Abstract
Upon infection, CD8+ T cells proliferate and differentiate into armed effector cells capable of eliminating the assaulting pathogen. Although the majority of the antigen-specific T cells will die as the immune response wanes, a few will survive indefinitely to establish the memory population and provide long-lived protection against reinfection. E protein transcription factors and their inhibitors, ID proteins, operate to balance expression of genes that control CD8+ T cell differentiation through this process. Here, we discuss the role of ID2 and ID3 in promoting the generation and survival of effector and memory populations, particularly highlighting their reciprocal roles in shaping the CD8+ T cell response unique to the inflammatory milieu. We further examine this coordinated control of gene expression in the context of additional transcription factors within the transcriptional network that programs CD8+ effector and memory T cell differentiation.
Introduction
In response to infection, a naive pathogen-specific CD8+ T cell undergoes a massive proliferative burst, during which one cell is capable of generating upwards of tens of thousands of progeny [1]. This expansion is concurrent with dramatic alterations in gene expression due to modifications in chromatin structure and expression of key transcription factors [2,3]. These changes also coincide with the acquisition of effector function including the capacity to secrete effector cytokines interferon (IFN)γ and tumor necrosis factor (TNF)α, and release cytolytic molecules such as perforin and granzymes to eliminate pathogen-infected cells [1,4,5]. This expanded population of CD8+ T cells is a heterogeneous mixture of cells that include short-lived memory and effector T cells ((which can be identified by high levels of the surface receptor killer cell lectin-like receptor G1 (KLRG1) and low levels of Interleukin-7 receptor (IL-7Rα), CD127)), as well as memory-precursor cells (contained within the KLRG1loCD127hi population) [5]. As indicated by their monikers, most short-lived effector cells will survive as a population for only a matter of days during the height of the immune response, after which they undergo a rapid contraction phase. The majority of the KLRG1loCD127hi effector population, which contains memory-precursor cells, also succumbs to programmed cell death after infection. However, ~5% of the effector cells endure and persist in greater numbers than their naive precursors, and are transcriptionally programed to seed the long-lived memory pool providing protection against re-infection [1,5-7].
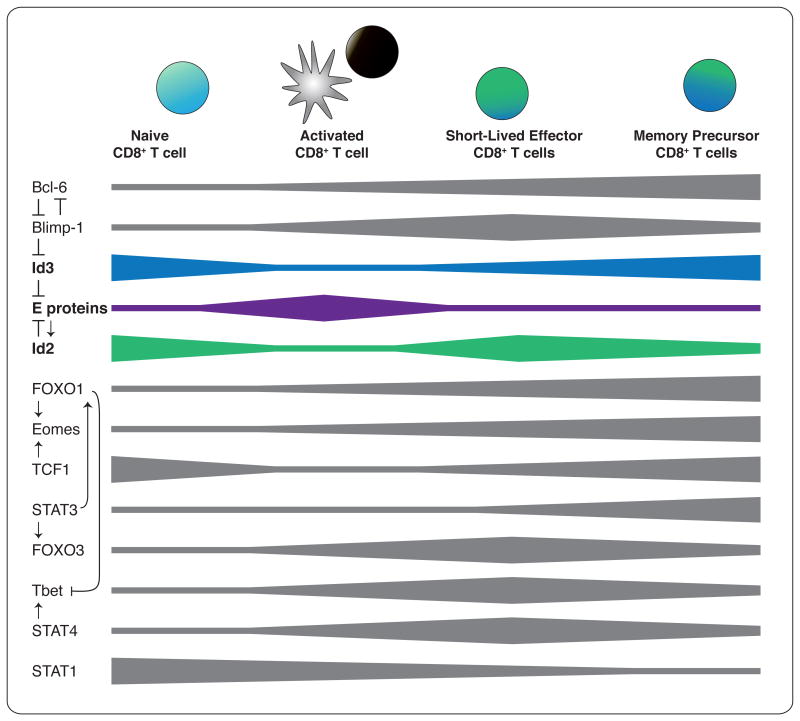
Of particular interest in the study of CD8+ T cell immunity are the transcriptional networks and targeted gene-expression changes that orchestrate the bifurcation of differentiation—mediating the short half-life of the effector cells versus the longevity of memory cells. While the integrated balance of expression and activity of T-BET, B lymphocyte-induced maturation protein-1 (BLIMP-1), signal transducer and activator of transcription (STAT) 4 and forkhead box O (FOXO) 3 have been shown to regulate effector cell differentiation, survival and contraction; eomesodermin (EOMES), B cell lymphoma-6 (BCL-6), T cell factor-1 (TCF-1), STAT3, and FOXO1 influence the generation and maintenance of memory cells [8-10] (Figure 1). Recently it was discovered that E and the inhibitor of DNA binding (ID) proteins also regulate the differentiation of both the shortlived effector and memory-precursor populations of CD8+ T cells [11-15]. This raises the possibility of an analogous role for these molecules in determining CD4+ T cell fate.
