Abstract
Objectives
Lung adenocarcinoma is considered a unique disease for Asian female non-smokers. We investigated whether plasma microRNA (miRNA) expression profiles are different by the EGFR status and are associated with survival outcomes of the patients.
Methods
Using real-time RT-PCR, we analyzed the expression of 20 miRNAs in the plasma of 105 female patients with lung adenocarcinoma. Kaplan-Meier survival analysis and Cox proportional hazards regression were performed to determine the association between miRNA expression and overall survival. Time dependent receiver operating characteristic (ROC) analysis was also performed.
Results
In the 20 miRNAs, miR-122 were found differently expressed between wild and mutant EGFR carriers (P=0.018). Advanced disease stage and tumor metastasis were independently associated with poor prognosis of patients with lung adenocarcinoma (P=0.010 and 1.0×10-4). Plasma levels of miR-195 and miR-122 expression were also associated with overall survival in the patients, especially in those with advanced stage (HR=0.23, 95%CI:0.07-0.84; and HR=0.22, 95%CI:0.06-0.77) and EGFR mutation (HR=0.27, 95%CI:0.08-0.96; and HR=0.23, 95%CI=0.06-0.81). Moreover, a model including miR-195, miR-122 may predict survival outcomes of female patients with lung adenocarcinoma (AUC=0.707).
Conclusions
Circulating miR-195 and miR-122 may have prognostic values in predicting the overall survival as well as predicting EGFR mutation status in non-smoking female patients with lung adenocarcinoma. Measuring plasma levels of miR-195 and miR-122 may especially be useful in EGFR mutant patients with lung adenocarcinoma.
Introduction
Lung cancer is one of the most lethal malignancies and the leading cause of cancer death in the world. The prevalence and mortality of the cancer is still rising. Lung cancer incidence in Asia varies from 14.3/100,000 in south central Asia to 50.4/100,000 in Japan and 61.4/100,000 in China[1]. Of the lung cancer cases diagnosed, approximately 85% are non-small cell lung cancer (NSCLC), which has poor prognosis and is difficult to treat. Despite years of research, the survival of NSCLC remains dismal, and the 5-year survival is only around 10% [2].
Although tobacco use is a well-established risk factor for lung cancer, the disease still occurs to patients who have no history of smoking. Recent evidence suggests that lung cancer incidence is rising among non-smokers, while the smoking population is declining in the developed countries [3,4]. Data from Asia also demonstrate that non-smokers constitute a significant proportion of NSCLC patients, who are quite different from smokers with NSCLC both in clinical and pathological characteristics. Non-smoking lung cancer has been considered a unique disease entity different from smoking one. Recently, it has been shown that patients who are responsive to the treatment of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are largely limited to Asian women who are non-smokers with adenocarcinomas. Patient cohort studies have investigated the differences in overall survival of lung cancer between smokers and non-smokers, and the results, however, were inconclusive due to possible confounding effects[5,6].
MicroRNAs are small non-coding RNAs with 18–25 nucleotides in length that regulate the activities of a large number of messenger RNAs and exert a wide range of biological functions, including behaving either as oncogenes or tumor suppressor genes. Accumulating evidence indicates that dysregulation of specific microRNAs contributes to a variety of diseases, most notably the development and progression of cancer, including lung cancer [7–9]. Recently, the potential for using circulating miRNAs as tumor biomarkers has been evaluated [10], but the results remain to be validated. In this study, we measured the expression of several microRNAs in the plasma of 105 non-smoking female lung adenocarcinoma patients and examined their associations with EGFR mutation and overall survival.
Materials and Methods
Patient enrollment
All patients in the study were recruited form Tianjin Medical University Cancer Hospital between January 2007 and June 2009. The patients were newly diagnosed and histologically confirmed primary non-small cell lung cancer. Patients with a previous medical history of cancer, radiotherapy or chemotherapy before surgery were excluded. The study was approved by an ethical review committee at Tianjin Medical University Cancer Hospital. After signing an informed consent form, all study participants were asked to complete a structured questionnaire with the help of trained research staff. The questionnaire collected information on demographic features and risk factors, including a family history of cancer and tobacco use (age at first use, years of smoking, cigarettes smoked per day, and age at quitting smoking if applicable). Clinical information such as histological type, tumor size, lymph node (LN) metastasis, disease stage, and post-surgical treatment including targeted therapy was extracted from medical records.
During the study recruitment, we enrolled total 478 patients in the study. For the current investigation, only female patients who were diagnosed with adenocarcinoma and never smoked cigarettes were included. The patients who lost to follow-up in the first year were excluded. All patients were followed after surgery through clinical visit and regular telephone contact. Survival time was calculated from date of diagnosis to the date of death or last follow-up in July, 2012.
Tissue and plasma samples
A blood sample (10 ml) was collected from each patient using an ethylene diamine tetraacetic acid (EDTA) vacutainer tube. Plasma was separated after centrifugation at 2,000 rpm for 20 min and then put into a liquid nitrogen tank for long-term storage until miRNA extraction and quantitative reverse transcription PCR (qRT-PCR). All matched tissue samples were histologically confirmed. These tissue samples were stored at -80°C.
EGFR exon 18-21 mutation sequencing
DNA for EGFR sequencing test was extracted from the surgical specimen using the pheno-chloroform method. PCR was performed on 100 ng DNA samples to identify mutations in the EGFR exons 18-21, and Table S1 shown the EGFR primers. Then, PCR products were purified from agarose gel after electrophoresis (1% wt/vol), using the EzgeneTM Gel/PCR Extraction Kit (Biomiga, San Diego, USA) according to the manufacture’s manual. Exons 18-21 of purified products were amplified in duplicate per exon and all variants were confirmed in both forward and reverse directions using the Big Dye Terminator Cycle Sequencing Kit (version 3.1, Applied Biosystems, Foster City, CA). The sequencing reaction was carried out in the ABI-3100 DNA sequencer (Applied Biosystems, Foster City, CA). All sequencing results were analyzed with the Sequencing Analysis software (SDS) version 5.4 (Applied Biosystems, Foster City, CA). Then, the sequences were compared with the standard sequences provided in the national center for biotechnology information (NCBI) database (http://ncbi.nlm.nih.gov/) or UCSC genome bioinformatics (http://genome.ucsc.edu/), and all the electropherograms were analyzed through visual inspection by an experienced researcher. The ‘already known’ EGFR mutations provided by the database were considered as mutation.
