted very little (R)-OH-AEP to Pi after a 30-min incubation with continuous air flushing (Fig. 5; Pi peak marked by asterisk). Incubation of the ferricyanide-oxidized PhnZ with 5.2 equivalents l-ascorbate regenerated the FeII/FeIII state (to ∼40%, as determined by EPR spectroscopy) and restored activity, and the resultant protein effected complete substrate conversion in less than 30 min (Fig. 5). These results demonstrate that the FeIII/FeIII form of PhnZ is inactive but can be converted to the active state by incubation with l-ascorbate. l-ascorbate reduces the FeIII/FeIII complex to the FeII/FeIII state but generates at most 4% of the fully reduced FeII/FeII state (as estimated by Mössbauer spectroscopy; SI Appendix, Fig. S9). The activity measurements suggest that the mixed-valent FeII/FeIII state is the catalytically active cofactor form, but substrate conversion by the small quantity of the FeII/FeII state in these samples could not be ruled out from these activity measurements alone.
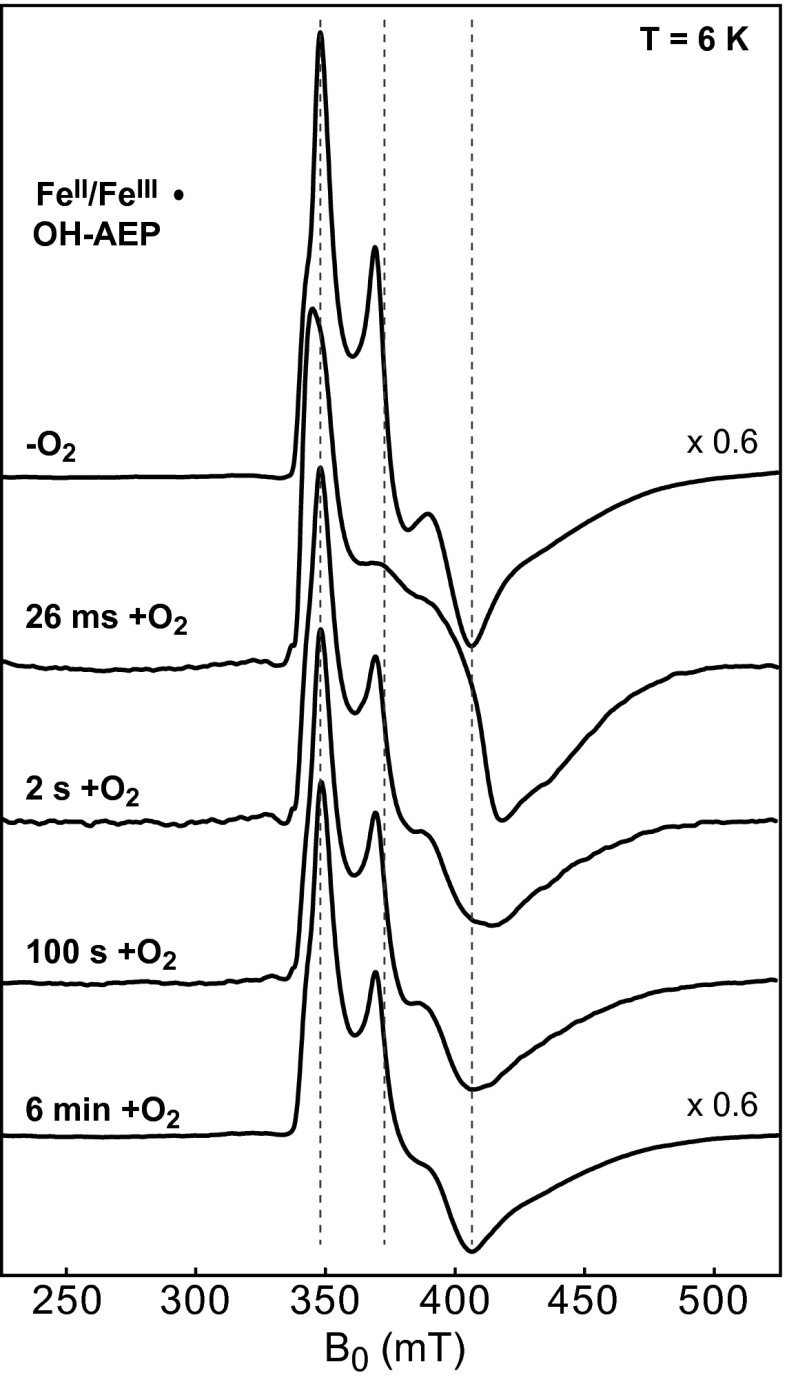
More definitive proof that the FeII/FeIII state is the active enzyme form was sought by FQ EPR experiments testing for a rapid, cyclic reaction of O2 with the FeII/FeIII cofactor in the substrate-bound complex, an observation crucial in establishing that the mixed-valent state is the catalytically active cofactor form in MIOX (17). Because oxidation of (R)-OH-AEP to glycine and phosphate is charge balanced with the four-electron reduction of O2 (Fig. 1), the catalytically active state should be regenerated upon completion of the reaction. Indeed, samples prepared by mixing a solution enriched in PhnZ-FeII/FeIII•(R)-OH-AEP with O2-saturated buffer and freeze quenching at reaction times as short as 0.026 s exhibit perturbations to the EPR spectrum