Figure 1. Interplay of transcription factor networks during CD8+ T cell activation and differentiation.
Width of bar indicates transcription factor activity and/or expression.
E proteins
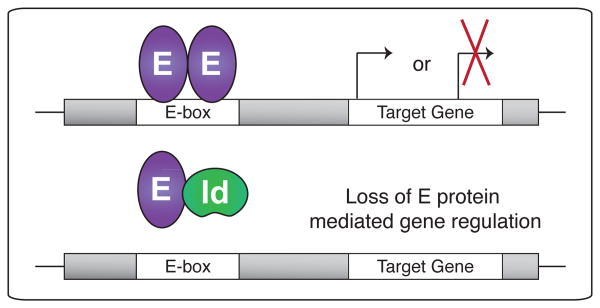
E proteins are transcription factors in the basic helix-loop-helix (bHLH) family that control many aspects of lymphocyte biology [16]. Four different E proteins, E12 and E47 (splice variants of E2A), E2-2 and HEB, are present in mammals. E proteins can interact as homo- and hetero-dimers via their HLH domains and bind specifically to DNA at E-box-consensus sequences acting as transcriptional activators or repressors (Figure 2) [16,17]. The ability of E proteins to bind DNA and regulate gene expression is inhibited by the highly related ID proteins, which share the HLH domain and thus form heterodimers with E proteins, but lack a DNA-binding domain, preventing E protein function (Figure 2) [18,19].
Figure 2. E protein activity is regulated by Id proteins.
E protein dimers bound to DNA can activate (top) or repress gene transcription. When E proteins heterodimerize with Id proteins, DNA binding is inhibited, also blocking target gene transcription (bottom).
E proteins are well-established regulators of thymocyte development and are required for proper control of progression, survival, proliferation and T cell receptor (TCR) rearrangements by T cell progenitors [16]. It is now clear that E proteins are also active in the early stages of mature T cell activation and induce expression of genes important for commitment to the memory lineage. E2A expression is upregulated by CD8+ T cells upon activation, and increased E protein DNA-binding activity is observed in antigen-specific CD8+ T cells early during infection (Figure 3) [20]. Deletion of E2A, E2-2, or HEB had minimal effects on the expansion and phenotype of CD8+ T cells responding to infection, indicating compensatory functions between E proteins in this context. However, deficiency in both E2A and HEB resulted in an increased frequency of KLRG1hi terminally-differentiated effectors [20]. Activated CD8+ T cells lacking E proteins exhibited altered gene-expression profiles with upregulation of genes linked to early effector populations and activation (CD28, Lymphocyte activation gene 3 (Lag3)) and a downregulation of genes associated with memory formation (II7r, Eomes) [20]. The genes identified to be differentially regulated upon loss of E proteins also possessed E2A-bound E-box sites in close proximity to their transcriptional start site (TSS), strongly suggesting direct regulation by E proteins [21]. Overall, these studies suggest that E proteins regulate transcription factors, cell-surface markers, and cytokine signaling early during CD8+ T cell activation to support memory-precursor formation [20].
