Abstract
Fungal infections are increasing worldwide, including in the aquatic environment. Microbiota that coexist with marine life can provide protection against fungal infections by secretion of metabolites with antifungal properties. Our laboratory has developed mass spectrometric methodologies with the goal of improving our functional understanding of microbial metabolites and guiding the discovery process of anti-infective agents from natural sources. GA40, a Bacillus amyloliquefaciens strain isolated from an octocoral in Panama, displayed antifungal activity against various terrestrial and marine fungal strains. Using matrix-assisted laser desorption/ionization-imaging mass spectrometry (MALDI-IMS), the molecular species produced by this microbe were visualized in a side-by-side interaction with two representative fungal strains, Aspergillus fumigatus and Aspergillus niger. The visualization was performed directly on the agar without the need for extraction. By comparison of spatial distributions, relative intensities and m/z values of GA40 secreted metabolites in the fungal interactions versus singly grown control colonies, we obtained insight into the antifungal activity of secreted metabolites. Annotation of GA40 metabolites observed in MALDI-IMS was facilitated by MS/MS networking analysis, a mass spectrometric technique that clusters metabolites with similar MS/MS fragmentation patterns. This analysis established that the predominant GA40 metabolites belong to the iturin family. In a fungal inhibition assay of A. fumigatus, the GA40 iturin metabolites were found to be responsible for the antifungal properties of this Bacillus strain.
Keywords: Aspergillus fumigatus, Aspergillus niger, Bacillus amyloliquefaciens, antifungal, MALDI-imaging mass spectrometry, MS/MS networking, lipopeptide, iturin, Muriceopsis bayeriana
INTRODUCTION
The occurrence of fungal infections is increasing worldwide and has impacted both terrestrial and aquatic environments (Fisher et al. 2011; Kupherschmidt 2012). A catastrophic infection by Batrachochytrium dendrobatidis (Longcore et al. 1999), a fungal amphibian pathogen, has been implicated in outbreaks of chytridiomycosis at pandemic levels causing the extinction of more than 200 amphibian species (Berger et al. 1998). The white-nose syndrome, first detected in 2006, has resulted in an unprecedented decline of the bat population in the Northeastern United States and is caused by a fungal infection with Geomyces destructans (Blehert et al. 2009; Lorch et al. 2011).
While these devastating fungal infections recently have captured significant interest, fungal disease in the marine environment has been an enduring phenomenon affecting marine species in their natural habitats as well as the aquaculture industry throughout the world. For instance, the massive destruction of sea fan corals in the Florida Keys (US) and throughout the Caribbean in the 1990s was attributed to terrestrially derived Aspergillus sydowii (Alker et al. 2001; Kim and Harvell 2004). This epizootic aspergillosis caused approximately 50% mortality in the population of the octocoral Gorgonia ventalina in 1997, but mortality decreased to 10% by 2003, and was less than 1% in 2007 (Bruno et al. 2011). Some studies suggest that the decline of the aspergillosis outbreak was due mainly to the emergence or survival of individuals resistant to the fungal infection among other factors (Bruno et al. 2011; Kim and Harvell 2004).
Corals are composed of symbiotic zooxanthellae algae and a complex microbial community associated with the coral tissue and mucus. This assemblage of organisms has been named the coral holobiont. It has been proposed that microbiota that inhabit corals play a critical role in the defense of their host against pathogens by secretion of specialized metabolites, including antifungal and antibiotic compounds, and signaling molecules (Golberg et al. 2011; Rypien et al. 2010; Shnit-Orland et al. 2012; Zhang et al. 2012). Moreover, the coral probiotic hypothesis suggests that the microbial component of the holobiont is involved in the development of resistance to coral pathogens (Reshef et al. 2006).
Identification of endogenous bacterial strains that produce antifungal compounds can lead to probiotic treatment of hosts against pathogenic fungal infections, a strategy that has been successfully applied in the fight against chytridiomycosis (Harris et al. 2009). In agricultural systems, antifungal-producing bacterial strains have been employed as biocontrol agents (Gardener and Fravel 2002), but in the marine environment this form of disease management is not well explored.
We isolated a Bacillus amyloliquefaciens strain GA40 from the octocoral Muriceopsis bayeriana collected at Galeta Point, Caribbean of Panama, home to a large number of indigenous and endemic microbial species. Strain GA40 was able to inhibit the growth of several fungal species from both terrestrial and marine origin including Aspergillus fumigatus, which is relevant to humans due to its ability to infect immunocompromised patients leading to aspergillosis (Latgé 1999). This strain also is common in the marine environment and leads to gliotoxin contamination in shellfish (Grovel et al. 2003).
To understand how coral-associated microbes interact with potentially pathogenic fungi at the molecular level, novel technologies are necessary. Matrix-assisted laser desorption/ionization-imaging mass spectrometry (MALDI-IMS) allows simultaneous visualization of hundreds of metabolites present in a microbial interaction with minimal sample manipulation and without the need for extraction from the agar growth medium (Watrous and Dorrestein 2011; Yang et al. 2009, 2012). Instead of focusing on one metabolite, IMS provides a snapshot of the complex exchange of metabolites that takes place when microbes live in close proximity, and can guide the isolation of metabolites of particular interest such as antifungal compounds. Annotation of the microbial metabolites is facilitated by MS/MS networking of microbial extracts or by using nanospray desorption electrospray ionization (nanoDESI), a novel mass spectrometry technique that can monitor metabolite production from live microbial colonies directly on agar (Watrous et al. 2012). Metabolites with similar MS/MS fragmentation patterns are grouped together by MS/MS networking. Within the network, identification of one member in a molecular family cluster facilitates annotation of the surrounding neighbors. Here, we demonstrate how MALDI-IMS and MS/MS networking were used together to study the interactions between the coral-associated Bacillus amyloliquefaciens GA40 and two representative fungi, Aspergillus fumigatus and Aspergillus niger, leading to the identification of iturin analogs as the predominant antifungal factors associated with this Bacillus strain.