signifying rapid consumption of the reactant complex and formation of a different EPR-active state. This state has apparent g values of 1.97, 1.67, and 1.55 (Fig. 6). By a reaction time of 2 s, the spectrum of the reactant complex has largely returned, suggesting that the transient state detected at 0.026 s is a reaction intermediate. Stopped-flow absorption data also suggest accumulation of an intermediate followed by regeneration of the reactant complex (SI Appendix, Fig. S10). This cyclic decay and reformation of the PhnZ-FeII/FeIII•(R)-OH-AEP complex upon addition of O2 mirror behaviors first seen with MIOX (17) and support the notion that this state of the cofactor is the active form. These results provide solid evidence that PhnZ is the second example of an HD protein that employs a mixed-valent FeII/FeIII cofactor for O2-driven oxidative bond cleavage. Presumably, PhnZ shares other mechanistic features with MIOX, including use of a superoxo-FeIII/FeIII intermediate to initiate substrate oxidation by abstraction of H• from C1. This mechanism might thus be more prevalent than has been recognized to date.
Fig. 6.
CW EPR spectra monitoring the reaction of the FeII/FeIII•(R)-OH-AEP complex with O2 in a FQ experiment. The spectrum of the reactant solution mixed with anaerobic buffer and hand frozen was scaled by a factor of 0.6 to account for the packing factor (dilution) of the FQ samples. Spectra of the FQ samples at times 0.026 s, 2 s, 100 s, and 6 min after reaction of the FeII/FeIII•(R)-OH-AEP complex with O2-saturated buffer (1.8 mM O2) at 5 °C. Experimental conditions: microwave frequency: 9.48 GHz; microwave power: 20 mW; and modulation amplitude: 1 mT. The vertical dashed lines denote the apparent principal g values of the FeII/FeIII•(R)-OH-AEP complex.