MicroRNA extraction
MiRNA was extracted from the plasma of lung adenocarcinoma patients using the QiaGen miRNeasy Mini Kit (QiaGen, Hilden, Germany) according to the manufacturer’s instructions. Since no miRNA has been established as a ‘house-keeping gene’ in the plasma, we added 25 fmol of synthetic C. elegans miRNA cel-miR-39 (Johnson & Johnson, Skillman, NJ, USA) in each plasma sample as an external control to monitor the quality of our RNA extraction and analysis.
TaqMan low density array (TLDA)
TaqMan low density array (TLDA) was performed screening for miRNAs that differently profiled in different EGFR status. We choose 8 advanced stage (iii or iv) non-smoking female NCLC patients, including 4 EGFR mutation status positive and 4 negative which were paired with age. Each of these 8 samples was analyzed with an A & B card for duplicate detection of a total of 667 miRNAs, which including endogenous and negative controls. Each reaction included 50 ng of total RNA, 4.5 μl of reverse transcription (RT) reaction mixture (including 0.8 μl 10×Megaplex RT Primer Pools A+B 10×), 0.2 μl of dNTPs (100 nM), 1.5 μl of MultiScribe Reverse Transcriptase (50 U/μl), 0.8 μl of RT Buffer (10×), 0.9 μl of MgCl2 (25 mM), 0.1 μl of RNase inhibitor (20 U/μl) and 0.2 μl of nuclease-free water. Pre-amplification was performed after the RT procedure to increase the sensitivity of TLDA by using the TaqMan PreAmp Master mix and the Megaplex PreAmp Primer Pools A+B (Applied Biosystems, Foster City, CA). All the reactions were carried out according to the protocols recommended by the manufacturer on 7900HT theremocycler (Applied Biosystems, Foster City, CA). miRNAs that had RQ change larger than 1.5 or smaller than -1.5 in all of the four pairs were selected as candidate and further validated in a cohort.
Reverse transcription
The extracted small RNA was reverse transcribed into its complementary DNA using the TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Each miRNA RT reaction included 1.5 μl of 10×RT buffer, 0.15 μl of 100 mM deoxynucleotide triphosphates, 1 μl of 50 U/μl MultiScribe Rever Transcriptase (Applied Biosystems, Foster City, CA), 0.19 μl of rebonuclease inhibitor, 3 μl of RT primer and 9.16 μl total RNA template. The reaction tube was incubated at 16°C for 30 minutes, 42°C for 30 minutes, 85°C for 5 minutes and finally 4°C for 5minutes. The RT reactions were multiple with 2 primers per RT.
Quantitative real-time polymerase chain reaction (qRT-PCR)
QRT-PCR was carried out on the 7900HT thermocycler (Applied Biosystems, Foster City, CA). Each sample was tested in triplicate using the TaqMan microRNA assay (TaqMan Universal PCR Master Mix ii, no AmpErase UNG) (Applied Biosystems, Foster City, CA) (Assay IDs were shown in Table S2) along with the TaqMan probes and primers (Applied Biosystems, Foster City, CA). 15μl of PCR master mix of the PCR reactions started at 95°C for 10 minutes, followed by 50 cycles of 95°C for 15 seconds and 60°C for 60 seconds. The average quantification cycle (Cq) and standard deviation were calculated with the SDS software, along with the coefficient of variation (CV) of each sample. Samples with CV>0.05 were tested again. In our miRNA test all the samples had a CV<0.01. Samples with Cq<15 or Cq>35 were excluded. Real-time PCR data were analyzed using the SDS software v2.3 (settings: automatic baseline; threshold, 0.2) and relative miRNA levels were calculated with the RQ Manager v1.2.1 (Applied Biosystems, Foster City, CA). In addition, the raw Cq data obtained from each sample were normalized to the mean expression of spike-in cel-miR-39 with the 2-ΔΔCq transition.
Statistical analysis
The associations of overall survival with miRNA expression and clinical factors were first estimated by the Kaplan-Meier survival analysis and log-rank test. The Cox proportional hazard regression models were then used to calculate the hazard ratios of miRNA and other risk factors. All the statistical analyses were performed using the SAS software version 9.2 (SAS Institute, Cary, NC, USA). P values less than 0.05 (two-sided) were considered statistically significant.
To assess the prediction accuracy of different miRNAs in the Cox regression model, time-dependent (ROC) curves for censored data and resulting area under the curve (AUC) were constructed according to Heagerty et al[11]. The Cox models with one, two or three selected miRNAs as covariates were fitted, and the risk scores were used to generate time-dependent sensitivity and specificity for the corresponding ROC curve at each observed event time. The AUC curve was plotted overtime to assess the prediction accuracy of the model in distinguishing subjects who have an event before detection from those who do not. Time-dependent ROC analyses were implemented using the R software (version 2.15.2) [12] and survival ROC package (version 1.0.3) [13] .
Results
Plasma miRNA expression and EGFR mutation
We profiled miRNAs extracted from the plasma of different EGFR mutation status lung adenocarcinoma patients (non-smoking female) using TLDA. Selected candidates identified in the initial screen were further validated in an extended study cohort of 105 non-smoking female lung adenocarcinomas. Table 1 showed the characteristics and clinical feature of the study cohort. Table 2 shows the expression of 20 miRNAs we tested in plasma. Since levels of expression varied widely among the miRNAs being analyzed, the raw Cq data for a specific miRNA were normalized to the mean raw Cq of this miRNA (or spike-in cel-miR-39) in all the 20 miRNA tested to get a ΔCq. Then ΔΔCq was defined as the difference between miRNA specific ΔCq and cel-miR-39 ΔCq. Expression levels were calculated using the 2-ΔΔCq transition. However, none of the miRNA levels were significantly different between patients with and without mutant EGFR expect for miR-122(P=0.018). Furthermore, dividing the patients into early and late stage (stage I/II versus III/IV), we found the associations of miR-16, miR-20b, miR-195, miR-122 and miR-486-3p with EGFR mutation status were evident in advanced stage (Table 3) (P=0.019, 0.047, 0.041, 0.033 or 0.017, respectively).
