Abstract
MicroRNAs (miRs) are small noncoding RNA sequences that negatively regulate the expression of target genes by posttranscriptional repression. miRs are dysregulated in various diseases, including cancer. let-7a miR, an antioncogenic miR, is downregulated in lung cancers. Our earlier studies demonstrated that let-7a miR inhibits tumor growth in malignant pleural mesothelioma (MPM) and could be a potential therapeutic against lung cancer. EphA2 (ephrin type-A receptor 2) tyrosine kinase is overexpressed in most cancer cells, including MPM and non-small-cell lung cancer (NSCLC) cells. Ephrin-A1, a specific ligand of the EphA2 receptor, inhibits cell proliferation and migration. In this study, to enhance the delivery of miR, the miRs were encapsulated in the DOTAP (N-[1-(2.3-dioleoyloxy)propyl]-N,N,N-trimethyl ammonium)/Cholesterol/DSPE (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[cyanur(polyethylene glycol)-2000])-PEG (polyethylene glycol)-cyanur liposomal nanoparticles (LNP) and ephrin-A1 was conjugated on the surface of LNP to target receptor EphA2 on lung cancer cells. The LNP with an average diameter of 100 nm showed high stability, low cytotoxicity, and high loading efficiency of precursor let-7a miR and ephrin-A1. The ephrin-A1 conjugated LNP (ephrin-A1–LNP) and let-7a miR encapsulated LNP (miR–LNP) showed improved transfection efficiency against MPM and NSCLC. The effectiveness of targeted delivery of let-7a miR encapsulated ephrin-A1 conjugated LNP (miR–ephrin-A1–LNP) was determined on MPM and NSCLC tumor growth in vitro. miR–ephrin-A1–LNP significantly increased the delivery of let-7a miR in lung cancer cells when compared with free let-7a miR. In addition, the expression of target gene Ras was significantly repressed following miR–ephrin-A1–LNP treatment. Furthermore, the miR–ephrin-A1–LNP complex significantly inhibited MPM and NSCLC proliferation, migration, and tumor growth. Our results demonstrate that the engineered miR–ephrin-A1–LNP complex is an effective carrier for the targeted delivery of small RNA molecules to lung cancer cells. This could be a potential therapeutic approach against tumors overexpressing the EphA2 receptor.
Keywords: liposomal nanoparticles, EphA2 receptor, microRNA, ephrin-A1, malignant pleural mesothelioma, non-small-cell lung cancer
Introduction
MicroRNAs (miRs) are short, 20–25 nucleotide sequences that serve as master regulators of gene expression. miRs bind to the complementary sequences in the 3′ untranslated regions of target messenger RNA and result in transcriptional repression of target gene expression.1,2let-7 is one of the first known genes identified as a regulator of developmental timing and proliferation of cells.3 let-7 miR functions as a tumor suppressor by silencing the Ras gene, a member of the small guanosine triphosphate (GTP)ase superfamily associated with cell proliferation, adhesion, and migration in lung cancer and malignant pleural mesothelioma (MPM) cells.4,5 Although gene-silencing therapy has been widely studied, clinical use of miR has been limited due to its high vulnerability and low cellular uptake in systemic administration. Therefore, developing an adequate carrier system that can provide protection and stability to miR and efficient targeted delivery to the cancer cells/tumor is crucial.
Liposomes are biodegradable and could be used to deliver high concentrations of therapeutics to the tumor tissue. Liposomes composed of the cationic lipid DOTAP (N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl-sulfate) have been shown to be the effective carrier for the anionic nucleotides RNA and DNA.6–10 Due to the overall cationic electrostatic charge on the lipid bilayers, cationic liposomes provide advantages that include high encapsulation efficiency of nucleotides and high cellular uptake. Cationic liposomes have been demonstrated to selectively accumulate in angiogenic endothelial cells in tumors and to be internalized by endocytosis after intravenous injection.11,12 However, it has been reported that in the presence of serum, the binding of serum proteins to the cationic liposomes leads to structural reorganization and aggregation or dissociation of cationic liposomes and influences the delivery.13–16 To prevent the aggregation induced by serum, cationic liposomes have been incorporated with PEGylated (polyethylene glycol-ylated) lipids to increase circulation lifetime and allow the accumulation in tumor tissue. However, the delivery efficiency and cellular uptake of PEGylated liposomes may be compromised.17–21
The Eph-ephrin signaling proteins comprise the largest known family of receptor tyrosine kinases. This family influences processes of cell migration and patterning through their interactions with each other and additional signals in the surrounding microenvironment that require cell–cell contact. Among this family, EphA2 (ephrin type-A receptor 2) and its ligand, ephrin-A1, play an important role as modulators of various processes during embryonic development.22,23 EphA2 is overexpressed in aggressive malignancies including lung cancer and MPM, but not significantly in normal tissue.24–29 Ephrin-A1 inhibits proliferation and migration of MPM cells by downregulation of EphA2 expression via binding to the EphA2 receptor on the cell membrane.30–32 In addition, ephrin-A1/Fc specifically binds to EphA2 in pancreatic adenocarcinoma cells and suppress tumor growth and invasion.33 Furthermore, earlier we reported that the tumor-suppressive properties of ephrin-A1 are due to the expression of let-7a miR.34
In the present study, let-7a miR was encapsulated in the liposomal nanoparticle (LNP) as a carrier to protect and deliver miR to lung cancer cells. In addition, to enhance the effectiveness of delivery, the ephrin-A1 protein was conjugated on the surface via coupling with PEGylated lipid to specifically target the EphA2 receptors on MPM and non-small-cell lung cancer (NSCLC). The combination therapy of miR and ephrin-A1 showed enhanced effectiveness when tested on MPM and NSCLC cells in vitro when compared with either ephrin-A1 or miR–LNP alone. Furthermore, MPM cells treated with miR–ephrin-A1–LNP showed reduced expression of Ras, the target gene. Importantly, the cell proliferation, migration, and clonogenic expansion of lung cancer cells were remarkably reduced. Our engineered LNP have potential for the delivery of miRs for therapeutic interventions against MPM and NSCLC for in vivo studies.
Materials and methods
Materials
DOTAP, cholesterol, DSPE-PEG-2000 (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[cyanur(polyethylene glycol)-2000]), and DSPE-PEG2000-cyanur were purchased from Avanti Polar Lipids, Inc (Alabaster, AL, USA). All analytical reagents used in this study were purchased from Sigma-Aldrich (St Louis, MO, USA) unless otherwise indicated. The miR precursor, miR inhibitor, and control miRs were obtained from Ambion/Life Technologies (Carlsbad, CA, USA).
Cell culture and treatment
CRL-2081 and CRL-5830 cells (MPM cell lines) and A549 cells (NSCLC cell line) were purchased from American Type Culture Collection (Manassas, VA, USA) and maintained in 100 mm dishes in RPMI 1640 (Sigma-Aldrich) containing 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL), as reported earlier.4,24 In brief, cells were incubated at 37°C in 5% CO2 and 95% air and were cultured to 80% confluence for 3–4 days and subcultured in 60 mm cell culture dishes (5 × 105 cells/dish) for various treatments accordingly. MPM and NSCLC in near confluence cultures were treated with or without 3.5 μg/mL recombinant mouse Ephrin-A1/Fc chimera (R&D Systems, Minneapolis, MN, USA) in serum-free RPMI 1640, for the indicated times as reported earlier.4 The cells were also treated with ephrin-A1 conjugated LNP (ephrin-A1–LNP) or let-7a miR encapsulated LNP (miR–LNP) or miR-control–LNP or only LNP. The cells were processed for total lysates and total RNA for immunoblotting and quantitative real-time polymerase chain reaction (qPCR), respectively.