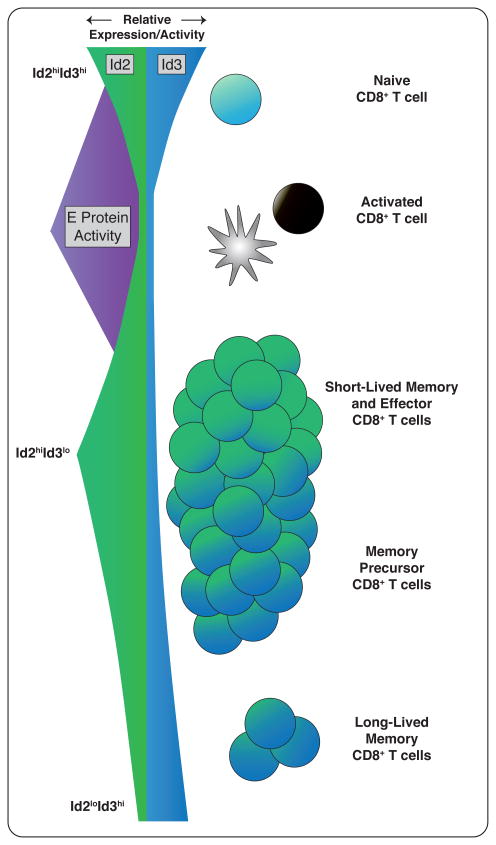
Figure 3. E and Id proteins show coordinated expression during CD8+ T cell activation and differentiation.
Id2 and Id3 levels are downregulated upon CD8+ T cell stimulation, coincident with an increase in E protein DNA-binding activity. Id2 expression increases at the peak of infection, promoting survival and terminal differentiation. Conversely, Id3 expression is downregulated at the peak of infection, but increases to mark memory precursors and maintain the long-lived memory pool.
Recent studies have provided a further link between E proteins and the regulation of other transcription factors central to cell-fate decisions of CD8+ T cells. E proteins are known to directly impact the expression of the FOXO transcription factors [21,22]. Interestingly, two family members, FOXO1 and FOXO3, have been recognized as important to memory formation [23-25]. While FOXO1 appears to be a key mediator of CD8+ T cells differentiating into long-lived memory cells [23], FOXO3 is suggested to function during the contraction phase of the T cell response [24-26]. E proteins have also been shown to regulate TCF-1 expression, a factor central in CD8+ T cell immunity [13,27]. TCF-1, also a likely E protein target, was shown to control Eomes expression and the differentiation, maintenance and function of CD8+ T cell memory [28]. From these studies, it is clear that E proteins play an important role in orchestrating the transcriptional network necessary for the generation of productive CD8+ T cell memory.
ID Proteins
There are four ID family members (ID1-ID4); with ID2 and ID3 emerging as the players relevant in shaping lymphocyte differentiation [16,18,19]. While ID protein-mediated regulation of E proteins is known to be crucial to lymphocyte development, the role of these factors in mature T cells is only beginning to be revealed. Importantly, a reciprocal relationship between ID2 and ID3 has been suggested in the differentiation of mature T cells during the response to infection [14]. Studies with knock-in reporter mice have shown that Id2 is upregulated in effector CD8+ T cells while induction of Id3 expression marked CD8+ T cells with memory potential [14].
ID2
Although downregulated early in infection, ID2 expression is upregulated at the peak and maintained in memory T cells, albeit at lower levels (Figure 3) [11,13,14,20]. ID2 plays important roles in the CD8+ T cell response to infection by mediating survival and differentiation of effector cells and repressing memory formation [11-13]. Naive CD8+ T cells lacking ID2 show normal activation, proliferation and differentiation into effector cells. However, they fail to accumulate through the course of infection with Listeria monocytogenes, Vesicular Stomatitis Virus and Lymphocytic Choriomeningitis Virus due to increased susceptibility to apoptosis [11]. The absence of cells to analyze following infection left it unclear how ID2-deficiency further affected CD8+ T cell responses. Two recent papers have begun to address this issue and show clear evidence that differentiation, as well as survival, are controlled by ID2 [12,13].