METHODS AND MATERIALS
Materials
All chemicals used for ISP2, YES and M1 media, Universal MALDI matrix, iturin A, Sephadex® LH-20, and Amberlite XAD were purchased from Sigma-Aldrich (St. Louis, MO, USA) except for Instant Ocean® aquarium salts (Pentair Aquatic Eco-Systems, Inc., Apopka, FL, USA). J.T. Baker® organic solvents were purchased from Avantor Performance Materials, Inc. (Center Valley, PA, USA).
Collection, Isolation and Identification of Bacillus amyloliquefaciens GA40
The octocoral Muriceopsis bayeriana (Sánchez 2001, 2007) was hand collected using SCUBA at 10 m depth from the waters surrounding Galeta Point, located on the Caribbean side of Panama in October 2009, and was identified based on its morphology by H. M. Guzman from the Smithsonian Tropical Research Institute. The sample was placed in a ziplock bag and transported to the laboratory within 1 h of collection. A piece of the coral was rinsed with autoclaved seawater to remove loosely attached bacteria, and a small portion was aseptically ground using a sterile mortar and before direct inoculation on agar plates with seawater-based nutrient medium (10 g of potato starch, 4 g of yeast extract, 2 g of peptone, 18 g of agar, 0.1 g of cyclohexamide in 1 liter of natural seawater) (Jensen et al. 2005). Agar plates were observed for bacterial growth at room temperature over a period of 1 mo. Strain GA40 was isolated from the collection plate and successively re-plated until a pure strain was obtained.
Taxonomic identification of the bacterial strain GA40 as B. amyloliquefaciens was carried out by sequencing 16S rRNA and cpn60 genes. The sequence of 16S rRNA was compared to RDP Naive Bayesian rRNA Classifier (Wang et al. 2007) and the non-redundant database of GenBank using BlastN (Altschul et al. 1990). The sequence of cpn60 partial segment was compared to cpnDB reference sequences (http://www.cpndb.ca). Phylogenetic trees were inferred by the Neighbor-Joining method and conducted in MEGA5 (Tamura et al. 2011). Reference specimens of the coral (GLGA-061009-02) and the bacterial strain (GA40) were deposited at the INDICASAT’s CDDB. Nucleotide sequences were deposited in GenBank under accession numbers KF157957 and KF157958.
Fungal Strains
Aspergillus fumigatus F0075 and Aspergillus versicolor F0073 are plant-endophytic fungi donated by Dr. Sarah Higginbotham (Panama ICBG program). Aspergillus niger NRRL 3, Aspergillus flavus NRRL 3357, Aspergillus oryzae RIB40, Aspergillus terreus FGSC A1156, Aspergillus nidulans FGSC A4, and Penicillium chrysogenum ATCC 28089 were donated by Stephanie Mounaud from the lab of Prof. W. Nierman (JCVI, Rockville, MD, USA). Aspergillus ustus CNK315 (from Hawaii, 1997), Trichoderma virens CNL910 (Garo et al. 2003), Acremonium species CNC890 (Belofsky et al. 2000), Fusarium species CNL292 (Belofsky et al. 1999) and Fusarium species CNT021F (from a sediment sample from San Onofre, CA, USA, 2005, 750 m depth) were gifts from the Paul Jensen lab (SIO, UC San Diego).
Fungal spore stocks were generated by inoculating the fungal slants on YES agar (Samson et al. 2002) (terrestrial strains) or M1 agar (10 g of potato starch, 4 g of yeast extract, 2 g of peptone, 18 g of agar, 28 g of Instant Ocean® in 1 liter deionized water) (marine strains) at 28 °C for 7 d. Sterile water was added on top of the fungal colonies, diluted to 20% glycerol, and stored at −80 °C in small aliquots (7-8 × 106 colony forming units (CFU) ml−1). No Tween detergent was used due to anticipated interference with mass spectrometry analysis.
Preparation of Bacterial and Fungal Samples for MALDI-IMS and Extract Analysis
GA40 was grown from a single colony in M1 liquid media overnight (OD600 = 1.07), diluted to a 20% glycerol stock, and stored in small aliquots at −80 °C (1.5 × 108 CFU ml−1).
GA40 (1 μl) was inoculated in a 5 mm streak at 5 mm distance from a spot of A. fumigatus inoculum (1 μl) on yeast extract/malt extract ISP2 agar (Yang et al. 2012) (10 ml) in 100 × 25 mm Petri dishes. Similarly, GA40 and A. niger were co-inoculated. All organisms also were inoculated on separate Petri dishes as controls. Samples were incubated for 48 h at 30 °C. Regions of agar containing both side-by-side inoculated organisms or the singly grown organism were cut to yield pieces ranging from 1 × 2 cm to 3 × 4 cm, which were placed on top of a MALDI MSP 96 anchor plate (Bruker Daltonics, Billerica, MA, USA). A photograph was taken and the aerial hyphae of A. fumigatus and A. niger subsequently were removed gently with a cotton swab dampened in acetonitrile (Moree et al. 2012). Another photograph was taken and a layer of Universal MALDI matrix (1:1 mixture of 2,5-dihydroxybenzoic acid and α-cyano-4-hydroxy-cinnamic acid) was applied to the sample using a 53 μm sieve. Samples were dried at 37 °C for a minimum of 5 h until they were completely dry and adhered to the MALDI plate.