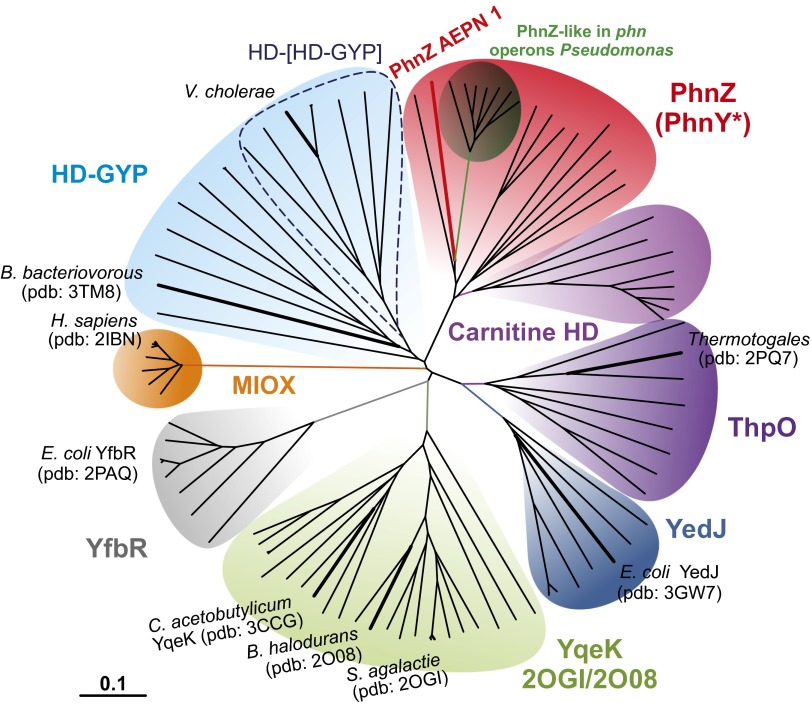
The large number of HD proteins that have not yet been biochemically characterized are likely to provide additional examples of mixed-valent diiron oxygenases (MVDOs) catalyzing yet-unknown reactions. To identify candidates with potential MVDO activity, we searched for proteins within the HD superfamily that, based on either the presence of the conserved H…HD…H…H…D sequence motif or structural information, contain a dimetal site. Interestingly, some of these enzymes have known phosphohydrolase activity (41, 42), illustrating that dimetal HD proteins can accommodate phosphohydrolase or oxygenase function and that the presence of the dimetal center is insufficient to assign a given HD protein as a new MVDO. In an approach to map the oxygenase/hydrolase boundaries within the dimetal HD family, we carried out a phylogenetic analysis of the known proteins. In the neighbor-joining radial phylogram shown in Fig. 7, dinuclear members of the superfamily are segregated into seven clades, with the mononuclear YfbR nucleotidases included as an “outgroup” (gray). Proteins of the HD-GYP clade (in light blue; SI Appendix, Fig. S11) are known to hydrolyze the phosphodiester bond of the bacterial second messenger, cyclic di-GMP, using a dinuclear cluster (41, 42). The metal ions in the active forms of these proteins are not known. The activity of HD-GYP from Borrelia burgdorferi was shown to be enhanced by the addition of MnII (42). By contrast, the crystal structure of HD-GYP from Bdellovibrio bacteriovorus was solved with a diiron cluster (7), and the HD-[HD-GYP] from Vibrio cholerae was suggested to be active in the diiron form (43). No functions have been assigned to the proteins in the YqeK clade (in light green; SI Appendix, Fig. S12), but they are very likely to be nucleotidases, because two different structures (PDB ID codes 2OGI and 2O08) have (d)NDPs bound to the dimetal centers. These proteins have a conserved Arg residue in a position spatially equivalent to that occupied by a functionally essential Arg in YfbR and other mononuclear HD phosphohydrolases such as the HIV-restricting triphosphohydrolase SAMHD1 (9, 12). The corresponding residues in MIOX and PhnZ are Asn and Gln. The presence of Arg at this position may thus be an indicator of a phosphohydrolase rather than MVDO function.
Fig. 7.
Phylogenetic tree of the dinuclear HD proteins. Representatives from the mononuclear YfbR phosphatases have been included as outgroup. The unrooted radial phylogenetic tree is based on analysis of the amino acid sequences of 97 representative proteins (SI Appendix, Table S2). Tree topography and evolutionary distance are given by the neighbor-joining method. The stability of the tree was verified by the bootstrap method with 1,000 replications. Similar trees were obtained by using the minimum evolution and maximum likelihood methods. The scale bar represents a difference of 0.1 substitutions per site. The “PhnZ-like” subclade (dark green) includes proteins that show a high degree of similarity to PhnZ and are encoded in gene clusters associated with phosphonate degradation pathways but lack a phnY-like gene.