Preparation of miR encapsulated liposomal nanoparticles (miR–LNP)
Liposomes were prepared by using combination of DOTAP, cholesterol, and DSPE-PEG 2000-cyanur. In brief, the lipids, DOTAP, cholesterol, and DSPE-PEG-2000-cyanur (1:1:0.1 molar ratio) were dissolved in ethanol (40–100 μg/μL) and then dispersed into aqueous solution (molecular grade) containing let-7a miR as described in previous reports.35 The weight ratio of the miR to LNP was 1:10. The LNP and miR were well dispersed and vortexed for the formation of stable miR–LNP complex. The encapsulation of miR in the LNP was determined by gel electrophoresis. Free miR, miR–LNP, and miR–ephrin-A1–LNP complex were electrophoresed on 1% agarose gel, and bands were visualized using Chemidoc-MP System (Bio-Rad Laboratories, Hercules, CA, USA).
Ephrin-A1 conjugation on LNP (ephrin-A1–LNP)
To acquire a stable conjugation of protein on LNP, ephrin-A1 was directly conjugated covalently to the liposome surface via the amine-reactive cyanur groups on the end of cyanuric chloride-activated DSPE-PEG(2000). To conjugate ephrin-A1 on the outer layer of liposomes, the cationic liposomes containing DSPE-PEG(2000)-cyanur were dispersed in the borate buffer saline containing the indicated amount of ephrin-A1 and incubated at room temperature for 24 hours with mild shaking.15,36 To acquire an appropriate ephrin-A1 concentration on LNP, the weight ratio of LNP to ephrin-A1 was 2:1. To determine the conjugation efficiency of ephrin-A1 on the LNP, the ephrin-A1–LNP dispersions were dialyzed using the dialysis device (Float-A-Lyzer® G2, 300kD, Spectrum Laboratories, Inc, Rancho Dominguez, CA, USA). After dialysis, solution inside the dialysis device was collected and the concentration of protein was determined by using the Pierce® BCA Protein Assay Kit (Thermo Fisher Scientific Inc, Rockford, IL, USA).
Transmission electron microscopy
The surface morphology of our synthesized LNP was examined by transmission electron microscopy (TEM) (model: Philips CM-100 Biotwin). In brief, a copper grid was precoated with poly-L-lysine aqueous solution for 10 minutes to provide a hydrophilic surface. After the precoating, a drop of liposome dispersions was placed on the gird for 15 minutes to allow the liposomes to attach, and excess solution was removed carefully by using Kim wipes on the edges. A negative staining agent, phosphotungstic acid, was added gently on the grid and then air-dried. To determine the cellular uptake of LNP in the CRL-2081 cells, the cells were cultured on the precoated grid and incubated with serum media at 37°C for 24 hours and then incubated with LNP for 40 minutes. The cells were washed with phosphate-buffered saline (PBS) before fixation and then observed under TEM.
Measurements of particle size distribution and zeta potential
The particle size distribution of LNP was measured by using the Nanotrac Particle Size Analyzer (Microtrac Nanotrac, Montgomeryville, PA, USA). The zeta potential of the LNP was determined using Smoluchowski mobility relations with Brookhaven ZetaPlus (Brookhaven Instruments Corporation, Holtsville, NY, USA). The miR–ephrin-A1–LNP was dialyzed to remove the excess ephrin-A1 and ionic molecules in dispersion solution before analysis.
Cytotoxicity of LNP
About 2,000 MPM and NSCLC cells were seeded in each well of a 96-well plate and incubated in 10% fetal bovine serum media at 37°C. After 24 hours of incubation, the cells were incubated with the desired amount of LNP. The lactate dehydrogenase (LDH) leakage was determined after 48 hours by using the CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega Corporation, Fitchburg, WI, USA), as reported earlier.37
Cellular uptake of LNP complexes by flow cytometry
The LNPs used for cellular uptake study were prepared by following the same procedure described earlier. A fluorescent tagged cholesterol that closely resembles the native cholesterol, 25 NBD-Cholesterol (25-[N-[(7-nitro-2–1, 3-benzoxadiazol-4-yl) methyl]amino]-27-norcholesterol; Avanti Polar Lipids, Inc) was used in LNP. About 5 × 105 cells were seeded in each well of the six-well plate and incubated at 37°C until confluency reached 70%–80%. Cells were incubated with 500 μL of 10 μg/mL LNP samples in serum-free media. After 40 minutes of incubation, the supernatant was discarded and the cells were washed carefully with PBS to remove excess LNP. Cells were collected and suspended in PBS for flow cytometry analysis using LSR-II analyzer (BD Biosciences, San Jose, CA, USA). LNP conjugated with bovine serum albumin (LNP-BSA) were used as negative control.
qPCR analysis
About 1 × 106 cells were seeded into the 60 mm petri dish. After 24 hours of incubation, the cells were transfected with the LNP or ephrin-A1–LNP or free miR or miR–ephrin-A1–LNP complexes or left untreated in serum-free media as control. Total RNA was extracted from the cells and the complementary DNA was generated using Enhanced Avian Reverse Transcriptase (Sigma-Aldrich). The primers and SYBR Green JumpStart Taq Ready Mix used to perform qPCR were purchased from Sigma-Aldrich. The primers used in the qPCR reaction: let-7a, sense: 5′-CCTGGATGTTCTCTTCACTG-3′, antisense: 5′-GCCTGGATGCAGACTTTTCT-3′. h18S, sense: 5′-AAACGGCTACCACATCCAAG-3′, antisense: 5′-TAACGAGGATCCATTGGAGG-3′. The primers used for Ras were reported earlier.4
Western blot analysis
Cells were cultured in 60 mm culture dishes (Corning, Tewksbury, MA, USA) to confluence and the cells were lysed in lysis buffer with the method reported earlier.4 Protein was estimated by using the Pierce® BCA Protein Assay Kit (Thermo Fisher Scientific Inc), and equal amounts of protein (20 mg/lane) were loaded. Proteins in the sample were separated in denaturing sodium dodecyl sulphate polyacrylamide gels (Bio-Rad Laboratories) and transferred electrophoretically onto polyvinylidene difluoride membrane (Immobilon-P; EMD Millipore, Billerica, MA, USA). The blots were blocked with 5% nonfat dry milk (Bio-Rad Laboratories) for 1 hour on a shaker at room temperature, and were incubated overnight at 4°C with respective antibodies, rabbit anti-human pan-Ras antibody (Cell Signaling Technology, Inc, Beverly, MA, USA), at 1:1,000 dilutions. After washing, they were incubated with the secondary antibody (horseradish peroxidase-conjugated goat antirabbit IgG Ab) at a dilution of 1:1,000 for 1 hour. Finally, protein expression was detected by enhanced chemiluminescence (GE Healthcare Bio-Sciences Corp, Piscataway, NJ, USA).
Effects of LNP complexes on cell proliferation
The effect of LNP complexes on MPM and NSCLC cell proliferation was determined by using the WST-1 reagent (Roche, Indianapolis, IN, USA), as reported earlier.4,37 Two thousand cells were seeded in each well of the 96-well plate and incubated in media containing LNP complexes. Cells were treated with free and LNP-encapsulated let-7a miR (miR–LNP, 20 nM), ephrin-A1 (ephrin-A1–LNP, 1 μg/mL–2μg/mL) and the miR–ephrin-A1–LNP combination (20 nM–2 μg/mL–4 μg/mL). The proliferation rates were determined after 48 hours of treatment. Every experiment was done in triplicate.
Three-dimensional tumor growth assay
Matrigel (BD, Franklin Lakes, NJ, USA) was diluted with serum-free media in the ratio (1:1) as reported earlier;4,37 200 μL of diluted Matrigel solution were added into each well of 48-well plate and then allowed to gel at 37°C for at least 1 hour. After gelation, cells at a density of 3,000 per well were plated in 200 μL of serum-free media. Another 200 μL of 2% serum media containing LNP complexes were added into each well. Randomly chosen fields from each well were photographed after 2 weeks. The tumor colonies were quantified by the number and size of colonies developed (diameter).