The sample plates were subjected to MALDI-TOF mass spectrometry (Microflex from Bruker Daltonics, Billerica, MA, USA) for IMS acquisition, and were run in positive reflectron mode, with 600-800 μm laser intervals in XY and a mass range of 100-2600 Da. The data were analyzed using FlexImaging 2.0 software. Detailed instrument parameters for collecting image data were described in Yang et al. (2009).
For extract analysis, 6 spots of GA40 (1 μl per spot, OD600 ~ 0.1) were inoculated on M1 or ISP2 agar (10 ml) in 100 × 25 mm Petri dishes and incubated at 30 °C for 48 h. Four replicate Petri dishes were extracted in 50 ml n-butanol overnight. The extracts were concentrated to dryness by rotary evaporation at 35 °C.
General MS Procedures for Extract Analysis
For the ion trap and Fourier transform ion cyclotron resonance (FT-ICR) MS data acquisition, each extract or compound was dissolved in 1:1 MeOH/H2O containing 1% formic acid, and subjected to nanoelectrospray ionization on a Triversa Nanomate (Advion Biosystems, Ithaca, NY, USA) using a back pressure of 0.3-0.5 psi and a spray voltage of 1.3-1.45 kV. MS and MS/MS spectra were acquired on a 6.42 T ThermoFinnigan LTQ-FT-ICR MS or a ThermoFinnigan LTQ-MS (Thermo-Electron Corporation, San Jose, CA, USA) running Tune Plus software version 1.0 and Xcalibur software version 1.4 SR1. The instrument was first autotuned on the m/z value 816 of cytochrome C (charge state 15). Ions of interest were isolated by the linear ion trap and fragmented by collision-induced dissociation (CID). The isolation window for the ion trap was ±1-2 mass units and for the FT-ICR ±2-3 mass units. Activation energy was set to 35% for both ion trap and FT-ICR.
Molecular MS/MS Network Generation
The MS/MS data of GA40 and commercial iturin A were clustered using the methodology described by Watrous et al. (2012) and visualized using Cytoscape (www.cytoscape.org). Algorithms assumed a precursor mass tolerance of 1.0 Da and a fragment mass tolerance of 0.3 Da with the cosine threshold set at 0.75. Nodes were colored according to origin, and the FM3 layout was used to separate the nodes into their respective clusters. Once the clusters containing the lipopeptides were located, individual nodes were selected and the MS/MS spectra were examined for sequence tags.
Isolation of Antifungal Compounds
Five tubes containing 5 ml of M1 liquid media were inoculated with single colonies of GA40 grown on M1 agar plates. The tubes were shaken overnight at 30 °C (OD600 = 1.03 average), added to 500 ml of liquid M1, and shaken for another 48 h at 30 °C (OD600 = 1.18). The broth was centrifuged at 3437 × g, and the supernatant was mixed with Amberlite XAD (25 g) and gently shaken for 1.5 h at 30 °C. The Amberlite was filtered, washed with water (3 times), and subsequently eluted with acetone followed by MeOH. The resulting extracts were concentrated by rotary evaporation to obtain 0.47 g crude material. The pellet obtained after centrifugation was treated with 1:1 dichloromethane/MeOH (200 ml) overnight at room temperature and centrifuged. The supernatant was concentrated by rotary evaporation (0.34 g of material), combined with the acetone/MeOH extract, applied to Sephadex LH-20 and eluted with MeOH at 0.2 ml min−1. Fractions were evaluated for fungal inhibition of A. fumigatus grown on M1 agar. Bioactive fractions were dissolved in MeOH, and a portion was purified by HPLC using an Agilent Infinity 1260 HPLC (Santa Clara, CA, USA) equipped with a diode array detector, a manual injector, and a Bio-Rad Model 2110 fraction collector (Hercules, CA, USA). A semi-preparative column (C-18, 150 × 10 mm, with 5 μm packing (Xterra from Waters Corporation, Milford, MA, USA) was used for purification at 25 °C with a mobile phase gradient of 10 to 99.9% acetonitrile/H2O containing 0.1% trifluoroacetic acid in 26 min with a 2.0 ml min−1 flow rate.
Comparison of Commercial Iturin A and Iturins in GA40 Extract
Iturin A was compared with the Sephadex LH-20 purified fractions from the GA40 extract that appeared to contain the iturin family of molecules using HPLC with an analytical column (C-18, 250 × 4.6 mm, with 5 μm packing (Luna from Phenomenex, Torrance, CA, USA) with a mobile phase gradient of 40 to 55% acetonitrile/H2O (0.1% trifluoroacetic acid) in 30 min which was ramped to 99.9% acetonitrile/0.1% trifluoroacetic acid in 10 min with a 1.0 ml min−1 flow rate.
Bioassay
Fungal growth inhibition by Bacillus GA40 was assessed with a variety of fungi including Aspergillus spp., Penicillium chrysogenea, Trichoderma virens, Acremonium and Fusarium spp., by spreading fungal spore stock (20 μl) on ISP2 or M1 agar (4 ml) plates in 50 × 25 mm Petri dishes using glass beads and letting the spore solution air dry. A 2 μl aliquot of GA40 was inoculated in the middle of the plate. After incubation at 30 °C for 48 h, diameters of clearance zones were measured to the nearest mm. Inhibition of fungal growth by purified iturin analogs was assessed by inoculating a 1 μl spot of A. fumigatus on M1 agar and placing the center of a 6 mm paper disk treated with test compound 1.4 cm from the spot. One dose was tested for each iturin isolated (R1= C11, C12 and C13) in a ratio equivalent to that present in the GA40 extract (85, 40, 15 μg). After incubation for 5 d at 30 °C, inhibition was assessed by measuring the clearance zone from the edge of the fungal colony to the nearest edge of the paper disk.