Proteins in the carnitine HD, ThpO, and YedJ clades are more likely to be MVDOs. Members of the clade that we have designated carnitine HD (in purple) are the most similar of all of the uncharacterized HD proteins to PhnZ. They are always encoded in tandem with a putative FeII- and αKG-dependent oxygenase sharing high similarity with γ-butyrobetaine hydroxylases (GbbHs), which are known to be involved in carnitine biosynthesis in other organisms (44, 45). The arrangement of tandem genes encoding an FeII/αKG-dependent hydroxylase and an HD protein is strikingly reminiscent of the PhnY/Z case and suggests that the carnitine HD proteins could be MVDOs cleaving a C-X bond in the product of the GbbH-like enzyme. The YedJ clade (in dark blue; SI Appendix, Fig. S13) comprises entirely uncharacterized proteins. This clade is more closely related to PhnZ than to the known hydrolases (HD-GYP and YfbR), its members are missing the aforementioned Arg residue of YfbR, and a biochemical screen for phosphohydrolase/nucleotidase activity found E. coli YedJ to lack such activity (9). Thus, these proteins are more likely MVDOs rather than phosphohydrolases, as they have been annotated. The clade (in violet; SI Appendix, Fig. S13) headed by a structurally characterized protein (PDB ID code 2PQ7) from an uncultured Thermotogales species, named by us ThpO (Thermotogales putative oxygenase), is very closely related to the YedJ clade and also comprises proteins of unknown function. Like the YedJ clade, the ThpO clade is phylogenetically closer to PhnZ than to the known hydrolases. The similarity relationships and the absence of the Arg residue of YfbR imply that ThpO is a MVDO. The two phosphates bound in the structure might hint at oxidative cleavage of a phosphonate, which would yield phosphate as a product, as in the reaction catalyzed by PhnZ.
Discussion
Our results demonstrate that the expanded catalytic repertoire of HD proteins originally suggested by the characterization of MIOX extends to other members capable of oxidative C-X bond cleavage using a mixed-valent diiron cofactor. The phylogenetic analysis of the dimetal HD proteins reveals that there are potentially many more MVDOs within the superfamily, illustrating how a common protein scaffold can accommodate multiple, very different catalytic functions. The association of some members of the dimetal family with hydrolase activity raises the interesting questions of whether the mixed-valent FeII/FeIII redox state is stabilized specifically in the MVDOs or inherently by the HD architecture and whether it might also support hydrolase function. If stabilization of the mixed-valent state proves to be specific to the MVDOs, it would provide a criterion for assigning functions to newly discovered HD proteins. Alternatively, should HD hydrolases prove also to stabilize the FeII/FeIII forms of their cofactors and, perhaps, even use them in catalysis (as the non-HD phosphatase uteroferrin does) (46), it would suggest that the ability of the HD architecture to stabilize the FeII/FeIII state, with its unique capacity for both Lewis acid and O2-activation facilities, might have been a crucial prerequisite for the divergent evolution of the HD superfamily into hydrolases and oxygenases. Hydrolases that are active in the mixed-valent state might in this case represent common ancestors or evolutionary intermediates between HD proteins with hydrolase and oxygenase function. Characterizing representative HD proteins of the various clades in the phylogenetic tree should allow these intriguing questions to be answered and should further enable identification of specific hallmarks of the two activities. Such determinants would then facilitate functional assignment of proteins in the largely uncharacterized HD superfamily.
Materials and Methods
A detailed description of materials and methods is provided in the SI Appendix, Materials and Methods. It describes the procedures employed in overexpression and purification of PhnZ, the enzymatic synthesis of OH-AEP from 2-AEP using PhnY, and PhnZ activity assays with detection by NMR spectroscopy and mass spectrometry. Finally, it details the preparation of FQ samples, Mössbauer and EPR spectroscopic methods, the crystallographic methods, and the phylogenetic analysis.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB1330784 (to J.M.B. and C.K.) and National Institutes of Health Pathway to Independence Award K99/R00 (to A.K.B.). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and Michigan Technology Tri-Corridor Grant 085P1000817.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4N6W and 4N71).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315927110/-/DCSupplemental.