Knell et al. determined that ID2 regulates CD8+ T cell responses at two levels by ensuring survival of effector cells as well as influencing effector to memory cell differentiation [12]. Consistent with the observation of defective survival, ID2-knockout effector CD8+ T cells displayed lower expression of the antiapoptotic molecules B cell lymphoma-2 (Bcl-2) and serine protease inhibitor 6 (Spi6), and higher levels of the proapoptotic factors Bcl2l11 (encoding Bcl-2-interacting mediator of cell death (BIM)) and cytotoxic T-lymphocyte antigen 4 (Ctla4) [11]. Closer examination identified >30 conserved E-box sites in the Bcl2l11 promoter, and as such, TCR activation led to increased E protein-binding at these E-box sites in CD8+ T cells [12]. Interestingly, loss of BIM expression rescued the accumulation defect observed for ID2-deficient CD8+ T cells, further defining a role for ID2 in effector cell survival [12]. Strikingly, we found that ID2-deficient and “rescued” ID2-deficient CD8+ T cells almost completely lacked a terminally differentiated KLRG1hi effector population during Listeria monocytogenes infection [12,14]. The effector cells that did form exhibited a cell-surface phenotype, cytokine and gene-expression profile consistent with memory-precursor cells [12]. In fact, microarray analysis revealed upregulation of several genes involved in memory cell formation or function including Id3, Tcf7 (encodes TCF-1), and Cxcr3 [12].
In a second study, ID2-deficient CD8+ T cells were found to have a significant defect in formation of the terminally-differentiated KLRG1hi population [13]. In this case, ID2-deficient CD8+ T cells accumulated normally during Influenza virus infection but not Listeria monocytogenes infection, suggesting that the specific inflammatory milieu or type of infection (systemic versus localized) may yield environments that differentially induce and/or require ID2 [13]. This study took advantage of the normal ID2-deficient CD8+ effector population following Influenza infection to study how loss of ID2 impacts effector and memory cell differentiation, and found an inverse correlation between ID2 levels and memory recall potential [13]. Consistent with this observation, the transcriptional program of the responding CD8+ T cells was influenced by ID2 expression; low or no ID2 resulted in high expression of characteristic memory cell-associated transcription factors, low expression of cytolytic molecules and altered integrin and chemokine receptors that promote localization in non-lymphoid tissues [13]. ID2 function appeared to be mediated through inhibition of E2A as the genes upregulated in the absence of ID2 largely contained E2A binding sites. Furthermore, shRNA knockdown of Tcfe2a expression partially compensated for ID2-deficiency, yielding increased expression of effector molecules and diminished expression of characteristic memory genes such as Tcf7, Id3 and suppressor of cytokine signalling (Socs)3 [13]. Conversely, Tbet, a key regulator of effector differentiation, was 2-fold reduced in ID2-deficient effector cells and rescue of expression by retroviral infection induced formation of KLRG1hi cells and somewhat restored granzyme and BLIMP-1 expression, suggesting ID2 is at least indirectly responsible for inducing Tbet which then supports the generation of short-lived effector CD8+ T cells [13]. Collectively, these studies demonstrate that ID2 mediates both survival of CD8+ T cells during the effector phase of infection and the differentiation of KLRG1hi cells, shaping the composition of the effector and memory populations.
ID3
ID3 is expressed at its highest level in naive CD8+ T cells and is rapidly downregulated upon activation. Expression of ID3 later increases during contraction of the effector response and coincides with the appearance of memory CD8+ T cells, effectively acting as a marker of memory-precursor cells (Figure 3) [14]. Defining a role for ID3 in T cell differentiation has been hindered by the complexity of the ID3-knockout mouse model. While viable, ID3 germline-knockout mice have a multifaceted phenotype that includes an outgrowth of Vγ1.1+Vδ6.3+ T cells in the thymus and spleen, excessive IFNγ and interleukin (IL)-4 production by these innate-like cells [26,29-32], as well as T cell-dependent Sjögren's autoimmune syndrome [33,34]. In light of this confounding phenotype, studies have utilized ID3 heterozygous GFP-reporter mice and mixed-bone marrow chimeras to circumvent extrinsic effects of ID3-deficiency. Importantly, at the height of the CD8+ T cell response, cells with little to no expression of ID3 were chiefly KLRG1hiCD127lo short-lived effector cells, while those expressing the highest level of ID3 were largely KLRG1loCD127hi memory-precursor T cells producing more IL-2, IFNγ and TNF [14]. This functional divergence based on ID3 expression was further supported by transfer of ID3hi and ID3lo populations sorted from the peak of infection, which revealed that the ID3hi cells preferentially differentiated into memory CD8+ T cells and that ID3 expression preceded other traditional phenotypic indicators of memory potential [14]. To investigate whether ID3 is a marker of memory-precursor CD8+ T cells at the transcriptional level, global gene-expression analyses was performed at effector and memory time points. Hallmark CD8+ T cell memory transcripts were expressed by ID3hi effector cells as early as day 5 of infection (IL7r, Sell, IL3, Bcl2 and Ccr7,) whereas transcripts indicative of terminal differentiation were downregulated (Gzmb, Klrg1, and Prdm1) [14]. Indeed, analysis of either ID3-wildtype:ID3-knockout mixed-bone marrow chimeras or adoptively transferred ID3-deficient melanoma-specific TCR-transgenic (pmel-1) CD8+ T cells, both revealed that loss of ID3 led to defects in the formation and survival of a long-lived memory, despite having little effect on population expansion [14,15]. Conversely, ectopic expression of ID3 in pmel-1 CD8+ T cells increased their accumulation following secondary viral challenge and enhanced survival after challenge with B16 melanoma compared to controls [15]. Taken together, this study concluded that the formation of a functional memory compartment is dependent on ID3 expression and forced expression of ID3 improved the secondary response through an increase in the number of effector-memory T cells.