RESULTS
Bacterial strain GA40 isolated from the octocoral M. bayeriana was identified as Bacillus amyloliquefaciens based on analysis of the partial 16S rRNA and cpn60 nucleotide sequences. The partial 16S rRNA gene of strain GA40 exhibited over 99% homology with most Bacillus species, while the partial cpn60 gene showed greater homology with B. amyloliquefaciens (99%) than other Bacillus species (less than 95%). Phylogenetic reconstruction including cpnDB reference sequences also showed the sequence of GA40 was mostly related with B. amyloliquefaciens (phylogenetic tree available in Online Resource Fig. 1).
The marine Bacillus GA40 displayed antifungal activity against various fungal species derived from both terrestrial (Table 1, entries 1-8) and marine environments (Table 1, entries 9-13) in a fungal lawn inhibition assay on M1 media (photographs in Online Resource Fig. 2). Interestingly, while in some cases no or weak fungal inhibition was observed in this lawn assay (A. fumigatus and A. flavus) (Fig. 1a, top row), robust inhibition of fungal growth was observed for these strains in an alternative experimental set up using a side-by-side inoculation of GA40 with the fungal strains (Fig. 1a, bottom row). A similar phenotype was observed for A. niger grown side-by-side with GA40, but in this case significant inhibition was also apparent in the lawn assay (Fig. 1a, column iii). For A. versicolor very little fungal growth was detected in the side-by-side interaction with GA40, which agreed with the large zone of inhibition in the lawn assay (Fig. 1a, column iv).
TABLE 1.
INHIBITION OF A VARIETY OF FUNGAL SPECIES BY BACILLUS AMYLOLIQUEFACIENS GA40 IN A LAWN ASSAY ON M1 AND ISP2 AGAR
| Fungal strain | Diameter of inhibition zone (mm) M1 agara |
Diameter of inhibition zone (mm) ISP2 agara |
|
|---|---|---|---|
| 1 | Aspergillus versicolor | 9.0±1.0 | 20.6±1.1 |
| 2 | Aspergillus flavus NRRL 3357 | inactive | Inactive |
| 3 | Aspergillus oryzae RIB40 | 7.0±0 | 11.0±1.0 |
| 4 | Aspergillus fumigatus | inactive | Inactive |
| 5 | Aspergillus niger NRRL 3 | 8.0±0.5 | 12.7±0.6 |
| 6 | Aspergillus terreus FGSC A1156 | 10.0±0b | 11.8±0.5b |
| 7 | Aspergillus nidulans FGSC A4 | 15.0±0 | 25.0±1.0c |
| 8 | Penicillium chrysogenum ATCC 28089 | 13.7±0.6 | 23.3±0.6c |
| 9 | Trichoderma virens CNL910 | 12.3±0.6 | NTd |
| 10 | Aspergillus ustus CNK315 | inactive | NT |
| 11 | Acremonium species CNC890 | 17.3±0.6 | NT |
| 12 | Fusarium species CNL292 | 14.0±0 | NT |
| 13 | Fusarium species CNT021F | 15.5±0.7 | NT |
The test fungi were prepared in a lawn on M1 agar or ISP2 agar and GA40 was inoculated in the center. After 48 h at 30 °C inhibition zones were measured in mm and reported as an average of N=3 ± standard deviation.
Fungal growth was not completely suppressed in the fungal clearance zone
Inhibition was measured at 96 hr because there was insufficient fungal growth at 48 h.
NT= not tested
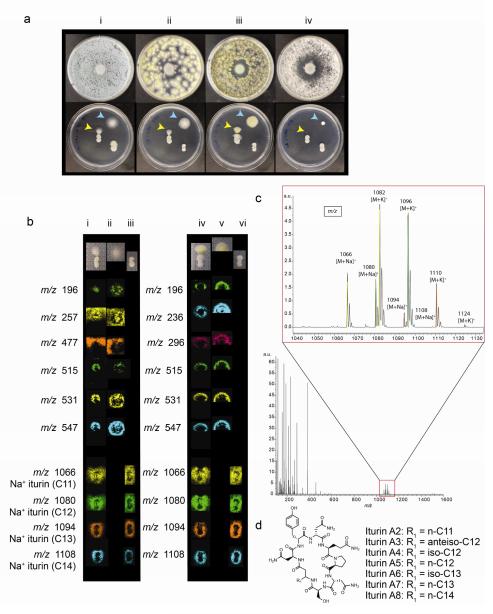
Fig. 1.