Table 1. Patient characteristics and clinical features.
| Variable | Number(%) n=105 | HR (95%CI) | HRa(95%CI) | Log-rank P |
|---|---|---|---|---|
| Age | ||||
| <58 | 51(48.57%) | 1.00 | 1.00 | 0.379 |
| >=58 | 54(51.43%) | 0.69(0.31-1.56) | 0.73(0.32-1.66) | |
| Stage | ||||
| Early(Ⅰ/Ⅱ) | 54(51.43%) | 1.00 | 1.00 | 0.010 |
| Late(Ⅲ/Ⅳ,) | 51(48.57%) | 2.99(1.24-7.22) | 3.13(1.23-7.98) | |
| EGFR | ||||
| Wild | 48(45.71%) | 1.00 | 1.00 | 0.068 |
| Mutant | 57(54.29%) | 2.22(0.92-5.36) | 1.64(0.63-4.26) | |
| Family Historyb | ||||
| No | 92(87.62%) | 1.00 | 1.00 | 0.184 |
| Yes | 13(12.38%) | 0.28(0.04-2.09) | 0.25(0.03-1.87) | |
| Disease Historyc | ||||
| No | 96(91.43%) | 1.00 | 1.00 | 0.946 |
| Yes | 9(8.57%) | 1.05(0.24-4.50) | 0.93(0.21-4.04) | |
| Distant metastasis | ||||
| No | 73(69.52%) | 1.00 | 1.00 | 1.000×10-4 |
| Yes | 32(30.48%) | 4.48(1.91-10.49) | 4.13(1.42-11.99) | |
| Lymph Node metastasis | ||||
| No | 52(49.52%) | 1.00 | 1.00 | 0.016 |
| Yes | 53(50.48%) | 2.82(1.17-6.82) | 2.02(0.71-5.72) | |
| Treatment regimen | ||||
| Surgery | 46(43.81%) | 1.00 | 1.00 | 0.374 |
| Surgery+Chemotherapy or Surgery+Radiation or Surgery+EGFR-TKIs | 59(56.19%) | 1.47(0.63-3.43) | 3.82(0.30-49.00) |
a: adjusted for stage, age and treatment regimen
b: family history of cancer
c: lung disease history
Table 2. Plasma levels of miRNA expression by EGFR mutation status.
| MiRNA expression | Median (range) | (2-ΔΔCq)a | P value |
|---|---|---|---|
| EGFR wild (n=48) | EGFR mutant (n=57) | ||
| miR-155 | 0.91(0.28-6.67) | 1.12(0.29-3.18) | 0.557 |
| miR-25 | 1.15(0.14-11.67) | 0.89(0.12-9.82) | 0.339 |
| miR-16 | 1.17(0.13-8.56) | 0.83(0.08-11.21) | 0.087 |
| miR-133a | 0.88(0.07-9.64) | 1.37(0.09-10.56) | 0.077 |
| miR-19a | 1.21(0.22-6.05) | 1.11(0.02-12.91) | 0.221 |
| miR-19b | 1.16(0.23-6.14) | 1.04(0.04-9.57) | 0.310 |
| miR-20a | 1.06(0.27-8.46) | 1.05(0.02-8.35) | 0.332 |
| miR-20b | 1.09(0.16-12.06) | 0.85(0.05-11.89) | 0.165 |
| miR-629 | 0.92(0.16-7.01) | 0.91(0.16-9.18) | 0.665 |
| miR-451 | 1.08(0.08-13.95) | 0.90(0.02-11.40) | 0.114 |
| miR-192 | 0.92(0.23-9.66) | 0.91(0.14-9.57) | 0.295 |
| miR-195 | 1.13(0.22-8.98) | 0.93(0.03-14.36) | 0.133 |
| miR-122 | 1.11(0.26-14.47) | 0.84(0.04-21.14) | 0.018 |
| miR-106b | 1.01(0.17-9.79) | 1.05(0.11-6.07) | 0.946 |
| miR-26b | 1.09(0.13-8.47) | 1.10(0.04-5.46) | 0.972 |
| miR-143 | 1.05(0.20-17.34) | 1.07(0.04-3.70) | 0.615 |
| miR-374 | 1.27(0.11-10.52) | 1.11(0.02-7.93) | 0.528 |
| miR-374-5p | 1.11(0.08-7.98) | 0.99(0.01-7.93) | 0.820 |
| miR-486-3p | 1.13(0.14-8.72) | 0.75(0.06-9.97) | 0.093 |
| miR-590-5p | 1.06(0.06-4.15) | 1.04(0.09-8.52) | 0.885 |
a: Expression level were calculated using the 2- ΔΔCq method.
miR-AΔCq = miR-A raw Cq- miR-Amean Cq,
miR-A ΔΔCq= miR-A ΔCq- cel-miR-39 ΔCq
Table 3. Plasma levels of miRNA expression by EGFR mutation status (Stage iii/iv).
| miRNA expression | Median (range) | (2-ΔΔCq)a | P value |
|---|---|---|---|
| EGFR wild (n=19) | EGFR mutant (n=32) | ||
| miR-155 | 0.73(0.39-2.34) | 1.03(0.29-3.02) | 0.568 |
| miR-25 | 0.88(0.26-11.67) | 0.62(0.12-7.35) | 0.078 |
| miR-16 | 1.18(0.26-7.46) | 0.75(0.08-7.08) | 0.019 |
| miR-133a | 0.90(0.14-8.31) | 1.16(0.09-10.56) | 0.229 |
| miR-19a | 1.24(0.23-4.13) | 0.99(0.02-6.61) | 0.081 |
| miR-19b | 0.93(0.27-4.72) | 0.92(0.04-5.99) | 0.201 |
| miR-20a | 1.08(0.27-4.15) | 0.87(0.02-8.35) | 0.128 |
| miR-20b | 1.09(0.34-6.83) | 0.71(0.05-9.09) | 0.047 |
| miR-629 | 0.93(0.31-7.01) | 0.78(0.16-5.07) | 0.170 |
| miR-451 | 1.05(0.14-13.95) | 0.63(0.02-11.40) | 0.069 |
| miR-192 | 0.96(0.26-9.66) | 0.74(0.14-6.94) | 0.091 |
| miR-195 | 1.21(0.23-5.38) | 0.62(0.03-8.81) | 0.041 |
| miR-122 | 1.03(0.32-9.77) | 0.65(0.04-8.53) | 0.033 |
| miR-106b | 0.92(0.17-7.79) | 0.86(0.11-6.07) | 0.992 |
| miR-26b | 1.01(0.13-5.16) | 1.07(0.04-5.46) | 0.961 |
| miR-143 | 1.09(0.20-2.58) | 1.03(0.04-3.51) | 0.884 |
| miR-374 | 1.29(0.11-2.93) | 1.01(0.03-7.93) | 0.749 |
| miR-374-5p | 1.09(0.08-3.37) | 1.00(0.01-7.93) | 0.839 |
| miR-486-3p | 1.08(0.32-8.72) | 0.64(0.06-9.97) | 0.017 |
| miR-590-5p | 1.07(0.28-3.91) | 1.00(0.09-3.95) | 0.445 |
a: Expression level were calculated using the 2- ΔΔCq method.