References
- 1.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 2.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLOS Comput Biol. 2009;5(12):e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23(12):469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 4.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: Protein homology by domain architecture. Genome Res. 2002;12(10):1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin MY, Koonin EV. Divergence and convergence in enzyme evolution. J Biol Chem. 2012;287(1):21–28. doi: 10.1074/jbc.R111.241976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown PM, et al. Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism. Proc Natl Acad Sci USA. 2006;103(41):15032–15037. doi: 10.1073/pnas.0605143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. MBio. 2011;2(5):e00163–11. doi: 10.1128/mBio.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galperin MY, Natale DA, Aravind L, Koonin EV. A specialized version of the HD hydrolase domain implicated in signal transduction. J Mol Microbiol Biotechnol. 1999;1(2):303–305. [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerman MD, Proudfoot M, Yakunin A, Minor W. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: The 5′-deoxyribonucleotidase YfbR from Escherichia coli. J Mol Biol. 2008;378(1):215–226. doi: 10.1016/j.jmb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE. 2011;6(8):e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 13.Charalampous FC. Biochemical studies on inositol. V. Purification and properties of the enzyme that cleaves inositol to D-glucuronic acid. J Biol Chem. 1959;234(2):220–227. [PubMed] [Google Scholar]
- 14.Moskala R, Reddy CC, Minard RD, Hamilton GA. An oxygen-18 tracer investigation of the mechanism of myo-inositol oxygenase. Biochem Biophys Res Commun. 1981;99(1):107–113. doi: 10.1016/0006-291x(81)91719-8. [DOI] [PubMed] [Google Scholar]
- 15.Bollinger JM, Jr, Diao Y, Matthews ML, Xing G, Krebs C. myo-Inositol oxygenase: A radical new pathway for O(2) and C-H activation at a nonheme diiron cluster. Dalton Trans. 2009;(6):905–914. doi: 10.1039/b811885j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing G, et al. A coupled dinuclear iron cluster that is perturbed by substrate binding in myo-inositol oxygenase. Biochemistry. 2006;45(17):5393–5401. doi: 10.1021/bi0519607. [DOI] [PubMed] [Google Scholar]
- 17.Xing G, et al. Oxygen activation by a mixed-valent, diiron(II/III) cluster in the glycol cleavage reaction catalyzed by myo-inositol oxygenase. Biochemistry. 2006;45(17):5402–5412. doi: 10.1021/bi0526276. [DOI] [PubMed] [Google Scholar]
- 18.Xing G, et al. Evidence for C-H cleavage by an iron-superoxide complex in the glycol cleavage reaction catalyzed by myo-inositol oxygenase. Proc Natl Acad Sci USA. 2006;103(16):6130–6135. doi: 10.1073/pnas.0508473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McSorley FR, et al. PhnY and PhnZ comprise a new oxidative pathway for enzymatic cleavage of a carbon-phosphorus bond. J Am Chem Soc. 2012;134(20):8364–8367. doi: 10.1021/ja302072f. [DOI] [PubMed] [Google Scholar]
- 20.Martinez A, Tyson GW, Delong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12(1):222–238. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- 21.Villarreal-Chiu JF, Quinn JP, McGrath JW. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front Microbiol. 2012;3:19. doi: 10.3389/fmicb.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metcalf WW, van der Donk WA. Biosynthesis of phosphonic and phosphinic acid natural products. Annu Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White AK, Metcalf WW. Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- 24.Quinn JP, Kulakova AN, Cooley NA, McGrath JW. New ways to break an old bond: The bacterial carbon-phosphorus hydrolases and their role in biogeochemical phosphorus cycling. Environ Microbiol. 2007;9(10):2392–2400. doi: 10.1111/j.1462-2920.2007.01397.x. [DOI] [PubMed] [Google Scholar]
- 25.Kamat SS, Raushel FM. The enzymatic conversion of phosphonates to phosphate by bacteria. Curr Opin Chem Biol. 2013;17(4):589–596. doi: 10.1016/j.cbpa.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Chang WC, Mansoorabadi SO, Liu HW. Reaction of HppE with substrate analogues: Evidence for carbon-phosphorus bond cleavage by a carbocation rearrangement. J Am Chem Soc. 2013;135(22):8153–8156. doi: 10.1021/ja403441x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shanklin J, Achim C, Schmidt H, Fox BG, Münck E. Mössbauer studies of alkane omega-hydroxylase: Evidence for a diiron cluster in an integral-membrane enzyme. Proc Natl Acad Sci USA. 1997;94(7):2981–2986. doi: 10.1073/pnas.94.7.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox BG, et al. Mössbauer, EPR, and ENDOR studies of the hydroxylase and reductase components of methane monooxygenase from Methylosinus trichosporium OB3b. J Am Chem Soc. 1993;115(9):3688–3701. [Google Scholar]