Wound healing assay
About 1 × 105 cells were seeded into each well of six-well plate. After 48 hours of incubation, cells reached 100% confluency and then the cell monolayers were scratched by 200 μL pipette tips to create a wound on cell monolayer and washed gently with PBS to remove cell debris, as reported earlier.37 The wounded cell cultures were incubated in fresh media containing 10% fetal bovine serum and the respective LNP complexes. The wound areas were photographed every 12 hours and the experiment was carried out in triplicate. The cell migration rate was determined by measuring the distance of cells invaded over time from the edge of wound. The invading distance was calculated as follows: relative invading distance is equal to invading distance of sample/invading distance of control sample; relative invading distance of control sample is considered as 1.
Statistical analysis
Differences among the treatment groups were statistically analyzed by Student’s t-test using SigmaStat3.5 (Systat Software, Inc, Chicago, IL, USA). The values were considered significant at P<0.05. Data reported is the mean ± standard error of the mean from at least three separate experiments.
Results
Characterization of LNP
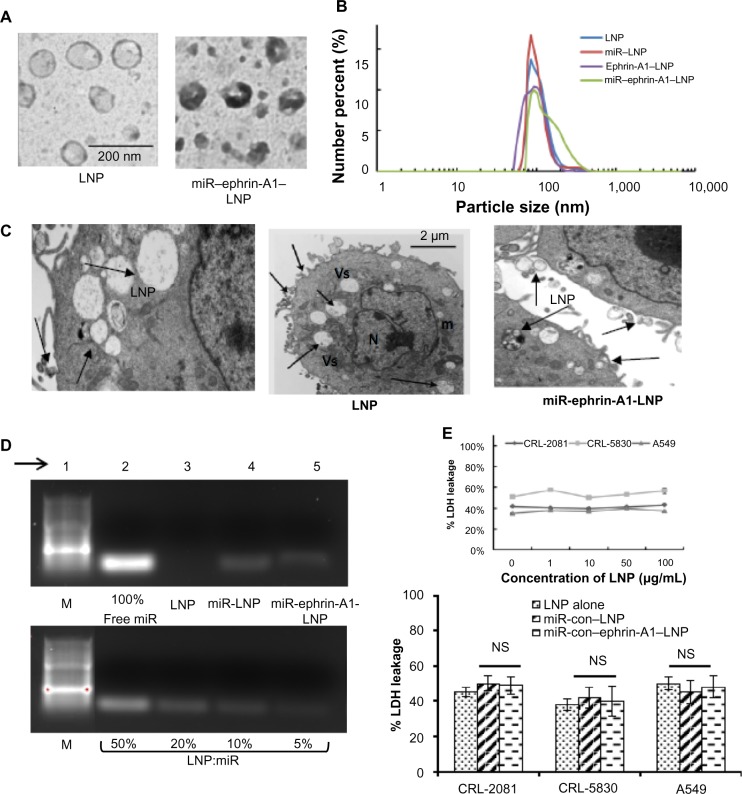
LNP were prepared using, DOTAP, cholesterol, and DSPE-PEG 2000-cyanur (1:1:0.1 molar ratios). The LNP shape and size were confirmed by TEM. The TEM image of LNP and miR–ephrin-A1–LNP showed a spherical shape with a smooth morphology, and an average diameter of about 100 nm (Figure 1A). Ephrin-A1–LNP and miR–LNP showed the mean diameter ranged between 60.00 ± 2.4 nm and 200.24 ± 5.8 nm. The conjugation of miR and ephrin-A1 did not affect the size distribution of LNP (Figure 1B). The mean zeta potential of LNP was +46 ± 1.3 mV, and after conjugation with ephrin-A1 on the surface it was −16 ± 1.1 mV with about 80% conjugation efficiency. In addition, the zeta potential of miR-ephrin-A1–LNP was −15 ± 2.2. The polydispersity index was also evaluated and the values are given in Table 1. The amount of ephrin-A1 on LNP was around 0.45 mg per milligram of LNP. The cellular uptake of LNP by MPM (CRL-2081) is shown in the TEM images (Figure 1C). The cells readily uptake LNP, as noted in the vesicles of the cell. The cells transfected with miR–ephrin-A1–LNP also showed efficient uptake when compared with cells transfected with LNP alone. The encapsulation efficiency of let-7a miR, measured by the centrifugal filtration method, was above 95%. The encapsulation efficiency of let-7a miR was confirmed by gel electrophoresis (Figure 1D). The centrifugal filtration method was used to measure the total loading (including encapsulated miR and the miR that are absorbed on the surface via charge attraction) of miR in the LNPs; the gel electrophoresis method was used for the measurement of encapsulated miR percentage. In the gel electrophoresis, the encapsulation efficiency noted was approximately 90%, which implies that around 10% of the miR was absorbed on the LNP surface instead of being encapsulated inside the LNPs when compared to free miR (100%). Besides, it shows that the ephrin-A1 conjugation process caused no leakage of miR from the LNPs (Figure 1D, lane 5 of upper panel); encapsulation of miR with LNP at various ratios (50%, 20%, 10% and 5%) are shown in the lower panel for comparison. At 10% weight ratio, maximum miR encapsulation was noted. The stability of LNP was determined by serum stability assay (data not shown).
Figure 1.
Characterization of engineered LNPs.
Notes: (A) Transmission electron microscopy image of LNPs. (B) Particle size distribution of LNP and miR–encapsulated LNP, ephrin-A1 conjugated LNP, and miR–ephrin-A1–LNP. (C) Cellular uptake of LNP as determined by transmission electron microscopy. The left hand panel is at a higher magnification. Arrows point to LNP with vesicles; bar represents 2 μm, middle panel. (D) The encapsulation of miR in LNPs was determined by using gel electrophoresis. In order to compare the encapsulation of miR, free miR (100% miR, upper panel) is shown versus 50%, 20%, 10%, and 5% of miR that encapsulated with LNPs, lower panel. The encapsulation efficiency of miR in LNPs was around 90% and remained high after protein conjugation; arrow points to the lane number in the gel. (E) LDH leakage after incubation of lung cancer cells with LNPs. Engineered LNPs showed low cytotoxicity on all three lung cancer cells tested, upper panel. LNP at an even higher concentration (100 μg/mL) failed to show any significant cytotoxicity in CRL-2081, CRL-5830, and A549 cells. The encapsulation of miR–ephrin-A1–LNP and ephrin-A1–LNP did not induce any cytotoxic effect on the cells, lower panel. Data represents mean ± SEM of three separate experiments.
Abbreviations: Con, control; LDH, lactate dehydrogenase; LNP, liposomal nanoparticle; m, mitochondria; M, marker; miR, microRNA; N, nucleus; NS, not significant; SEM, standard error of the mean; Vs, vesicles.
Table 1.
Engineered LNPs polydispersity index
| Particles | Mean zeta-potential (pH) | Mn (nm) | Mv (nm) | PI |
|---|---|---|---|---|
| LNP | +46 ± 1.3 (~7) | 109 | 203 | 1.86 |
| Ephrin-A1–LNP | −16 ± 1.1 (~7) | 105 | 234 | 2.22 |
| miR–ephrin-A1–LNP | −15 ± 2.2 (~7) | 120 | 280 | 2.33 |
Abbreviations: LNP, liposomal nanoparticle; miR, microRNA; Mn, mean number particle size; Mv, mean volume particle size; PI, polydispersity index (Mv/Mn).
We determined the cytotoxicity of LNP in MPM (CRL-2081, CRL-5830) and NSCLC (A549) cells by using LDH leakage assay. LNP up to a high concentration of 100 μg/mL showed no significant cytotoxicity on all the three cell lines tested (Figure 1E, upper panel). In addition, the LNP were encapsulated with control miR sequence and the cytotoxicity was determined. miR-con–LNP or miR-con-ephrin-A1–LNP did not show any cytotoxic effect on LLCs when compared with LNP alone. These results suggest that the engineered LNPs possess high cellular uptake efficiency with low cytotoxic effect.