miR-AΔCq = miR-A raw Cq- miR-Amean Cq,
miR-A ΔΔCq= miR-A ΔCq- cel-miR-39 ΔCq
Patient characteristics and clinical features
This study was based on 105 non-smoking female lung adenocarcinoma patients with a median follow-up of 23 months (range 4-58 months). Of all the patients enrolled, there were 24 deaths and 6 losses to follow-up. The mean age of patients at diagnosis was 58.60±9.65 years and the range was between 34 and 80 years. The distributions of patient characteristics and clinical features were summarized in Table 1. Log-rank test suggested that patients with advanced disease stage(stage iii/iv), distant metastasis, lymph node involvement lung adenocarcinoma had a significantly shorter overall survival compared to those without these features (P=0.010, 1.0×10-4, or 0.016, respectively). Cox regression analysis showed that the risk for death was associated with distant metastasis (HR=4.13, 95%CI=1.42-11.99) or lymph node metastasis (HR=2.02, 95%CI=0.71-5.72) after adjusting for age, stage, treatment regimen. However, no differences in survival time were observed between patients with different age, EGFR mutation status, a family history of cancer, a history of lung diseases and treatment regimen.
Plasma miRNA expression and non-smoking female lung adenocarcinoma survival
The relationships between 20 miRNA expression in plasma and the survival of non-smoking female lung adenocarcinoma patients are shown in Table 4. We grouped the patients into two classes (low versus high expression) based on the median level of expression in each miRNA. Our survival analysis found that among the 20 miRNAs miR-19a, miR-195 and miR-122 were significantly associated with overall survival (P=0.040, 0.011 and 0.012, respectively). High expression of miR-19a, miR-195 and miR-122 in plasma were associated with lower risks of death compared to low expression. The hazards ratios (HRs) were 0.41, 0.30 and 0.33, respectively. These associations, except for miR-19a, remained significant after adjusting for stage, age, treatment regimen (including surgey, surgey+chemotherapy, surgey+radiation, surgey+EGFR-TKIs).
Table 4. miRNA expression and survival in non-smoking female lung adenocarcinoma.
| miRNAs | group | Number n =105 | HR (95% CI) | HR a (95% CI) | Log-rank P |
|---|---|---|---|---|---|
| miR-155 | LE | 52 | 1.00 | 1.00 | 0.533 |
| HE | 53 | 1.29(0.58-2.89) | 1.38(0.61-3.13) | ||
| miR-25 | LE | 53 | 1.00 | 1.00 | 0.160 |
| HE | 52 | 0.55(0.23-1.29) | 0.68(0.28-1.62) | ||
| miR-16 | LE | 52 | 1.00 | 1.00 | 0.184 |
| HE | 53 | 0.57(0.24-1.33) | 0.61(0.26-1.47) | ||
| miR-133a | LE | 52 | 1.00 | 1.00 | 0.280 |
| HE | 53 | 1.57(0.69-3.62) | 1.86(0.79-4.35) | ||
| miR-19a | LE | 52 | 1.00 | 1.00 | 0.040 |
| HE | 53 | 0.41(0.17-0.99) | 0.41(0.17-1.00) | ||
| miR-19b | LE | 52 | 1.00 | 1.00 | 0.095 |
| HE | 53 | 0.49(0.21-1.16) | 0.51(0.22-1.22) | ||
| miR-20a | LE | 53 | 1.00 | 1.00 | 0.218 |
| HE | 52 | 0.59(0.25-1.39) | 0.68(0.28-1.63) | ||
| miR-20b | LE | 52 | 1.00 | 1.00 | 0.180 |
| HE | 53 | 0.56(0.24-1.32) | 0.63(0.26-1.51) | ||
| miR-629 | LE | 52 | 1.00 | 1.00 | 0.218 |
| HE | 53 | 0.60(0.26-1.37) | 0.61(0.27-1.39) | ||
| miR-451 | LE | 52 | 1.00 | 1.00 | 0.335 |
| HE | 53 | 0.67(0.29-1.53) | 0.79(0.34-1.86) | ||
| miR-192 | LE | 51 | 1.00 | 1.00 | 0.093 |
| HE | 54 | 0.49(0.21-1.15) | 0.55(0.23-1.31) | ||
| miR-195 | LE | 53 | 1.00 | 1.00 | 0.011 |
| HE | 52 | 0.30(0.11-0.80) | 0.31(0.11-0.83) | ||
| miR-122 | LE | 53 | 1.00 | 1.00 | 0.012 |
| HE | 52 | 0.33(0.13-0.82) | 0.34(0.13-0.88) | ||
| miR-106b | LE | 53 | 1.00 | 1.00 | 0.090 |
| HE | 52 | 0.49(0.21-1.14) | 0.49(0.20-1.21) | ||
| miR-26b | LE | 52 | 1.00 | 1.00 | 0.831 |
| HE | 53 | 0.92(0.41-2.06) | 0.98(0.41-2.31) | ||
| miR-143 | LE | 53 | 1.00 | 1.00 | 0.497 |
| HE | 52 | 1.32(0.59-2.99) | 1.29(0.54-3.06) | ||
| miR-374 | LE | 52 | 1.00 | 1.00 | 0.892 |
| HE | 53 | 1.06(0.47-2.39) | 1.03(0.44-2.43) | ||
| miR-374-5p | LE | 52 | 1.00 | 1.00 | 0.887 |
| HE | 53 | 0.94(0.42-2.12) | 0.92(0.39-2.18) | ||
| miR-486-3p | LE | 53 | 1.00 | 1.00 | 0.096 |
| HE | 52 | 0.48(0.20-1.17) | 0.55(0.23-1.36) | ||
| miR-590-5p | LE | 52 | 1.00 | 1.00 | 0.052 |
| HE | 53 | 0.44(0.19-1.03) | 0.43(0.18-1.02) |
LE: Low Expression; HE: High Expression. LE and HE were divided by median expression of specific miRNA.