(a) Fungal lawn inhibition by Bacillus amyloliquefaciens GA40 compared to side-by-side inoculation of GA40 and fungal strains Top row: Fungal lawns of (i) Aspergillus fumigatus, (ii) Aspergillus niger NRRL 3, (iii) Aspergillus flavus NRRL 3357, and (iv) Aspergillus versicolor with GA40 inoculated in the center on ISP2, 30°C for 48 h. Bottom row: Side-by-side inoculation of GA40 (bottom, 0.5 cm streak) at 0.5 cm distance from fungal spot inoculation (top, indicated by a yellow (light) arrow). Controls are inoculated on the right, GA40 on the bottom and fungal strains (indicated by a blue (dark) arrow) on the top. (b) MALDI-IMS of the interaction between GA40 and A. fumigatus (left) compared to interaction of GA40 vs. A. niger (right) including the controls (i) GA40 on the bottom and A. fumigatus on the top (ii) A. fumigatus control (iii) GA40 control (iv) GA40 on the bottom and A. niger on the top (v) A. niger control (half of a colony was imaged) (vi) GA40 control (c) Average MS spectrum displaying all metabolite ions observed in the interaction of GA40 and A. fumigatus with a zoom-in on the m/z 1040-1140 region (d) Examples of structures of iturin molecules isolated from Bacillus species (Besson et al. 1976, 1984)
MALDI-IMS was used to visualize the metabolite exchange that occurs when Bacillus GA40 was grown in proximity to either A. fumigatus or A. niger in a side-by-side inoculation. MALDI-IMS of side-by-side microbial interactions is more informative than lawn inoculations because metabolite origin and directionality of metabolite secretion can be more clearly detected. Although we have successfully used MALDI-IMS on a variety of agar media (Yang et al. 2012), ISP2 agar is preferred due to limited background mass spectrometry signals from the media. In addition, when ISP2 agar is dried after treatment with Universal MALDI matrix, it typically adheres firmly to the MALDI anchor plate. M1, a seawater based media, has more significant background signals due to a high salt concentration that can suppress ionization of metabolites, and is more prone to flaking once the anchor plate is exposed to high vacuum. We found that the overall fungal inhibitory response for GA40 was similar on M1 and ISP2 (Table 1 and Online Resource Fig. 2). GA40 was more prone to swarming on ISP2 than M1, which contributed to larger zones of fungal inhibition observed on ISP2.
The MALDI-IMS of the side-by-side interaction between GA40 and two representative fungi, A. fumigatus and A. niger, on ISP2 media revealed a large number of metabolites produced by both GA40 and the fungi (Fig. 1b). The mass spectra of all the ions observed in the microbial interaction of GA40 and A. fumigatus during the IMS run were averaged (Fig. 1c). A close-up of the m/z 1040-1140 region displays a set of ions assigned as [M+Na]+ and [M+K]+ with +14 Da differences; only the corresponding [M+Na]+ ions are shown in the IMS (Fig. 1b). Based on these parent masses and searches in the AntiMarin Database (www:http//chem.canterbury.ac.nz/marinlit/) for lipopeptides reported for Bacillus, we hypothesized that these metabolites belonged to the iturin molecular family (Fig. 1d) (Besson et al. 1976).
Within the fungal metabolites, different distribution patterns were apparent, for example m/z 477 (A. fumigatus) was secreted in the media and m/z 196 (A. fumigatus) was confined to the nucleus of the fungal colony itself. The set of metabolites m/z 515, 531, 547, from A. fumigatus and A. niger, was concentrated at the periphery of the colony. At the interaction site with GA40, these metabolites were barely detected indicating that they were consumed, that their secretion by the fungus was inhibited, or that their ionization was suppressed. These metabolites were not further identified.
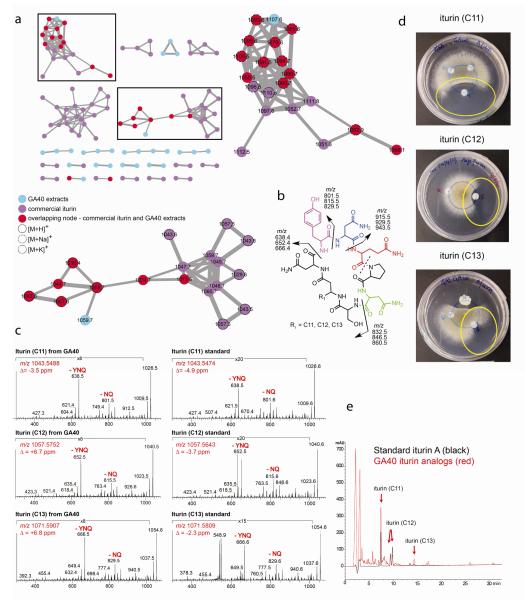
The same set of ions observed in IMS of GA40 and A. fumigatus, m/z 1066, 1080, 1094, and 1108, were detected in an n-butanol extract of GA40 and grouped together using MS/MS network analysis (Fig. 2a). Based on MS fragmentation patterns, high mass accuracy, and comparison to authentic iturin A, the GA40 molecular species clustered with the authentic iturin A, and hence were annotated as iturins (Fig. 2a, b, c and Online Resource Fig. 3-7) (Besson et al. 1976, Williams and Brodbelt 2004).
Fig. 2.
(a) MS/MS network of n-BuOH extract from Bacillus amyloliquefaciens GA40 (grown on ISP2 and M1, 48 hr, 30 °C) with commercial iturin A showing a zoom-in on iturin containing MS/MS clusters (GA40 signals shown in blue, iturin A signals in purple and signals that occur in both GA40 and commercial iturin A in red) (b) General structure of iturin showing major fragments observed in MS/MS spectra (in accordance with Gueldner et al. 1988; Williams et al. 2004) (c) Comparison of IT-MS/MS spectra from iturin (R1= C11-C13) isolated from GA40 with iturin (R1= C11-C13) present in standard iturin A (d) Radial inhibition of Aspergillus fumigatus by iturin (R1= C11-C13) isolated from GA40 (e) Overlay of HPLC chromatogram from standard iturin A and combined iturin fractions isolated from GA40 (Yokota et al. 2012)
Iturins have antifungal properties (Besson et al. 1984; Falardeau et al., 2013). To determine if iturins were responsible for the antifungal activity observed in the interaction of GA40 with A. fumigatus, a bioassay-guided fractionation was carried out. The fractions that showed antifungal activity in a radial growth inhibition assay (Fig. 2d) corresponded to the GA40 ions m/z 1066 (8 mm clearance zone to the closest edge of the paperdisk), 1080 (2 mm clearance zone), and 1094 (1 mm clearance zone). These ions were the sodium adducts of iturins with different fatty acid side chain lengths (R1= C11, C12 C13) observed in IMS (Fig. 1b), and were members of the iturin cluster in the MS/MS network (Fig. 2a). The MS/MS fragmentation patterns and HPLC retention times of the GA40-produced ions exactly matched authentic iturin A (Fig. 2c, 2e).