Cellular uptake of ephrin-a1 conjugated LNP
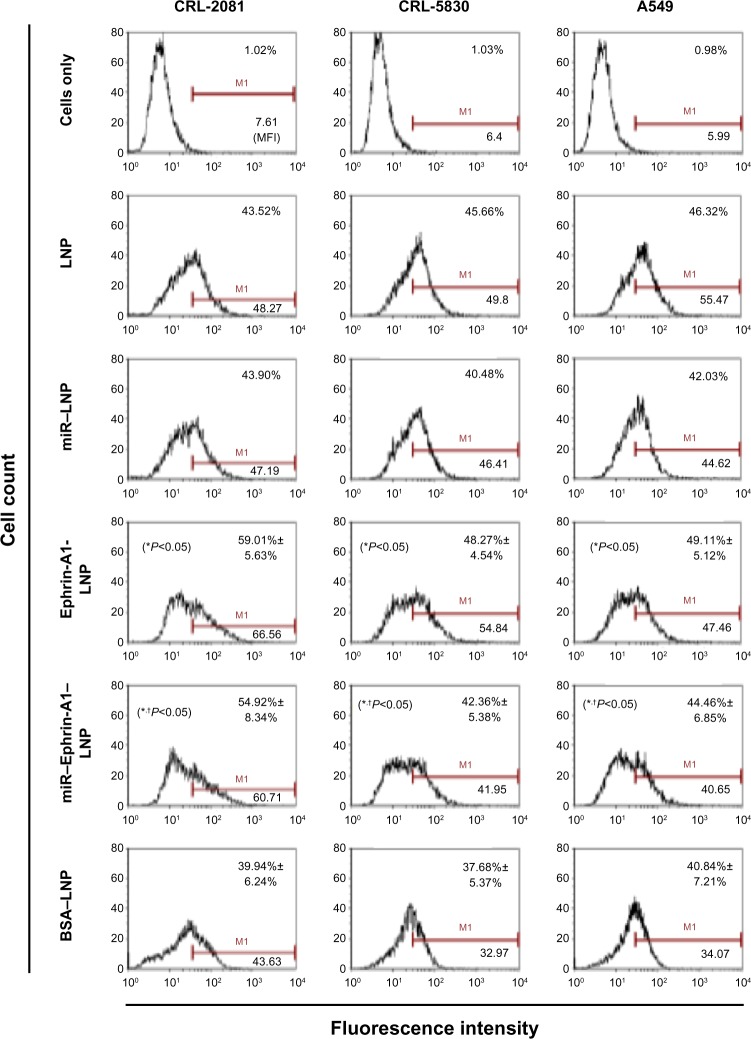
MPM and NSCLC cells were incubated for 40 minutes with LNP and cellular uptake was determined by flow cytometry. Ninety-eight percent of the viable cells have shown the uptake of LNP (Figure 2). LNP conjugated with BSA (nonspecific protein), showed lower cellular uptake efficiency, and total intensity was reduced. However, the LNP conjugated with ephrin-A1 (ephrin-A1–LNP) showed significantly higher cellular uptake efficiency compared with the LNP-BSA. In cell line CRL-2081, ephrin-A1–LNP showed highest percentage cellular uptake and mean fluorescence intensity compared with LNP alone, which indicates that the specific ligand targeting is more prominent than the surface charge. However, the LNP with positively charged surface showed a marginal increase in uptake efficiency in CRL-5830 when compared with ephrin-A1–LNP. The cellular uptake of miR–LNP was significantly low when compared with miR–ephrin-A1–LNP. Ligand conjugated LNP complex increased the efficiency of the miR delivery to the tumor cells when compared with miR–LNP. These data suggest that the cells that overly express the targeting EphA2 effectively uptake the ephrin-A1–LNP or miR–ephrin-A1–LNP when compared with the cells that show lower expression for the targeting receptor EphA2.
Figure 2.
Cellular uptake of LNP and protein conjugated LNP in lung cancer cells as determined by flow cytometry analysis.
Notes: Cellular uptake of LNPs was evaluated after 40 minutes of incubation of cell cultures with LNP. The cellular uptake efficiency of the cationic LNPs was high in all the lung cancer cells (CRL-2081, CRL-5830 and A549 cells). The ephrin-A1 conjugation on the LNP surface showed enhanced cellular uptake via ligand targeting on lung cancer cells. The ephrin-A1–LNP showed the highest percent cellular uptake and intensity in CRL-2081 cells when compared with CRL-5830 and A549 cell lines. However, ephrin-A1 conjugation on the LNPs enhanced the uptake efficiency compared with the LNPs conjugated with nonspecific proteins (BSA–LNP). miR–ephrin-A1–LNP cellular was significantly higher when compared with miR–LNP. Data presented is mean ± SEM of three independent experiments. *P<0.05 when compared with BSA–LNP; †P<0.05 when compared with miR–LNP alone.
Abbreviations: BSA, bovine serum albumin; LNP, liposomal nanoparticle; miR, microRNA; SEM, standard error of the mean.
Transfection efficiency of miR–ephrin-A1–LNP complex
To evaluate the transfection efficiency of miR–ephrin-A1–LNP, the expression levels of let-7a miR in the lung cancer cells were investigated after 24 hours of transfection (Figure 3A). The cells transfected with miR–ephrin-A1–LNP showed significant increase in let-7a miR mRNA expression compared to the cells transfected with miR–LNP. The miR–ephrin-A1–LNP complex showed the most effective transfection efficiency of let-7a miR in CRL-2081 cells, with several-fold increased expression. In CRL-5830 and A549 cells treated with miR–ephrin-A1–LNP, the levels of let-7a miR mRNA were also increased when compared to free miR. In addition, the target gene Ras mRNA and protein expression were also determined in CRL-2081. The mRNA expression of H-Ras isoform significantly inhibited in let-7a-ephrin-A1–LNP miR transfected cells when compared to other isoforms (N-Ras and K-Ras). Ephrin-A1–LNP and miR–let-7a–LNP complex transfection also showed a significant decrease in all the three isoforms of Ras when compared with control cells (Figure 3B). The transfection of let-7a-ephrin-A1-LNP miR complex significantly repressed the Ras expression when compared to let-7a–LNP miR alone. The expression of Ras was also inhibited in cells transfected with miR–let-7a–LNP when compared to control (Figure 3C). Taken together, these results indicate that the synthesized LNP complexes have great potential to inhibit the expression of target gene and thereby inhibit lung cancer cell proliferation and migration. Thus, these LNP complexes may be effective to inhibit tumor growth in future in vivo delivery systems.
Figure 3.
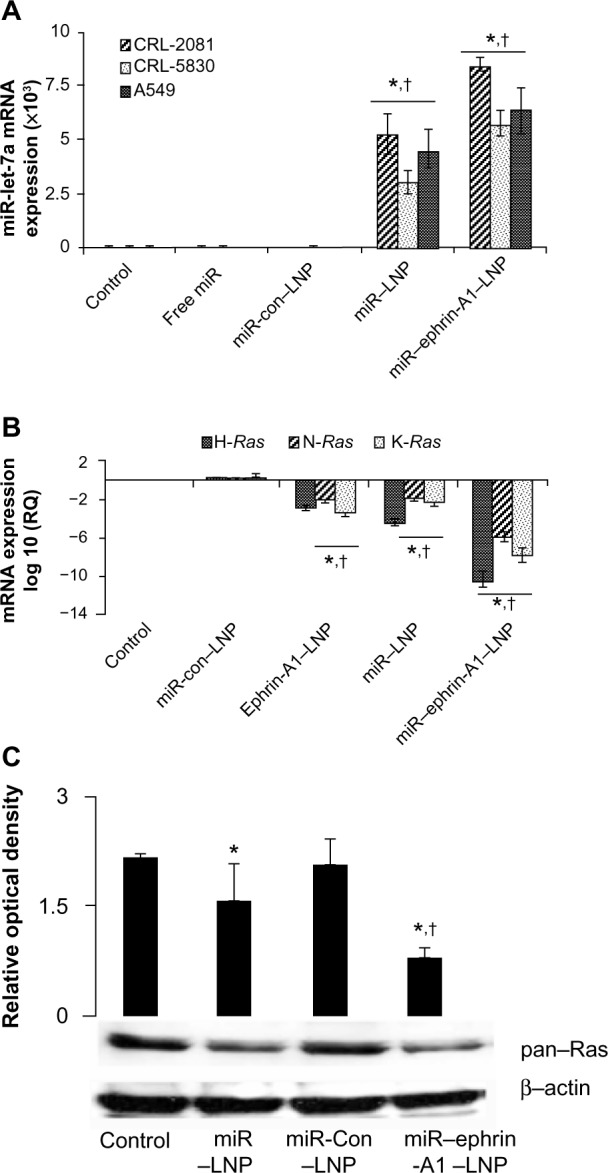
miR–LNP and miR–ephrin-A1–LNP complexes enhance let-7a miR expression in lung cancer cells.