adjusted for stage, age and treatment
Furthermore, dividing the patients into early and late stage (stage I/II versus III/IV), we found the associations of miR-19a, miR-195 and miR-122 with survival were evident in advanced stage (Table 5). Among the patients with stage iii or iv diseases, high expression of miR-19a, miR-195 and miR-122 were significantly associated with a low risk of death (adjusted HR=0.26, 95%CI: 0.08-0.83; adjusted HR=0.23, 95%CI: 0.07-0.84, and adjusted HR=0.22, 95%CI: 0.06-0.77, respectively). Interestingly, we also found significant relationships between miRNA expression and survival among patients with mutant EGFR (Table 6). Since patients with EGFR mutation were likely to receive target therapy, which could be a confounding factor, we performed the Cox regression analysis adjusted for the treatment regimen in addition to stage and age. Our results indicated that high expression of miR-19a, miR-19b, miR-195, miR-122 and miR-590-5p in plasma were associated with better overall survival among non-smoking female lung adenocarcinoma patients who had EGFR mutation.
Table 5. miRNA expression and survival in non-smoking female lung adenocarcinoma by stage.
| Stage | miRNAs | group | Number | HR (95% CI) | HR a (95% CI) | Log-rank P |
|---|---|---|---|---|---|---|
| Ⅰ/Ⅱ | miR-19a | LE | 27 | 1.00 | 1.00 | 0.543 |
| HE | 27 | 0.58(0.13-2.59) | 0.51(0.11-2.37) | |||
| miR-195 | LE | 26 | 1.00 | 1.00 | 0.229 | |
| HE | 28 | 0.38(0.07-1.96) | 0.40(0.08-2.04) | |||
| miR-122 | LE | 24 | 1.00 | 1.00 | 0.599 | |
| HE | 30 | 0.67(0.15-3.01) | 0.82(0.17-3.93) | |||
| Ⅲ/Ⅳ | miR-19a | LE | 25 | 1.00 | 1.00 | 0.047 |
| HE | 26 | 0.34(0.11-1.04) | 0.26(0.08-0.83) | |||
| miR-195 | LE | 27 | 1.00 | 1.00 | 0.037 | |
| HE | 24 | 0.29(0.08-1.00) | 0.23(0.07-0.84) | |||
| miR-122 | LE | 29 | 1.00 | 1.00 | 0.019 | |
| HE | 22 | 0.25(0.07-0.88) | 0.22(0.06-0.77) |
LE: Low Expression; HE: High Expression. LE and HE were divided by median expression of specific miRNA.
adjusted for stage, age and treatment
Table 6. miRNA expression and survival in non-smoking female lung adenocarcinoma by EGFR mutation status.
| EGFR | miRNAs | group | Number | HR (95% CI) | HR a (95% CI) | Log-rank P |
|---|---|---|---|---|---|---|
| Wild | miR-19a | LE | 23 | 1.00 | 1.00 | 0.686 |
| HE | 25 | 0.74(0.16-3.30) | 0.73(0.16-3.34) | |||
| miR-19b | LE | 22 | 1.00 | 1.00 | 0.857 | |
| HE | 26 | 1.15(0.26-5.13) | 1.20(0.27-5.41) | |||
| miR-195 | LE | 22 | 1.00 | 1.00 | 0.261 | |
| HE | 26 | 0.40(0.08-2.09) | 0.41(0.08-2.19) | |||
| miR-122 | LE | 21 | 1.00 | 1.00 | 0.671 | |
| HE | 27 | 0.73(0.16-3.26) | 0.84(0.16-4.37) | |||
| miR-590-5p | LE | 23 | 1.00 | 1.00 | 0.673 | |
| HE | 25 | 0.73(0.16-3.25) | 0.68(0.15-3.11) | |||
| mutant | miR-19a | LE | 29 | 1.00 | 1.00 | 0.026 |
| HE | 28 | 0.30(0.10-0.92) | 0.28(0.09-0.88) | |||
| miR-19b | LE | 30 | 1.00 | 1.00 | 0.033 | |
| HE | 27 | 0.32(0.10-0.97) | 0.28(0.09-0.91) | |||
| miR-195 | LE | 31 | 1.00 | 1.00 | 0.034 | |
| HE | 26 | 0.28(0.08-0.99) | 0.27(0.08-0.96) | |||
| miR-122 | LE | 32 | 1.00 | 1.00 | 0.008 | |
| HE | 25 | 0.22(0.06-0.75) | 0.23(0.06-0.81) | |||
| miR-590-5p | LE | 29 | 1.00 | 1.00 | 0.021 | |
| HE | 28 | 0.31(0.11-0.89) | 0.28(0.09-0.85) |
LE: Low Expression; HE: High Expression. LE and HE were divided by median expression of specific miRNA.
adjusted for stage, age and treatment
By using Cox proportional hazards regression models we got three model to help calculate the risk score of patient survival. One covariate model was conducted by miR-195 expression (Risk Score=-1.21×miR-195exp), while two covariates model including miR-195 and miR-122 expression (Risk Score=-1.04×miR-195exp-0.96×miR-122exp), in three covariates model besides miR-195 and miR-122, miR-143 was also included (Risk Score = -1.26×miR-195exp-1.34×miR-122exp+1.09×miR-143exp). In order to evaluate the accuracy of survival prediction in the 105 non-smoking female lung cancer patients, we plot the time-dependent ROC curve. The AUC (500 days) was 0.588 for the one-covariate Cox regression model (Figure 1A), 0.675 for a two-covariate Cox model (Figure 1B) and 0.707 for a three-covariate model (Figure 1C). In addition, since the accuracy of each model may change over time, we plotted the AUC curves overtime under each Cox model (Figure 2). In general, we found that the three-covariate Cox model was the most predictive one for non-smoking female lung adenocarcinoma patients as it achieved more than 70% of AUC after 500 days.