- 29.Lynch JB, Juarez-Garcia C, Münck E, Que L., Jr Mössbauer and EPR studies of the binuclear iron center in ribonucleotide reductase from Escherichia coli. A new iron-to-protein stoichiometry. J Biol Chem. 1989;264(14):8091–8096. [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 31.Thorsell AG, et al. Structural and biophysical characterization of human myo-inositol oxygenase. J Biol Chem. 2008;283(22):15209–15216. doi: 10.1074/jbc.M800348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu KE, et al. Spectroscopic detection of intermediates in the reaction of dioxygen with the reduced methane monooxygenase/hydroxylase from Methylococcus capsulatus (Bath) J Am Chem Soc. 1994;116(16):7465–7466. [Google Scholar]
- 33.Tong WH, et al. Mechanism of assembly of the diferric cluster-tyrosyl radical cofactor of Escherichia coli ribonucleotide reductase from the diferrous form of the R2 subunit. J Am Chem Soc. 1996;118(8):2107–2108. [Google Scholar]
- 34.Broadwater JA, Ai J, Loehr TM, Sanders-Loehr J, Fox BG. Peroxodiferric intermediate of stearoyl-acyl carrier protein delta 9 desaturase: Oxidase reactivity during single turnover and implications for the mechanism of desaturation. Biochemistry. 1998;37(42):14664–14671. doi: 10.1021/bi981839i. [DOI] [PubMed] [Google Scholar]
- 35.Pereira AS, et al. Direct spectroscopic and kinetic evidence for the involvement of a peroxodiferric intermediate during the ferroxidase reaction in fast ferritin mineralization. Biochemistry. 1998;37(28):9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- 36.Murray LJ, et al. Characterization of the arene-oxidizing intermediate in ToMOH as a diiron(III) species. J Am Chem Soc. 2007;129(46):14500–14510. doi: 10.1021/ja076121h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korboukh VK, Li N, Barr EW, Bollinger JM, Jr, Krebs C. A long-lived, substrate-hydroxylating peroxodiiron(III/III) intermediate in the amine oxygenase, AurF, from Streptomyces thioluteus. J Am Chem Soc. 2009;131(38):13608–13609. doi: 10.1021/ja9064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandelia ME, et al. Substrate-triggered addition of dioxygen to the diferrous cofactor of aldehyde-deformylating oxygenase to form a diferric-peroxide intermediate. J Am Chem Soc. 2013;135(42):15801–15812. doi: 10.1021/ja405047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davydov RM, et al. EPR study of the mixed-valent diiron sites in mouse and herpes simplex virus ribonucleotide reductases. Effect of the tyrosyl radical on structure and reactivity of the diferric center. Biochemistry. 1997;36(30):9093–9100. doi: 10.1021/bi9700375. [DOI] [PubMed] [Google Scholar]
- 40.Makris TM, Chakrabarti M, Münck E, Lipscomb JD. A family of diiron monooxygenases catalyzing amino acid beta-hydroxylation in antibiotic biosynthesis. Proc Natl Acad Sci USA. 2010;107(35):15391–15396. doi: 10.1073/pnas.1007953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan RP, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA. 2006;103(17):6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Sultan SZ, et al. Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect Immun. 2011;79(8):3273–3283. doi: 10.1128/IAI.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miner KD, Klose KE, Kurtz DM., Jr An HD-GYP cyclic-di-guanosine monophosphate phosphodiesterase with a non-heme diiron-carboxylate active site. Biochemistry. 2013;52(32):5329–5331. doi: 10.1021/bi4009215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindstedt G, Lindstedt S. Cofactor requirements of γ-butyrobetaine hydroxylase from rat liver. J Biol Chem. 1970;245(16):4178–4186. [PubMed] [Google Scholar]
- 45.Leung IK, et al. Structural and mechanistic studies on γ-butyrobetaine hydroxylase. Chem Biol. 2010;17(12):1316–1324. doi: 10.1016/j.chembiol.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Antanaitis BC, Aisen P. EPR signal, purple color, and iron binding in porcine uteroferrin. J Biol Chem. 1982;257(4):1855–1859. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.