Coordination of ID2 and ID3
ID2 and ID3 appear to have distinct roles in CD8+ T cell differentiation; however, it is the timing of their expression that is required to regulate E protein activity and orchestrate the transition from naive to effector or memory cell (Figure 3). Interestingly, in the absence of ID2, ID3 is significantly upregulated in effector cells; however, the reverse does not occur and ID2 remains unchanged in ID3-deficient CD8+ T cells, indicating that ID2 regulates expression of ID3 [14]. Consistent with this, E-box binding sites have been identified in the Id3 locus [13,21,35]. Thus, factors that control Id2 expression will likely also inversely impact Id3 expression via ID2-mediated inhibition of E protein activity.
Though the regulation of ID3 by ID2 needs to be explored further, the direct repression of ID3 by BLIMP-1 has been demonstrated [15]. BLIMP-1 expression is inversely correlated with that of ID3 as well, with chromatin immunoprecipitation (ChIP) analysis showing BLIMP-1 bound to the Id3 promoter region [15]. Since ID3 is required for the persistence of long-term memory cells, BLIMP-1 presumably represses Id3 transcription and prevents the over-accumulation of memory T cells. In support of this idea, ID3 retroviral overexpression, where BLIMP-1 cannot efficiently repress Id3, resulted in a blunted contraction phase and increased persistence of effector cells into the memory phase [15].
Extrinsic factors also influence the balance of ID2 and ID3 levels within a differentiating CD8+ T cell, dictating its fate. In support of this, cytokine signaling through STAT4 and 5 was demonstrated to be important for the modulation of ID2 and ID3 expression by CD8+ T cells. Using the ID2- and ID3-reporter mice, we found that activation of CD8+ T cells in the presence of IL-2, IL-12 and IL-21 upregulated Id2 expression but lead to downregulation of Id3. Furthermore, ChIP analysis identified STAT-binding sites in the Id2 promoter [14]. Such environmental control of ID protein expression may provide an explanation for infection-specific differences observed in ID2-mediated CD8+ T cell survival [11,13]. During Influenza infection, loss of ID2 does not impair the accumulation of antigen-specific cells as compared to other systemic, highly inflammatory infections [11,12]. The cytokine profile of Influenza infection may not induce significant upregulation of ID2; however, at the same time, ID3 expression would not be substantially decreased, so it may function in a compensatory manner to control the transcriptional program. This would include providing protection from elevated BIM levels and apoptosis. Thus, the information provided by the infection itself may instruct the differentiating T cells to generate effector and memory populations optimal for the response.
The regulation of ID2 and ID3 expression and their relative levels over time within a responding T cell is likely a major determinant of its fate. Early during infection, E protein expression is upregulated, coinciding with downregulation of ID2 and ID3. This allows E proteins to target gene expression, inducing formation of memory precursors [11,14,15,20]. As the T cell response peaks, ID2 protein levels increase, possibly induced through cytokine signaling. E protein activity is then inhibited, permitting the survival and differentiation of late CD8+ effector cells [13,20]. Reciprocally, cells expressing ID3 and low levels of ID2 are memory precursors (Figure 3). Although both ID2 and ID3 are thought to similarly function by repressing E protein activity, there is a clear distinction in the role they play in CD8+ T cell differentiation. This could be a result of variation in E protein binding, such as a preference for specific homo- or heterodimers or the affinity with which the interaction takes place. Further studies clarifying the downstream binding partners of ID2 and ID3 will be necessary to interpret their individual contributions to CD8+ T cell responses.