Figure 1. ROC curves for the model score, AUC based on Cox model.
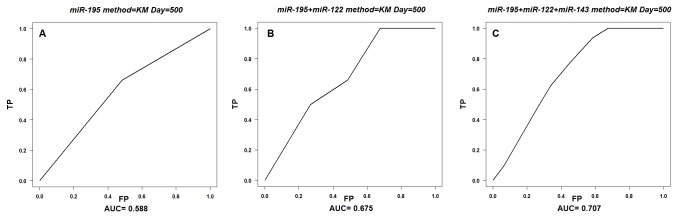
A: one covariate model ROC curves based on the risk score of miR-195; B: two covariate model ROC curves based on the risk score of miR-195 and miR-122; C: three covariate model ROC curves based on the risk score of miR-195, miR-122 and miR-143.
Figure 2. Time-dependent ROC curves of three covariate models.
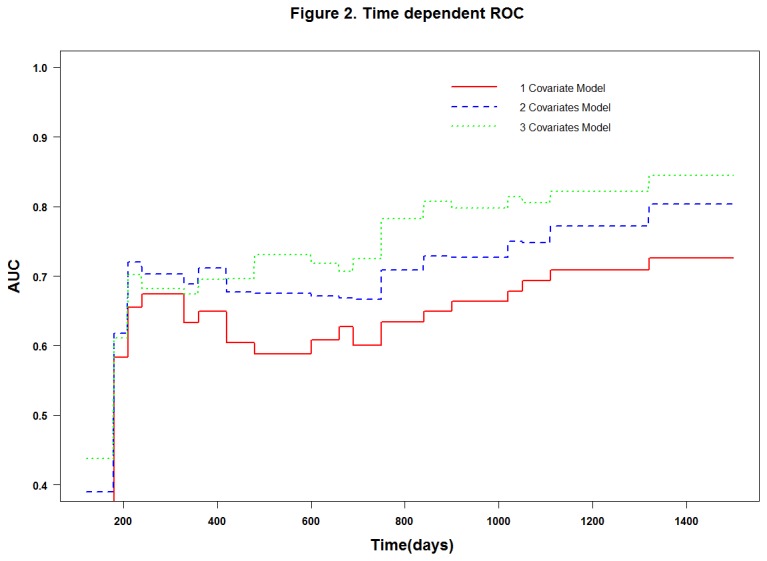
AUC(t) based on one, two and three covariate model overtime under Cox model, when after 500 days three covariate Cox model was the most predictive one, which achieved more than 70% of AUC.
Discussion
According to the estimation by WHO, 25% of lung cancer occurs to never smokers worldwide [14]. Lung cancer is the top 10 cause of death among non-smokers in the world, and the 9th leading cause of cancer death in women according to the data from the European cancer registries[1]. Non-smoking female lung cancer is considered a unique disease entity. The genetic and environmental attributes to this malignancy, however, remain unclear [15]. Our study showed that advanced disease stage and tumor metastasis(distant metastasis and lymph node metastasis) were independent risk factors for the poor prognosis of non-smoking female lung adenocarcinoma patients. These findings were consistent with the literature[16]. Furthermore, our additional analyses indicated that plasma levels of miR-122 expression were associated with EGFR mutation status along with the overall survival of these patients. Especially in those with advanced disease stage, miR-195 and miR-122 expression were found to be associated with EGFR mutation and overall survival of lung adenocarcinoma. Moreover, for the patients with EGFR mutation, miR-19a, miR-19b and miR-590-5p expression were also found to be associated with overall survival in addition to miR-195 and miR-122.
Disease stage at diagnosis is known to be a major prognostic determinant[16], but differences in survival still exist among patients with similar disease stage, which is explained by marked heterogeneity in the genetic and molecular background of the disease[17]. The pathogenesis of lung cancer differs substantially from patient to patient, including their smoking status. Distinct genomic profiles regarding oncogene mutation (EGFR) and patient response to targeted therapy indicate the possible cellular and molecular differences. The possible effect of sex hormones on lung cancer is considered another distinct molecular aspect because adenocarcinoma is diagnosed more frequently in Asian women who do not smoke than in men. A recent study also suggests that different molecular profiles of estrogen receptor subtypes α and β may affect the survival outcomes of lung cancer [18]. All these indicate sexual hormones may influence the disease process.
Advance in genomics, epigenomics and molecular pathology has led to the identification of new biomarkers for cancer detection and outcome prediction. MicroRNA is an expansion to the existing pool of oncogenes and tumor suppressor genes. Emerging evidence indicates that alterations of miRNA expression may affect the initiation and progression of cancer. A number of miRNAs have been reported to be associated with clinical outcomes of patients with cancer, including chronic lymphocytic leukemia[19], breast cancer [20], and lung cancer[21,22]. Although our knowledge on the role of miRNA expression in cancer is still limited, evidence suggests that miRNAs may serve as a new class of clinical biomarker for cancer prognosis. For example, reduced expression of let-7 in lung cancer has been found to be associated with poor survival [22]. The prognostic values of let-7, along with miR-155 and let-7a-2 in lung cancer were also demonstrated in another study [23].
A large number of miRNAs have been found to be stably expressed in human serum and plasma. Circulating miRNAs and their expression profiles have been proposed to be useful biomarkers in the diagnosis and prognosis of cancer. Hu et al.first found that the expression of four serum miRNAs(miR-486, miR-30d, miR-1 and miR-499) were significantly associated with overall survival of NSCLC patients[24]. Over-expression of serum miR-21 was linked to the poor survival of NSCLC and associated with lymph node metastasis and advanced stage [25]. In addition, several other miRNAs have been reported to be associated with clinical outcomes of NSCLC patients, including miR-30e-3p, let-7f[26], miR-126, miR-183[27], miR-125b[28], and miR-17-5p[29]. Recently, Wang et al. reported a group of TGF-β signaling pathway-related serum miRNA as predictors of survival in advanced NSCLC, which including miR-16 [30].