DISCUSSION
The emergence of both multidrug resistant pathogenic bacterial strains and a worldwide increase in pathogenic fungal infections necessitate the discovery of new antimicrobials. The waters of Panama are home to a large number of unique plants, animals and microbial species that provide a rich source of natural products, which could be mined in the search for molecules to combat infectious diseases. Besides focusing on single molecules with anti-infective properties, we are interested in the potential of bacterial strains to act as probiotics or biocontrol agents against pathogenic microorganisms by studying their collective arsenal of chemical defense agents. With the development of MALDI-IMS methodology, it is possible to obtain a comprehensive overview of the metabolites produced by microbes in media with minimal sample manipulation, thus reducing the risk of observing artifacts or losing critical signals (Yang et al. 2012).
Healthy corals harbor a variety of microbial associates that play a key role in the defense of the host through production of secondary metabolites including antifungals, antibiotics, and signaling molecules. We cultured organisms from Muriceopsis bayeriana that was collected in the waters surrounding Galeta Point at the Caribbean of Panama. Bacillus amyloliquefaciens GA40 displayed robust antifungal activity against a variety of terrestrial and marine strains when tested in a fungal lawn assay or a side-by-side interaction on marine media (Table 1). Many of the tested terrestrial strains including A. fumigatus also are abundant in the aquatic environment. The metabolites including the antifungals produced by GA40 when grown alone or in close proximity to fungal strains were visualized with MALDI-IMS. MALDI-IMS indicated that many metabolites were produced by the bacterial and fungal species in the interaction of GA40 with either A. fumigatus or A. niger, as evidenced by the selection of the most intense signals displayed in Fig. 1b. Intensity of signals can vary between experiments and, therefore, interacting organisms and controls were imaged on one plate in one experiment.
A set of bacterial metabolites was secreted into the agar by GA40 when grown alone or when grown with the fungi. The spatial distribution for each ion was similar, while the intensity varied. Spatial distribution and relative intensities can suggest whether metabolites are secreted by the organisms in the media, confined to the colony, induced, or consumed by neighboring organisms. This information can further fuel hypotheses regarding metabolite activity. These metabolites were abundant at the interaction site and were detected well into the space occupied by the fungi. The pattern of +14 Da mass shifts that was observed in this set of ions is characteristic of different fatty acid chain lengths that are common to non-ribosomal lipopeptides, but also can be caused by methylation or Val–Leu/Ile substitutions.
Much like a fingerprint, the MS/MS fragmentation pattern of each metabolite is unique, yet metabolites that belong to the same molecular family will have very similar MS/MS fragmentation patterns and will form a cluster in the network. In MS/MS networking, molecules are subjected to fragmentation and visualized as nodes (circles), while the relatedness of each node is defined by an edge (lines). The thickness of the edge defines the degree of similarity of the MS/MS spectra. A cluster of ions corresponding to the major ions observed in IMS was identified as the iturin molecular family. By including these data in a network with MS/MS fragmentation of commercial iturin A, we confirmed the identification of this metabolite family cluster (Fig. 2a).
Even though the retention times and MS/MS match, it should be noted that neither the stereochemistry nor the regiochemistry of the fatty acid side chains in these GA40 iturins are confirmed. By linking MS/MS information with the corresponding biosynthetic gene cluster, it was confirmed that these molecules belong to the iturin family of molecules (Nguyen et al. In press).
Bacillus species have been mined for microbial biopesticides and have yielded several commercially produced bacterial biocontrol agents (Favel 2005). Bacillus species that produce iturin lipopeptides, including B. amyloliquefaciens. have received attention as potent biocontrol agents against plant pathogens (, Arguelles-Arias et al. 2009; Balhara et al. 2011; Falardeau et al., 2013; Gueldner et al. 1988). Moreover, marine Bacillus species associated with gorgonian octocorals produce active lipopeptides that contribute to the regulation of microbial populations that adhere to the coral surface (Jiang et al. 2013).
Our results support the hypothesis that coral-associated bacteria are an important part of the defensive strategies employed by the holobiont against pathogen infections. In particular, we demonstrate how the octocoral-associated Bacillus GA40 secretes lipopeptides of the iturin family as specialized metabolites at the interaction site with fungi. The bacterial secretion could prevent invasion or colonization of the holobiont by fungi.
In conclusion, MALDI-IMS in conjunction with MS/MS networking is a powerful mass spectrometric technique that can map the molecular space of microbes grown alone, adjacent to other microorganisms, or as part of complex microbial interactions. The data may suggest hypotheses regarding activity of metabolites, may guide decisions about whether and how to carry out a large fermentation and isolation scheme, may focus the isolation process on specific anti-infective agents, and may provide information on the metabolites produced by organisms under consideration as potential biocontrol agents. Here, we show how these techniques facilitated the identification of the iturin lipopeptide family as the compounds responsible for the antifungal properties of the coral-associated Bacillus amyloliquefaciens GA40.
Supplementary Material
Acknowledgments
We thank the Prof. W. Nierman lab (JCVI) and the Paul Jensen lab (SIO, UCSD) for donation of strains, and Dr. Hector M. Guzman for taxonomical identification of the coral. We acknowledge the Government of Panama (ANAM) for granting permission to make the collections of the coral. This work was supported by NIH AI095125 and S10RR029121, by the NIH Fogarty International Center International Cooperative Biodiversity Groups program TW006634, and by the Government of Panama SENACYT COL09-047 and COL08-061.