Notes: (A) CRL-2081, CRL-5830, and A549 cells were transfected with miR–ephrin-A1–LNP, miR–LNP, miR-con–LNP, or free miR, or were left untransfected (control) for 24 hours. The let-7a miR mRNA expression was measured by quantitative real-time PCR. Data is presented as the relative quantification compared with control (cells cultured in medium alone) samples as the baseline value. Data represent the mean ± SEM of three separate experiments. 18S RNA was probed as endogenous control to normalize the expression of let-7a miR. *P<0.001, cells transfected with miR–ephrin-A1–LNP versus control; †P<0.001 when compared with miR-con–LNP. (B) H-Ras, N-Ras, and K-Ras mRNA expression as analyzed by quantitative real-time PCR. CRL-2081 were transfected with miR–ephrin-A1–LNP, miR–LNP, ephrin-A1–LNP, or miR-con–LNP, or were left untransfected (control) for 24 hours. Data presented are the mean ± SEM of three separate experiments. 18S RNA was used as endogenous control to normalize the expression of let-7a miR. *P<0.001, CLR-2081 transfected with miR–ephrin-A1–LNP versus control; †P<0.05 when compared with miR-con–LNP. (C) Ras expression in CRL-2081 as detected by Western blot analysis. CRL-2081 were transfected with miR–ephrin-A1–LNP, miR-con–LNP, miR–LNP, or left untransfected to determine ras expression. The β-actin was detected to demonstrate equal loading of protein. Upper panel represents relative optical density. Data presented are the mean ± SEM of three separate experiments. *P<0.001, CLR-2081 transfected with miR–ephrin-A1–LNP versus control; †P<0.05 when compared with miR-con–LNP.
Abbreviations: con, control; LNP, liposomal nanoparticle; miR, microRNA; PCR, polymerase chain reaction; RQ, relative quantification; SEM, standard error of the mean.
Effects of LNP complexes on MPM and NSCLC cell proliferation
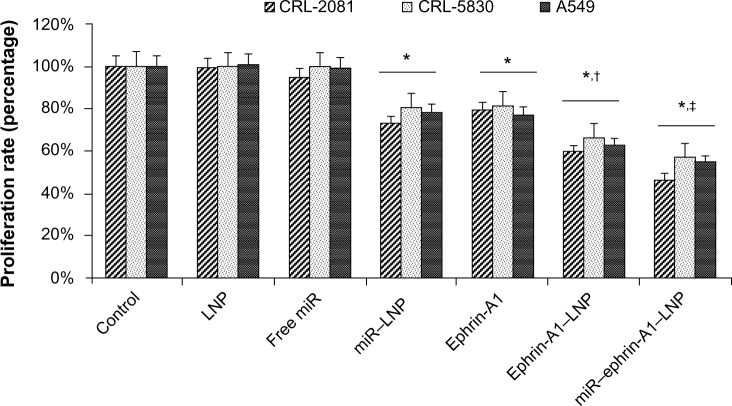
The effects of LNP complexes on cell proliferation of LCCs (lung cancer cells) were determined by using WST-1 assay. Cells treated with free let-7a miR without liposomal carriers showed no significant effect on cell proliferation. Lung cancer cells treated with the let-7a miR encapsulated LNP (miR–LNP) showed about 20%–30% reduction in cell proliferation (Figure 4). In addition, compared to cells treated with ephrin-A1, the ephrin-A1–LNP treatment showed relatively higher inhibitory effect on cell proliferation. Moreover, most striking was the reduction in cell proliferation seen when comparing miR–ephrin-A1–LNP results with those of LNP alone. The MPM cells (CRL-2081) showed highest reduction of proliferation rate (53.29% ± 4.21%) when compared with the other two cell lines. These results suggest that the conjugation of LNP significantly enhanced the effect of miR–ephrin-A1–LNP complex and showed maximum inhibition of cellular proliferation when compared with free miR.
Figure 4.
Effect of LNP complexes on lung cancer cell proliferation.
Notes: Lung cancer cell lines CRL-2081, CRL-5830, and A549 were treated with LNP complexed with miR or ephrin-A1 and compared with free miR and ephrin-A1 after 48 hours of treatment. The miR–ephrin-A1–LNP showed enhanced inhibitory effect on cell proliferation. Data presented is mean ± SEM of three separate experiments. *P<0.05 when compared with control; †P<0.05 when compared with LNP; ‡P<0.05 when compared with miR–LNP complex.
Abbreviations: Con, control; LNP, liposomal nanoparticle; miR, microRNA; PCR, polymerase chain reaction; RQ, relative quantification; SEM, standard error of the mean.
miR–ephrin-A1–LNP complex inhibited tumor growth
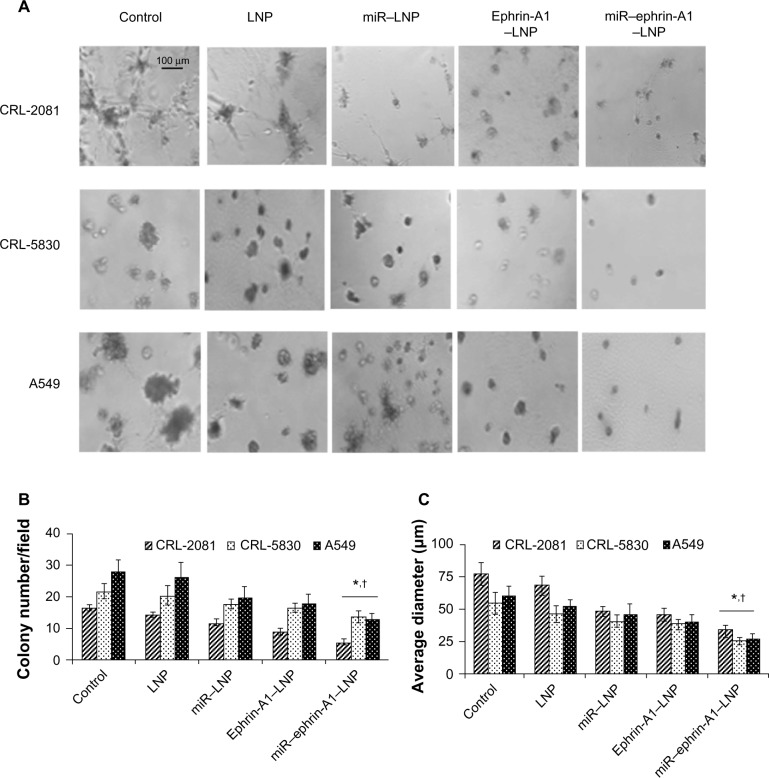
In the tumor growth assay, the LNP complexes effectively reduced the number and size of tumor colonies of CRL-2081, CRL-5830, and A549 cells in Matrigel (Figure 5). Among these LNP complexes, miR–ephrin-A1–LNP showed the most effective inhibition on tumor growth. LNP alone showed little effect on tumor growth in Matrigel. In the highly aggressive cell line CRL-2081, treatment with miR–ephrin-A1–LNP complex significantly reduced the number and size of tumor colonies when compared with LNP alone (Figure 5B and C). In addition, a remarkable reduction in size of tumor colonies as determined by average diameter was noted in all the lung cancer cells upon miR–ephrin-A1–LNP treatment versus miR–LNP or ephrin-A1–LNP treatment. These results indicate that the engineered miR–ephrin-A1–LNP complex has the potential to inhibit the growth of tumor colonies in Matrigels.