E and ID proteins in CD4+ T cells
In contrast to CD8+ T cell differentiation where memory subsets have been defined in substantial detail, the gene-expression and phenotypic changes that CD4+ effector T cells undergo during memory formation is less clear. Since effector CD4+ T cells can differentiate into multiple helper (Th) populations [36], additional complexity exists and analysis of memory formation from each effector subset needs to be established. As in the case of CD8+ T cell responses, relative ID2 and ID3 levels may act as novel markers of early CD4+ T cell memory-precursors, in addition to regulating gene-expression programs that govern effector versus memory cell formation. Recently, it was demonstrated that Id2 was highly expressed in the Th1 population, whereas Id3 transcript was almost exclusively expressed in the T follicular helper (Tfh) population after infection [37]. Another study demonstrated that ID3-deficiency resulted in aberrant formation of effector-memory-like CD4+ T cells suggesting that ID3 is important for the maintenance of the naïve state. Furthermore, ID3-deficiency led to the upregulation of Tfh markers at the RNA (Bcl6) and protein level (CXCR5, ICOS and PD-1), as well as elevated IFNγ and IL-4 production following stimulation [35]. Further, ID2 and ID3 have been examined in models of CD4+ T cell-mediated autoimmunity. ID2 was shown to be an important factor in the development of experimental autoimmune encephalomyelitis with high ID2 expression in the most encephalitogenic CD4+ T cells [38]. ID2-deficiency led to decreased numbers of effector IL17A+IFNγ+CD4+ T cells due to reduced proliferation, and similar to CD8+ T cells, increased apoptosis. ID2 also appeared to be important in mediating cytokine production in this system by regulating expression of the repressor SOCS3 [38]. ID3 has also been implicated in the TGFβ1-dependent reciprocal regulation of T regulatory (Treg) and Th17 development [39-41]. In the absence of ID3, Tregs do not properly develop as a result of an inability to inhibit GATA-3 binding to the Foxp3 promoter; instead, Th17 differentiation is increased both in vitro and in vivo in a model of asthma [39]. These studies suggest that the balance between ID2 and ID3 will also be important in the fate decisions of mature CD4+ T cells as they respond to infection and differentiate into distinct effector and memory populations.
Conclusion
The idea that pairs of transcription factors function in a coordinated and, at times, opposing fashion has been proposed for T-BET/EOMES, BLIMP-1/BCL-6, ZEB1/ZEB2 and STAT3/STAT4 [8]. Similarly, it is becoming clear that E and ID proteins work to balance the generation of short-term effector-memory versus long-lived memory populations. Over the course of the CD8+ T cell response, ID2 and ID3 also demonstrate reciprocal expression and function, suggesting a complex interplay in their regulation of E protein activity that is not yet fully understood. Ultimately, it is how these pairs function together to direct the transcriptional programs required for mature T cell differentiation that is of greatest interest. Future studies identifying the direct gene targets for each of these transcriptional regulators in the context of global gene-expression changes will be critical for assembling the comprehensive transcriptional network governing CD8+ T cell immunity.
Highlights.
E and ID proteins control many aspects of CD8+ T cell differentiation.
ID2 mediates effector cell survival and differentiation to memory.
ID3 marks cells as memory precursors and maintains the longevity of these cells.
ID2/ID3 coordination, controlled by the infection milieu, shapes the CD8+ T cell response.