Few studies have investigated the prognosis of NSCLC in non-smoking female patients. In order to improve the management of these patients, we focus our study on non-smoking female lung adenocarcinoma in searching for molecular markers that can identify patients who have poor prognosis after surgery. Circulating miRNAs are good candidates for non-invasive blood-based test in prediction of lung cancer prognosis. The results of our study provided some evidence that plasma levels of miR-195 and miR-122 expression were associated with EGFR status along with overall survival of non-smoking female lung adenocarcinoma patients, especially in those with advanced stage or EGFR mutation positive. MiR-195 is located on chromosome 17p13.1, and belongs to the miR-16/15/195/424/497 family which shares the same 3’UTR binding seed sequence. A number of studies have shown that this family plays an important role in tumorigenesis. MiR-195 has been found in several types of cancer, and recently the ectopic expression of miR-195 was found in hepatocellular carcinoma and colorectal cancer cells [31,32]. Xu et al. reported that miR-195 could block the G(1)/S transition by interfering with the Rb-E2F signaling pathway through targeting multiple molecules (cyclin D1, CDK6, and E2F3)[31]. Furthermore, over-expression of miR-195 in the Rb-E2F pathway which acts as a vital check point in cell cycle progression, can promote cell proliferation, which may subsequently facilitate the development of cancer. Similarly, Liu et al. revealed that the miR-15/16/195 directly targeted cyclin D1 and CDK6, and miR-195 expression increased the proportion of G1 cells in a lung cancer cell line[32]. In addition, circulating levels of miR-195 may be a biomarker for minimal invasion of breast cancer[33]. Our results indicated that reduced expression of miR-195 in plasma could lead to better survival of non-smoking female lung adenocarcinoma patients, especially for those with advanced stage. Despite the consistent findings, the mechanism that underlies the effect of miR-195 on lung cancer is still unclear. Our study was one of the few investigations which focused on non-smoking female lung adenocarcinoma. Our finding of association between circulating miR-195 and non-smoking female lung adenocarcinoma survival warrants further elucidation.
MiR-122, located on chromosome 18q21.3, is a liver-specific miRNA representing two thirds of hepatic miRNAs. This miRNA was reported to facilitate the replication of hepatitis C virus[34] and regulate the metabolism of lipids[35] and expression of hepatic circadian genes[36]. Notably, down-regulation of miR-122 has been associated with human hepatocellular carcinoma development and progression [37]. We investigated the relationship between miR-122 and EGFR mutation status along with survival in non-smoking female lung adenocarcinoma, and found that reduced expression of miR-122 in plasma was associated with EGFR mutation and favorable survival, especially in patients with advanced disease. To date, neither the mechanisms underlying miR-122 regulation nor the regulatory networks of miR-122 have been investigated. Recently, a few targets of miR-122 have been investigated including cyclin G1, serum response factor, insulin-like growth factor 1 receptor (IGF-1R)[37–39]. MiR-122 may have influence on survival of lung adenocarcinoma by targeting these gene or through the GSK-3b–C/EBPa–miR-122–IGF-1R regulatory circuitry[40].
Interestingly, we identified a group of miRNAs (including miR-19a, miR-19b, miR-195, miR-122 and miR-590-5p) that could be predictive of survival outcomes of non-smoking female lung adenocarcinoma patients who have EGFR mutations in Exons 18-21. It has been suggested that the carcinogenic pathway of lung cancer may differ by smoking status. The differences between smoking and non-smoking lung adenocarcinoma are found at cellular and molecular levels [41], including distinct profiles of oncogenic mutations (e.g., EGFR). Our results suggest that miR-19a, miR-19b, miR-195, miR-122 and miR-590-5p may predict the prognosis of lung adenocarcinoma with mutant type of EGFR in non-smoking females. Furthermore, these results provide new evidence that EGFR mutant lung adenocarcinoma may have distinct miRNAs associated with overall survival. In this group of miRNAs, miR-195 and miR-122 was also differently expressed in EGFR wild and EGFR mutant groups. Which indicated that miR-195 and miR-122 may possibly be potential non-invasive biomarker in EGFR mutation prediction and prognosis prediction of non-smoking female lung adenocarcinomas. These findings need to be confirmed in large well-designed clinical studies.
To our knowledge, this study represents the first effort to characterize the relationships of plasma miRNAs with EGFR mutation and lung adenocarcinoma survival in non-smoking women. We measured plasma levels of 20 miRNAs and analyzed their associations with lung adenocarcinoma survival. Our study also evaluated the association in patients stratified by clinical stage and status of EGFR mutation, which was valuable for assessing possible interaction. We also used different analytic strategies like survival ROC analysis to confirm our results. However, our study also had a few limitations. First, although we performed subgroup analysis according to clinical stage and EGFR status and calculation of adjusted HRs, confounding factors may still exist in our study. Second, we found associations between overall survival and plasma miRNA expression with little knowledge on their biologic functions. Functional analyses of these miRNAs are needed to establish their biological relevance. Third, this is a relatively small study. Our findings need to be validated in large well-powered studies independently.
In conclusion, our study showed that circulating miRNAs were associated with EGFR status and overall survival of lung adenocarcinoma. High expression of miR-195 and miR-122 in plasma could help predict EGFR mutation status and overall survival of advanced stage female non-smokers with lung adenocarcinoma. Furthermore, the association between plasma miRNAs and overall survival of lung adenocarcinoma may differ by the status of EGFR mutation.
Supporting Information
EGFR Exon18-21 PCR Primers. Table S1shown the EGFR primers. DNA for EGFR sequencing test was extracted from the surgical specimen using the pheno-chloroform method. PCR was performed on 100 ng DNA samples to identify mutations in the EGFR exons 18-21.
(DOCX)
Assay IDs for the microRNA assays (Applied Biosystems, Foster City, CA). Each sample was tested in triplicate using the TaqMan microRNA assays, the assay IDs were shown in Table S2 along with the TaqMan probes and primers (Applied Biosystems, Foster City, CA).