Figure 5.
The LNP complexes reduce lung cancer tumor growth in three-dimensional Matrigels in vitro.
Notes: The lung cancer cells were incubated in serum free medium (control) or treated either with ephrin-A1–LNP or miR–ephrin-A1–LNP or LNP alone, and tumor formation was determined in Matrigel. (A) The miR–ephrin-A1–LNP was highly effective on tumor growth inhibition of CRL-2081, CRL-5830 and A549 cells in Matrigel. (B and C) The quantification data of tumor colonies and size. The data presented are the mean ± SEM of three independent experiments performed at separate times; values were considered significant at *P<0.001 when compared with control and †P<0.001 when compared with miR–LNP complex and miR–ephrin-A1–LNP.
Abbreviations: LNP, liposomal nanoparticle; miR, microRNA.
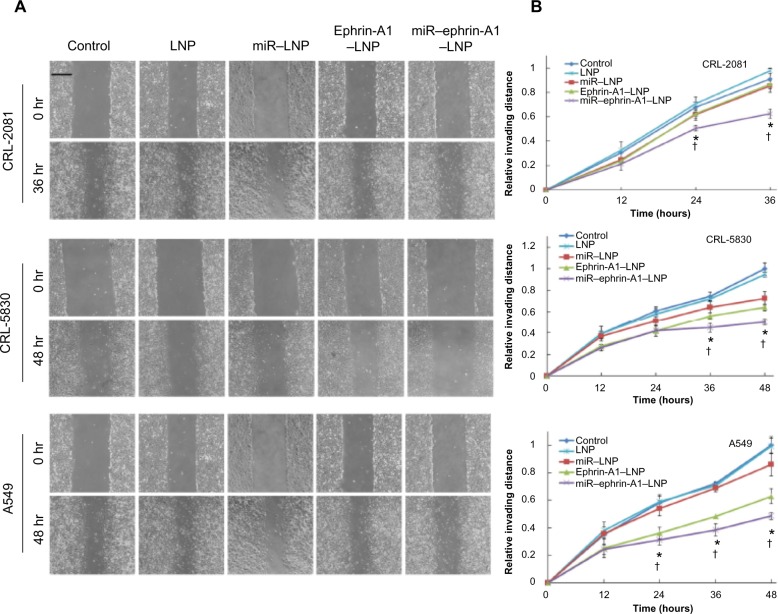
miR–ephrin-A1–LNP complex inhibited MPM and NSCLC cell migration
Cell migration was determined by performing the wound healing assay. Cell migration decreased in LNP complex-treated lung cancer cells (CRL-2081, CRL-5083, and A549 cells) while the LNP alone showed no significant effects on cell migration (Figure 6A). In all the cell lines tested, miR–ephrin-A1–LNP showed the most effective inhibition on cell migration compared to miR–LNP and ephrin-A1–LNP. The CRL-2081 and CRL-5830 cells treated with miR–ephrin-A1–LNP showed a 35% and 50% reduction in cell migration compared to the untreated cells (Figure 6B). The cell migration was observed for 36 hours in CRL-2081 as it reached confluency in control and LNP-alone treated cells, whereas in CRL-5830 and A549 cell lines the cell migration rate was noted until 48 hours. Taken together these results suggest that miR–ephrin-A1–LNP complex is highly effective in attenuation of MPM and NSCLC migration.
Figure 6.
LNP complexes inhibit invasion of lung cancer cells.
Notes: Lung cancer cell invasion was determined by wound healing assay. (A) Optical images of wounded cell monolayers were taken at 0 hours and 36 hours (CRL-2081) or 48 hours (CRL-5830 and A549) after wounding the cultures. The cell migration rates of CRL-2081, CRL-5830, and A549 were determined by measuring the distance cells invaded over time from the edge of wound. (B) The invading distances are shown in relative distance to the control cultures (relative invading distance = invading distance of sample/invading distance of control sample; relative invading distance of control sample is 1). The miR–ephrin-A1–LNP complex effectively reduced the recovery rate of wound area on the monolayer. The data presented is the mean ± SEM of three independent experiments performed at separate times; values were considered significant at *P<0.001 when compared with control and †P<0.001 when compared with miR–LNP complex and miR–ephrin-A1–LNP.
Abbreviations: LNP, liposomal nanoparticle; miR, microRNA; SEM, standard error of the mean.
Discussion
Malignant pleural mesothelioma is still a frustrating clinical challenge with no effective treatment. Various therapeutic approaches, including surgery, radiotherapy, chemotherapy, and traditional gene therapy, showed limited effectiveness on improving survival rate of patients.38 We earlier reported that let-7a miR over expression inhibits MPM tumor growth. In addition, we also demonstrated that let-7a miR targets Ras gene in MPM cells and inhibits tumor growth.4 In lung cancer, the expression of let-7 miR is low when compared to normal tissue.39 The significance of reduced expression of let-7 miR and tumor growth was further indicated in the transgenic NSCLC mouse model.40 Furthermore, ectopic expression of let-7 inhibited the growth of lung cancer cells and lung cancer xenografts.41 Taken together, these studies provide evidence that let-7 miR is a potential therapeutic miR against lung cancer.
To develop an effective therapeutic regimen using let-7a miR for malignant mesothelioma and other lung cancers is a challenge. Because miRs are highly unstable molecular sequences, they rapidly degrade in circulation and show a poor uptake by cancer cells due to their negative charge. A novel approach is highly required to protect the miRs and to deliver them to tumor sites. An ideal carrier system for miRs should provide high loading efficiency and protection from degradation as well as be able to target the cancer cells/tumor tissue while in circulation. Several strategies have been applied to deliver miRs into tumor cells and tumor sites. In a transgenic mouse model of hepatocellular carcinoma, systemic administration of miR-26a was performed using adeno-associated virus.42 In a lung-metastasis mouse model, liposome-polycation-hyaluronic acid nanoparticles were used to deliver miR-34a.43 In this report, we engineered a novel liposomal delivery carrier for small nucleotide molecules by using cationic lipids DOTAP and DSPE-PEG containing cyanuric groups. To specifically target lung cancer cells that overly express the EphA2 receptor, its ligand ephrin-A1 was conjugated on the surface of LNP to develop miR–ephrin-A1–LNP complex. The cationic lipid has been widely used in liposomes for the purpose of DNA or RNA delivery due to its high efficiency of retaining negatively charged biomolecules. However, for the proteins that are overall negatively charged, using liposomes containing DOTAP for the delivery of proteins has not been widely accepted. It is possible that the positively charged liposome surfaces may not be able to provide a stable ionic absorption with proteins, and denaturation of proteins can be caused by the interactions with liposomes. Thus, in order to obtain a stable attachment of proteins on liposome surface for targeting cancer cells, DSPE-PEG-cyanur was added into the liposomes to chemically conjugate with proteins to form a stable complex and minimize the denaturation of proteins. In addition, the phospholipid derivatives of PEG have been shown to increase the particle stability of unilamellar liposomes in plasma and minimize interactions with other biological macromolecules.8
Blood circulation and tumor tissue accumulation of liposomes are important factors to affect the targeting efficiency of liposomes to tumors and the therapeutic activity of delivered molecules.44 These pharmacokinetic properties are not only affected by the size of liposomes, but also surface charge, lipid composition, and the targeting agents on liposome surface.45,46 The diameter of 100 nm has been generally considered the most ideal size of liposomes for efficient delivery, thus the LNPs in this report were controlled in this size range using the ethanol dilution method allowing high encapsulation of miR. In contrast, the conventional lipid film hydration method following the size extrusion cannot provide an appropriate miR delivery because size extrusion through a filter membrane causes low encapsulation efficiency, and for complex formation, miR molecules are too small to form a stable complex. The total loading efficiency of miR with LNP exceeded 95%. The electrophoresis results indicate that the encapsulation efficiency was high at 1:10 miR–LNP complex formulation.