Acknowledgments
We thank the Goldrath laboratory members for critical review of the manuscript including A. Phan and Dr. A. Best. This work was supported by grants from the NIH to A.W.G. and by the Leukemia and Lymphoma Society (A.W.G. and K.D.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 3.Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, Dustin ML, Goldrath AW. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 5.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med. 2001;7:913–919. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 7.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 8.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12:649–661. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Cruz LM, Rubinstein MP, Goldrath AW. Surviving the crash: transitioning from effector to memory CD8+ T cell. Semin Immunol. 2009;21:92–98. doi: 10.1016/j.smim.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. This study was the first to identify a role for ID2 in ensuring survival of effector CD8+ T cell as well as in controlling the transition to memory. [DOI] [PubMed] [Google Scholar]
- 12*.Knell J, Best JA, Lind NA, Yang E, D'Cruz LM, Goldrath AW. Id2 influences differentiation of killer cell lectin-like receptor G1(hi) short-lived CD8+ effector T cells. J Immunol. 2013;190:1501–1509. doi: 10.4049/jimmunol.1200750. This study with Ref. [13] demonstrates that ID2 is important for regulating the effector versus memory CD8+ T cell differentiation programs and is required for the formation of the terminally-differentiated KLRG1hi CD8+ T cell population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, Speed TP, Busslinger M, Nutt SL, Belz GT. Id2-Mediated Inhibition of E2A Represses Memory CD8+ T Cell Differentiation. J Immunol. 2013;190:4585–4594. doi: 10.4049/jimmunol.1300099. See annotation to Ref. [12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14**.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D'Cruz LM, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ Tcell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. This study with Ref. [15] establishes the requirement for ID3 in the formation and maintenance of long-lived CD8+ T cell memory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, Reger RN, Palmer DC, Borman ZA, Muranski P, et al. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12:1230–1237. doi: 10.1038/ni.2153. See annotation to Ref. [14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 17.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 18.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 20*.D'Cruz LM, Lind KC, Wu BB, Fujimoto JK, Goldrath AW. Loss of E protein transcription factors E2A and HEB delays memory-precursor formation during the CD8+ T- cell immune response. Eur J Immunol. 2012;42:2031–2041. doi: 10.1002/eji.201242497. This study finds increased E protien expression and activity early during CD8+ T cell responses and suggests this favors rapid formation of memory-precurosors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welinder E, Mansson R, Mercer EM, Bryder D, Sigvardsson M, Murre C. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci USA. 2011;108:17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Hess Michelini R, Doedens AL, Goldrath AW, Hedrick SM. Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med. 2013;210:1189–1200. doi: 10.1084/jem.20130392. This study reveals a critical role for FOXO1 in the CD8+ T cell effecter versus memory precursor lineage decision which may provide a link between E protein regulation and CD8+ T cell memory formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan JA, Kim EH, Plisch EH, Peng SL, Suresh M. FOXO3 regulates CD8 T cell memory by T cell-intrinsic mechanisms. PLoS Pathog. 2012;8:e1002533. doi: 10.1371/journal.ppat.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzelepis F, Joseph J, Haddad EK, Maclean S, Dudani R, Agenes F, Peng SL, Sekaly RP, Sad S. Intrinsic role of FoxO3a in the development of CD8+ T cell memory. J Immunol. 2013;190:1066–1075. doi: 10.4049/jimmunol.1200639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikawa T, Kawamoto H, Wright LY, Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- 30.Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent ″innate″ gammadelta T cells. PLoS One. 2010;5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T- like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Dai M, Zhuang Y. A T cell intrinsic role of Id3 in a mouse model for primary Sjogren's syndrome. Immunity. 2004;21:551–560. doi: 10.1016/j.immuni.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Guo Z, Li H, Han M, Xu T, Wu X, Zhuang Y. Modeling Sjogren's syndrome with Id3 conditional knockout mice. Immunol Lett. 2011;135:34–42. doi: 10.1016/j.imlet.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. This study determines that Id3 plays an important role in maintaining the naïve state of peripheral CD4+ T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pepper M, Jenkins MK. Origins of CD4(+) effector and central memory T cells. Nat Immunol. 2011;12:467–471. doi: 10.1038/ni.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, Crotty S. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol. 2013;190:4014–4026. doi: 10.4049/jimmunol.1202963. This study identifies differential expression of ID2 and ID3 in CD4+ T cell subsets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YY, Jones-Mason ME, Inoue M, Lasorella A, Iavarone A, Li QJ, Shinohara ML, Zhuang Y. Transcriptional regulator Id2 is required for the CD4 T cell immune response in the development of experimental autoimmune encephalomyelitis. J Immunol. 2012;189:1400–1405. doi: 10.4049/jimmunol.1200491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruyama T, Li J, Vaque JP, Konkel JE, Wang W, Zhang B, Zhang P, Zamarron BF, Yu D, Wu Y, et al. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat Immunol. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 41.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]