REFERENCES
- ALKER AP, SMITH GW, KIM K. Characterization of Aspergillus sydowii, a fungal pathogen of Caribbean sea fan corals. Hydrobiologia. 2001;460:105–111. [Google Scholar]
- ALTSCHUL SF, GISH W, MILLER W, MYERS EW, LIPMAN DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- ARGUELLES-ARIAS A, ONGENA M, HALIMI B, LARA Y, BRANS A, JORIS B, FICKERS P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 2009;8:63. doi: 10.1186/1475-2859-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALHARA M, RUIL S, DHANKHAR S, CHHILLAR AK. Bioactive compounds hold up- Bacillus amyloliquefaciens as a potent bio-control agent. Nat. Prod. J. 2011;1:20–28. [Google Scholar]
- BELOFSKY GN, JENSEN PR, FENICAL W. Sansalvamide A: A new cytotoxic cyclic depsipeptide produced by a marine isolate of a fungus of the genus Fusarium. Tetrahedron Lett. 1999;40:2913–2916. [Google Scholar]
- BELOFSKY GN, ANGUERA M, JENSEN PR, FENICAL W, KÖCK M. Oxepinamides A±C and Fumiquinazolines H± I: Bioactive metabolites from a marine isolate of a fungus of the genus Acremonium. Chem. Eur. J. 2000;6:1355–1360. doi: 10.1002/(sici)1521-3765(20000417)6:8<1355::aid-chem1355>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- BERGER L, SPEARE R, DASZAK P, GREEN DE, CUNNINGHAM AA. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forest of Australia and Central America. Proc. Natl. Acad. Sci. USA. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BESSON F, PEYPOUX F, MICHEL G, DELCAMBE L. Characterization of iturin A in antibiotics from various strains of Bacillus subtilis. J. Antibiot. (Tokyo) 1976;29:1043–1049. doi: 10.7164/antibiotics.29.1043. [DOI] [PubMed] [Google Scholar]
- BESSON F, PEYPOUX F, QUENTIN MJ, MICHEL G. Action of antifungal peptidolipids from Bacillus subtilis on the cell membrane of Saccharomyces cerevisiae. J. Antibiot. (Tokyo) 1984;37:172–177. doi: 10.7164/antibiotics.37.172. [DOI] [PubMed] [Google Scholar]
- BLEHERT DS, HICKS AC, BEHR M, METEYER CU, BERLOWSKI-ZIER BM, BUCKLES EL, COLEMAN JTH, DARLING SR, GARGAS A, NIVER R, OKONIEWSKI JC, RUDD RJ, STONE WB. Bat white-nose syndrome: an emerging fungal pathogen? Science. 2009;323:227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- BRUNO JF, ELLNER SP, VU I, KIM K, HARVELL CD. Impacts of aspergillosis on sea fan coral demography: modeling a moving target. Ecol. Monograph. 2011;81:123–139. [Google Scholar]
- FALARDEAU J, WISE C, NOVITSKY L, AVIS TJ. Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. J. Chem. Ecol. 2013;this volume doi: 10.1007/s10886-013-0319-7. [DOI] [PubMed] [Google Scholar]
- FAVEL D. Commercialization and implementation of biocontrol. Annu. Rev Phytopathol. 2005;43:A337–359. doi: 10.1146/annurev.phyto.43.032904.092924. [DOI] [PubMed] [Google Scholar]
- FISHER MC, HENK DA, BRIGGS CJ, BROWNSTEIN JS, MADOFF LC, MCCRAW SL, GURR SJ. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2011;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARDENER BBM, FRAVEL DR. Biological control of plant pathogens: Research, commercialization, and application in the USA. Plant Health Prog. 2002 May 10; doi:10.1094/PHP-2002-0510-01-RV. [Google Scholar]
- GARO E, STARKS CM, JENSEN PR, FENICAL W, LOBKOVSKY E, CLARDY J. Trichodermamides A and B, cytotoxic modified dipeptides from the marine-derived fungus Trichoderma virens. J. Nat. Prod. 2003;66:423–426. doi: 10.1021/np0204390. [DOI] [PubMed] [Google Scholar]
- GOLBERG K, ELTZOV E, SHNIT-ORLAND M, MARKS RS, KUSHMARO A. Characterization of quorum sensing signals in coral-associated bacteria. Microb. Ecol. 2011;61:783–792. doi: 10.1007/s00248-011-9848-1. [DOI] [PubMed] [Google Scholar]
- GROVEL O, POUCHUS YF, VERBIST J-F. Accumulation of gliotoxin, a cytotoxic mycotoxinfrom Aspergillus fumigatus, in blue mussel (Mytilus edulis) Toxicon. 2003;42:297–300. doi: 10.1016/s0041-0101(03)00146-6. [DOI] [PubMed] [Google Scholar]
- GUELDNER RC, REILLY CC, PUSEY PL, COSTELLO CE, ARRENDALE RF, COX RH, HIMMELSBACH DS, CRUMLEY FG, CUTLER HG. Isolation and identification of iturins as antifungal peptides in biological control of peach brown rot with Bacillus subtilis. J. Agric. Food Chem. 1988;36:366–370. [Google Scholar]
- HARRIS RN, BRUCKER RM, WALKE JB, BECKER MH, SCHWANTES CR, FLAHERTY DC, LAM BA, WOODHAMS DC, BRIGGS CJ, VREDENBURG VT, MINBIOLE KPC. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skinfungus. ISME J. 2009;3:818–824. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- JIANG P, ZHANG X, XU X, HE F, QI S. Diversity and chemical defense role of culturable non-actinobacterial bacteria isolated from the South China Sea gorgonians. J. Microbiol. Biotechnol. 2013;23:437–443. doi: 10.4014/jmb.1208.08010. [DOI] [PubMed] [Google Scholar]