During the process of conjugation, the attachment of ephrin-A1 on LNP may be mainly via chemical conjugation and partially via physical absorption, since theoretically ephrin-A1 is net negatively charged with pH >7. However, after a continuous vigorous vortex and dialysis in water, a high loading efficiency (about 70%) of ephrin-A1 on LNP was noted, and it was mainly by a stable conjugation. The stable conjugation was detected when the weight ratio of LNP to ephrin-A1 was 2:1. After conjugation with ephrin-A1, the particle size distribution shifted about 10 nm to the high particle size region, which implied that the ephrin-A1 protein molecules were conjugated on the surface instead of being surrounded by LNP to form a complex. The change of zeta potential of LNP after ephrin-A1 conjugation, from approximately +23 mV to −10 m V, also suggested that the ephrin-A1 proteins were successfully conjugated on the LNP surface. The conjugation of ephrin-A1 on LNP surface attenuated the surface charge from positive to closer to neutral. This enables the ephrin-A1–LNP complex to be more stable and less susceptible to damage by plasma proteins when systemically applied. The cytotoxicity of the cationic liposomes containing DOTAP was also attenuated by the addition of PEG-containing lipids. The low cytotoxicity of LNP allows it to be widely applied into delivery systems for various biomolecules. Cellular uptake of LNP can be affected by particle size, surface charge, lipid composition, and targeting reagent on the surface.39
In the MPM (CRL-2081) cells incubated with ephrin-A1–LNP, the fluorescence intensity increased to 1.45 fold when compared to the cells incubated with LNP alone (Figure 2). It indicates that the ephrin-A1 on the LNP surface enhanced the binding of ephrin-A1–LNP to cells even while the surface charge was attenuated by the proteins conjugated on the surface. However, the dominant factor in determining the cellular uptake of LNP may vary in different cell types with different morphology.
Ephrin-A1, can inhibit the proliferation and migration of lung cancer cells via binding to overly expressed EphA2 receptors.31,32,34 Thus the ephrin-A1 on the LNP serves not only as a specifically targeting molecule to lung cancer cells but also a therapeutic reagent for cancer treatments. In the cell proliferation assay, cells treated with ephrin-A1–LNP showed a lower cell proliferation than the cells treated with ephrin-A1 alone. This suggests ephrin-A1 has an enhanced inhibiting effect on cell proliferation after LNP conjugated conjugation. It is important that the ability of ephrin-A1 on inhibiting cell proliferation was not only being retained but was enhanced after the conjugation process. This enhancement may be the result of the fast diffusion of ephrin-A1–LNP toward the cell membrane or faster binding kinetics due to the high bioavailability of surface-bound ligands in the milieu.
Transfection of MPM with let-7a miR precursor has been reported to inhibit cell proliferation and tumor growth.4 The purpose of using ephrin-A1–LNP to deliver miR is to enhance the transfection efficiency on lung cancer cells by cell surface receptor EphA2 targeted delivery. In addition, our study provides ephrin-A1 and let-7a miR combination therapy for MPM and NSCLC. The miR–ephrin-A1–LNP complex is highly effective in inhibiting cell proliferation, cell migration, and tumor growth compared to ephrin-A1–LNP and miR–LNP. In addition, the enhanced inhibitory effect of miR–ephrin-A1–LNP implies that the let-7a miR were successfully transfected in the lung cancer cells via the ephrin-A1–LNP delivery. Furthermore, in the wound healing assay, the miR–ephrin-A1–LNP resulted in greater reduction on cell migration rate than did miR–LNP and ephrin-A1–LNP treatments. Although the positive charge on LNP surface was attenuated by ephrin-A1 conjugation, which usually results in slower diffusion of liposomes into the cell membrane, the cellular uptake of ephrin-A1–LNP was enhanced by the specific receptor-ligand affinity of ephrin-A1 and EphA2 expressing cancer cells. This suggests that the combination treatment of ephrin-A1 and let-7a miR was highly effective on inhibiting cell migration and the conjugation of ephrin-A1 on the surface of LNP improved the transfection efficiency for the encapsulated miRs. In the qPCR analysis, miR–ephrin-A1–LNP induced a significant increase of let-7a miR expression level in all the lung cancer cell lines (CRL-2081, CRL-5830, and A549) tested. These data demonstrate that the ephrin-A1–LNP complex provided an effective delivery of let-7a miR into MPM and NSCLC cells. In addition, all the three isoforms of Ras contains putative binding site in their 3′ untranslated regions for let-7a miR, and the treatment of miR–ephrin-A1–LNP complex significantly inhibited the mRNA expression of all three isoforms of Ras and this repression of Ras was significantly higher when compared to miR–LNP. These results further indicate that the engineered miR–ephrin-A1–LNP was highly effective in inhibiting cell proliferation, migration, and tumor growth of all studied lung cancer cells in vitro.
In this report we introduced a novel membrane receptor-targeting liposomal carrier for let-7a miR delivery to lung cancer cells. The LNPs conjugated with the targeting ligand ephrin-A1 served as a stable carrier for efficient delivery of let-7a miR to MPM and NSCLC. In addition, ephrin-A1 on the liposome surface was highly effective to inhibit cell proliferation, migration, and tumor growth of the lung cancer cells in vitro. On the basis of these results, the combination treatment of let-7a miR and ephrin-A1 by the miR–ephrin-A1–LNP complex is promising in that it has significant potential for in vivo targeted delivery applications that aim to provide an enhanced therapeutic effectiveness for malignancies that overly express EphA2.
Acknowledgments
This work was supported by RC1 grant # 09KW-08 from Florida Department of Health and Department of Medicine, University of Florida (Gatorade funds) to Najmunnisa Nasreen. We thank Zita Burkhalter and Linda Wright for technical assistance.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Khodayari N, Mohammed KA, Goldberg EP, Nasreen N. EphrinA1 inhibits malignant mesothelioma tumor growth via let-7 microRNA-mediated repression of the RAS oncogene. Cancer Gene Ther. 2011;18(11):806–816. doi: 10.1038/cgt.2011.50. [DOI] [PubMed] [Google Scholar]
- 5.Miao H, Wei BR, Peehl DM, et al. Activation of EphA receptor tyrosine kinase inhibits the Ras/MAPK pathway. Nat Cell Biol. 2001;3(5):527–530. doi: 10.1038/35074604. [DOI] [PubMed] [Google Scholar]
- 6.McLean JW, Fox EA, Baluk P, et al. Organ-specific endothelial cell uptake of cationic liposome-DNA complexes in mice. Am J Physiol. 1997;273(1 Pt 2):H387–H404. doi: 10.1152/ajpheart.1997.273.1.H387. [DOI] [PubMed] [Google Scholar]
- 7.Felgner PL, Ringold GM. Cationic liposome-mediated transfection. Nature. 1989;337(6205):387–388. doi: 10.1038/337387a0. [DOI] [PubMed] [Google Scholar]
- 8.Felgner PL, Gadek TR, Holm M, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Templeton NS, Lasic DD, Frederik PM, Strey HH, Roberts DD, Pavlakis GN. Improved DNA: liposome complexes for increased systemic delivery and gene expression. Nat Biotechnol. 1997;15(7):647–652. doi: 10.1038/nbt0797-647. [DOI] [PubMed] [Google Scholar]
- 10.Pires P, Simões S, Nir S, Gaspar R, Düzgünes N, Pedroso de Lima MC. Interaction of cationic liposomes and their DNA complexes with monocytic leukemia cells. Biochim Biophys Acta. 1999;1418(1):71–84. doi: 10.1016/s0005-2736(99)00023-1. [DOI] [PubMed] [Google Scholar]
- 11.Yeeprae W, Kawakami S, Suzuki S, Yamashita F, Hashida M. Physicochemical and pharmacokinetic characteristics of cationic liposomes. Pharmazie. 2006;61(2):102–105. [PubMed] [Google Scholar]
- 12.Thurston G, McLean JW, Rizen M, et al. Cationic liposomes target angiogenic endothelial cells in tumors and chronic inflammation in mice. J Clin Invest. 1998;101(7):1401–1413. doi: 10.1172/JCI965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JP, Huang L. Time-dependent maturation of cationic liposome-DNA complex for serum resistance. Gene Ther. 1998;5(3):380–387. doi: 10.1038/sj.gt.3300596. [DOI] [PubMed] [Google Scholar]
- 14.Crook K, Stevenson BJ, Dubouchet M, Porteous DJ. Inclusion of cholesterol in DOTAP transfection complexes increases the delivery of DNA to cells in vitro in the presence of serum. Gene Ther. 1998;5(1):137–143. doi: 10.1038/sj.gt.3300554. [DOI] [PubMed] [Google Scholar]
- 15.Bendas G, Krause A, Bakowsky U, Vogel J, Rothe U. Targetability of novel immunoliposomes prepared by a new antibody conjugation technique. Int J Pharm. 1999;181(1):79–93. doi: 10.1016/s0378-5173(99)00002-2. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Anchordoquy TJ. The role of lipid charge density in the serum stability of cationic lipid/DNA complexes. Biochim Biophys Acta. 2004;1663(1–2):143–157. doi: 10.1016/j.bbamem.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong TK, Girouard LG, Anchordoquy TJ. Effects of PEGylation on the preservation of cationic lipid/DNA complexes during freeze-thawing and lyophilization. J Pharm Sci. 2002;91(12):2549–2558. doi: 10.1002/jps.10255. [DOI] [PubMed] [Google Scholar]
- 18.Shi F, Wasungu L, Nomden A, et al. Interference of poly(ethylene glycol)-lipid analogues with cationic-lipid-mediated delivery of oligonucleotides; role of lipid exchangeability and non-lamellar transitions. Biochem J. 2002;366(Pt 1):333–341. doi: 10.1042/BJ20020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyvönen Z, Rönkkö S, Toppinen MR, Jääskeläinen I, Plotniece A, Urtti A. Dioleoyl phosphatidylethanolamine and PEG-lipid conjugates modify DNA delivery mediated by 1,4-dihydropyridine amphiphiles. J Control Release. 2004;99(1):177–190. doi: 10.1016/j.jconrel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Gabizon A, Papahadjopoulos D. Liposome formulations with prolonged circulation time in blood and enhanced uptake by tumors. Proc Natl Acad Sci U S A. 1988;85(18):6949–6953. doi: 10.1073/pnas.85.18.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvie P, Wong FM, Bally MB. Use of poly(ethylene glycol)-lipid conjugates to regulate the surface attributes and transfection activity of lipid-DNA particles. J Pharm Sci. 2000;89(5):652–663. doi: 10.1002/(SICI)1520-6017(200005)89:5<652::AID-JPS11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Flenniken AM, Gale NW, Yancopoulos GD, Wilkinson DG. Distinct and overlapping expression patterns of ligands for Eph-related receptor tyrosine kinases during mouse embryogenesis. Dev Biol. 1996;179(2):382–401. doi: 10.1006/dbio.1996.0269. [DOI] [PubMed] [Google Scholar]
- 23.Daniel TO, Stein E, Cerretti DP, St John PL, Robert B, Abrahamson DR. ELK and LERK-2 in developing kidney and microvascular endothelial assembly. Kidney Int Suppl. 1996;57:S73–S81. [PubMed] [Google Scholar]
- 24.Nasreen N, Mohammed KA, Antony VB. Silencing the receptor EphA2 suppresses the growth and haptotaxis of malignant mesothelioma cells. Cancer. 2006;107(10):2425–2435. doi: 10.1002/cncr.22254. [DOI] [PubMed] [Google Scholar]
- 25.Kinch MS, Moore MB, Harpole DH. Predictive value of the EphA2 receptor tyrosine kinase in lung cancer recurrence and survival. Clin Cancer Res. 2003;9(2):613–618. [PubMed] [Google Scholar]
- 26.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61(5):2301–2306. [PubMed] [Google Scholar]
- 27.Walker-Daniels J, Coffman K, Azimi M, et al. Overexpression of the EphA2 tyrosine kinase in prostate cancer. Prostate. 1999;41(4):275–280. doi: 10.1002/(sici)1097-0045(19991201)41:4<275::aid-pros8>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 28.Easty DJ, Herlyn M, Bennett DC. Abnormal protein tyrosine kinase gene expression during melanoma progression and metastasis. Int J Cancer. 1995;60(1):129–136. doi: 10.1002/ijc.2910600119. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki T, Kato H, Fukuchi M, Nakajima M, Kuwano H. EphA2 overexpression correlates with poor prognosis in esophageal squamous cell carcinoma. Int J Cancer. 2003;103(5):657–663. doi: 10.1002/ijc.10860. [DOI] [PubMed] [Google Scholar]
- 30.Noblitt LW, Bangari DS, Shukla S, et al. Decreased tumorigenic potential of EphA2-overexpressing breast cancer cells following treatment with adenoviral vectors that express EphrinA1. Cancer Gene Ther. 2004;11(11):757–766. doi: 10.1038/sj.cgt.7700761. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura R, Kataoka H, Sato N, et al. EPHA2/EFNA1 expression in human gastric cancer. Cancer Sci. 2005;96(1):42–47. doi: 10.1111/j.1349-7006.2005.00007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasreen N, Mohammed KA, Lai Y, Antony VB. Receptor EphA2 activation with ephrinA1 suppresses growth of malignant mesothelioma (MM) Cancer Lett. 2007;258(2):215–222. doi: 10.1016/j.canlet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. Ligation of EphA2 by Ephrin A1-Fc inhibits pancreatic adenocarcinoma cellular invasiveness. Biochem Biophys Res Commun. 2004;320(4):1096–1102. doi: 10.1016/j.bbrc.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 34.Woodle MC, Scaria P, Ganesh S, et al. Sterically stabilized polyplex: ligand-mediated activity. J Control Release. 2001;74(1–3):309–311. doi: 10.1016/s0168-3659(01)00339-x. [DOI] [PubMed] [Google Scholar]
- 35.Piao L, Zhang M, Datta J, et al. Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Mol Ther. 2012;20(6):1261–1269. doi: 10.1038/mt.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakowsky H, Richter T, Kneuer C, et al. Adhesion characteristics and stability assessment of lectin-modified liposomes for site-specific drug delivery. Biochim Biophys Acta. 2008;1778(1):242–249. doi: 10.1016/j.bbamem.2007.09.033. [DOI] [PubMed] [Google Scholar]
- 37.Lee HY, Mohammed KA, Peruvemba S, Goldberg EP, Nasreen N. Targeted lung cancer therapy using ephrinA1-loaded albumin microspheres. J Pharm Pharmacol. 2011;63(11):1401–1410. doi: 10.1111/j.2042-7158.2011.01306.x. [DOI] [PubMed] [Google Scholar]
- 38.Tsao AS, Wistuba I, Roth JA, Kindler HL. Malignant pleural mesothelioma. J Clin Oncol. 2009;27(12):2081–2090. doi: 10.1200/JCO.2008.19.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18(9):1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagayasu A, Uchiyama K, Kiwada H. The size of liposomes: a factor which affects their targeting efficiency to tumors and therapeutic activity of liposomal antitumor drugs. Adv Drug Deliv Rev. 1999;40(1–2):75–87. doi: 10.1016/s0169-409x(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 45.Sapra P, Allen TM. Ligand-targeted liposomal anticancer drugs. Prog Lipid Res. 2003;42(5):439–462. doi: 10.1016/s0163-7827(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 46.Krasnici S, Werner A, Eichhorn ME, et al. Effect of the surface charge of liposomes on their uptake by angiogenic tumor vessels. Int J Cancer. 2003;105(4):561–567. doi: 10.1002/ijc.11108. [DOI] [PubMed] [Google Scholar]