- JENSEN PR, GONTANG E, MAFNAS C, MINCER TJ, FENICAL W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol. 2005;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- KIM K, HARVELL CD. The rise and fall of a six-year coral-fungal epizootic. Am. Nat. 2004;164:S52–S63. doi: 10.1086/424609. [DOI] [PubMed] [Google Scholar]
- KUPHERSCHMIDT K. Attack of the clones. Science. 2012;337:636–638. doi: 10.1126/science.337.6095.636. [DOI] [PubMed] [Google Scholar]
- LATGÉ J-P. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONGCORE JE, PESSIER AP, NICHOLS DK. Batrachochytrium dendrobatidis gen. etsp. nov., a chytrid pathogenic to amphibians. Mycologia. 1999;91:219–227. [Google Scholar]
- LORCH JM, METEYER CU, BEHR MJ, BOYLES JG, CRYAN PM, HICKS AC, BALLMANN AE, COLEMAN JTH, REDELL DN, REEDER DM, BLEHERT DS. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature. 2011;480:376–378. doi: 10.1038/nature10590. [DOI] [PubMed] [Google Scholar]
- MOREE WJ, PHELAN VV, WU C-H, BANDEIRA N, CORNETT DS, DUGGAN BM, DORRESTEIN PC. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA. 2012;109:13811–13816. doi: 10.1073/pnas.1206855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NGUYEN DD, WU C-H, MOREE WJ, LAMSA A, MEDEMA MH, ZHAO X, GAVILAN RG, APARICIO M, ATENCIO L, JACKSON C, BALLESTEROS J, SANCHEZ J, WATROUS JD, PHELAN VV, VAN DE WIEL C, KERSTEN RD, MEHNAZ S, DE MOT R, SHANK EA, CHARUSANTI P, NAGARAJAN H, DUGGAN BM, MOORE BS, BANDEIRA N, PALSSON B. ø., POGLIANO K, GUTIÉRREZ M, DORRESTEIN PC. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. USA. 2013 doi: 10.1073/pnas.1303471110. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RESHEF L, KOREN O, LOYA Y, ZILBER-ROSENBERG I, ROSENBERG E. The coral probiotic hypothesis. Environ. Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- RYPIEN KL, WARD JR, AZAM F. Antagonistic interactions among coral-associated Bacteria. Environ. Microbiol. 2010;12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- SAMSON RA, HOEKSTRA ES, FRISVAD JC, FILTENBORG O, editors. Introduction to food- and airborne fungi. 6th edn Centraalbureau voor schimmelcultures; Utrecht, Netherlands: 2002. [Google Scholar]
- SÁNCHEZ JA. Systematic of the southwestern Caribbean Muriceopsis Aurivillius (Cnidaria: Octocorallia), with the description of a new species. Bull. Biol. Soc. Wash. 2001;10:160–180. [Google Scholar]
- SÁNCHEZ JA. A new genus of Atlantic octocorals (Octocoralia: gorgoniidae): Systematics of gorgoniids with asymmetric sclerites. J. Nat. Hist. 2007;41:493–509. [Google Scholar]
- SHNIT-ORLAND M, SIVAN A, KUSHMARO A. Antibacterial activity of Pseudoalteromonas in the coral holobiont. Microb. Ecol. 2012;64:851–859. doi: 10.1007/s00248-012-0086-y. [DOI] [PubMed] [Google Scholar]
- TAMURA K, PETERSON D, PETERSON N, STECHER G, NEI M, KUMAR S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. emi_2027 28..39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Q, GARRITY GM, TIEDJE JM, COLE JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATROUS JD, DORRESTEIN PC. Imaging mass spectrometry in microbiology. Nature Rev. Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATROUS J, ROACH P, ALEXANDROV T, HEATH BS, YANG JY, KERSTEN RD, VAN DER VOORT M, POGLIANO K, GROSS H, RAAIJMAKERS JM, MOORE BS, LASKIN J, BANDEIRA N, DORRESTEIN PC. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS SM, BRODBELT JS. MSn characterization of protonated cyclic peptides and metal complexes. J. Am. Soc. Mass Spectrom. 2004;15:1039–1054. doi: 10.1016/j.jasms.2004.03.015. [DOI] [PubMed] [Google Scholar]
- YANG YL, XU Y, STRAIGHT P, DORRESTEIN PC. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG JY, PHELAN VV, SIMKOVSKY R, WATROUS JD, TRIAL RM, FLEMING TC, WENTER R, MOORE BS, GOLDEN SS, POGLIANO K, DORRESTEIN PC. A primer on agar-based microbial imaging mass spectrometry. J. Bacteriol. 2012;194:6023–6028. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOKOTA K, YATSUDA M, MIWA E, HIGUCHI K. Comparative study on sample preparation methods for the HPLC quantification of iturin from culture supernatant of an antagonistic bacillus strain. J. ISSAAS. 2012;18:70–75. [Google Scholar]
- ZHANG X, SUN Y, BAO J, HE F, XU X, QI S. Phylogenetic survey and antimicrobial activity of culturable microorganisms associated with the South China Sea black coral Antipathes dichotoma. FEMS Microbiol. Lett. 2012;336:122–130. doi: 10.1111/j.1574-6968.2012.02662.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.