(DOCX)
Funding Statement
This study were supported by National Natural Science Foundation of China (No.81071914 and No.81372229, http://www.nsfc.gov.cn) and Natural Science Foundation of Tianjin (No.13JCYBJC23100, http://www.tstc.gov.cn). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55: 74-108. doi: 10.3322/canjclin.55.2.74. PubMed: 15761078. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Ward E, Hao Y, Xu J et al. (2008) Cancer statistics, 2008. CA Cancer J Clin 58: 71-96. doi: 10.3322/CA.2007.0010. PubMed: 18287387. [DOI] [PubMed] [Google Scholar]
- 3. Wakelee HA, Chang ET, Gomez SL, Keegan TH, Feskanich D et al. (2007) Lung cancer incidence in never smokers. J Clin Oncol 25: 472-478. doi: 10.1200/JCO.2006.07.2983. PubMed: 17290054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boffetta P, Järvholm B, Brennan P, Nyrén O (2001) Incidence of lung cancer in a large cohort of non-smoking men from Sweden. Int J Cancer 94: 591-593. doi: 10.1002/ijc.1507. PubMed: 11745449. [DOI] [PubMed] [Google Scholar]
- 5. Toh CK, Wong EH, Lim WT, Leong SS, Fong KW et al. (2004) The impact of smoking status on the behavior and survival outcome of patients with advanced non-small cell lung cancer: a retrospective analysis. Chest 126: 1750-1756. doi: 10.1378/chest.126.6.1750. PubMed: 15596669. [DOI] [PubMed] [Google Scholar]
- 6. Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G (2004) Improved survival in never-smokers vs current smokers with primary adenocarcinoma of the lung. Chest 126: 347-351. doi: 10.1378/chest.126.2.347. PubMed: 15302716. [DOI] [PubMed] [Google Scholar]
- 7. Chen X, Ba Y, Ma L, Cai X, Yin Y et al. (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997-1006. doi: 10.1038/cr.2008.282. PubMed: 18766170. [DOI] [PubMed] [Google Scholar]
- 8. Gao W, Shen H, Liu L, Xu J, Shu Y (2011) MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol 137: 557-566. doi: 10.1007/s00432-010-0918-4. PubMed: 20508945. [DOI] [PubMed] [Google Scholar]
- 9. Shen J, Todd NW, Zhang H, Yu L, Lingxiao X et al. (2011) Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest 91: 579-587. doi: 10.1038/labinvest.2010.194. PubMed: 21116241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105: 10513-10518. doi: 10.1073/pnas.0804549105. PubMed: 18663219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heagerty PJ, Zheng Y (2005) Survival model predictive accuracy and ROC curves. Biometrics 61: 92-105. doi: 10.1111/j.0006-341X.2005.030814.x. PubMed: 15737082. [DOI] [PubMed] [Google Scholar]
- 12. The R Project for Statistical Computing. Available: http://www.r-project.org/. Accessed 2013 October 26
- 13.http://cran.r-project.org/web/packages/survivalROC/index.html survivalROC. Available: Accessed 2013 Jan 13.
- 14. Lee YJ, Cho BC, Jee SH, Moon JW, Kim SK et al. (2010) Impact of environmental tobacco smoke on the incidence of mutations in epidermal growth factor receptor gene in never-smoker patients with non-small-cell lung cancer. J Clin Oncol 28: 487-492. doi: 10.1200/JCO.2009.24.5480. PubMed: 20008630. [DOI] [PubMed] [Google Scholar]
- 15. Sun S, Schiller JH, Gazdar AF (2007) Lung cancer in never smokers--a different disease. Nat Rev Cancer 7: 778-790. doi: 10.1038/nri2172. PubMed: 17882278. [DOI] [PubMed] [Google Scholar]
- 16. Brundage MD, Davies D, Mackillop WJ (2002) Prognostic factors in non-small cell lung cancer: a decade of progress. Chest 122: 1037-1057. doi: 10.1378/chest.122.3.1037. PubMed: 12226051. [DOI] [PubMed] [Google Scholar]
- 17. Herbst RS, Heymach JV, Lippman SM (2008). Lung Cancer - N Engl J Med 359: 1367-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stabile LP, Dacic S, Land SR, Lenzner DE, Dhir R et al. (2011) Combined analysis of estrogen receptor beta-1 and progesterone receptor expression identifies lung cancer patients with poor outcome. Clin Cancer Res 17: 154-164. doi: 10.1158/1078-0432.CCR-10-0992. PubMed: 21062926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M et al. (2005) A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353: 1793-1801. doi: 10.1056/NEJMoa050995. PubMed: 16251535. [DOI] [PubMed] [Google Scholar]
- 20. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R et al. (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065-7070. doi: 10.1158/0008-5472.CAN-05-1783. PubMed: 16103053. [DOI] [PubMed] [Google Scholar]
- 21. Yu SL, Chen HY, Chang GC, Chen CY, Chen HW et al. (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13: 48-57. doi: 10.1016/j.ccr.2007.12.008. PubMed: 18167339. [DOI] [PubMed] [Google Scholar]
- 22. Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H et al. (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64: 3753-3756. doi: 10.1158/0008-5472.CAN-04-0637. PubMed: 15172979. [DOI] [PubMed] [Google Scholar]
- 23. Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K et al. (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189-198. doi: 10.1016/j.ccr.2006.01.025. PubMed: 16530703. [DOI] [PubMed] [Google Scholar]
- 24. Hu Z, Chen X, Zhao Y, Tian T, Jin G et al. (2010) Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 28: 1721-1726. doi: 10.1200/JCO.2009.24.9342. PubMed: 20194856. [DOI] [PubMed] [Google Scholar]
- 25. Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ et al. (2012) High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol 29: 618-626. doi: 10.1007/s12032-011-9923-y. PubMed: 21516486. [DOI] [PubMed] [Google Scholar]
- 26. Silva J, García V, Zaballos A, Provencio M, Lombardía L et al. (2011) Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J 37: 617-623. doi: 10.1183/09031936.00029610. PubMed: 20595154. [DOI] [PubMed] [Google Scholar]
- 27. Lin Q, Mao W, Shu Y, Lin F, Liu S et al. (2012) A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non-small-cell lung cancer by stem-loop RT-PCR. J Cancer Res Clin Oncol 138: 85-93. doi: 10.1007/s00432-011-1068-z. PubMed: 22009180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuxia M, Zhennan T, Wei Z (2012) Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol 138: 2045-2050. doi: 10.1007/s00432-012-1285-0. PubMed: 22806310. [DOI] [PubMed] [Google Scholar]
- 29. Chen Q, Si Q, Xiao S, Xie Q, Lin J et al. (2013) Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol 30: 353. doi: 10.1007/s12032-012-0353-2. PubMed: 23263848. [DOI] [PubMed] [Google Scholar]
- 30. Wang Y, Gu J, Roth JA, Hildebrandt MA, Lippman SM et al. (2013) Pathway-Based Serum microRNA Profiling and Survival in Patients with Advanced Stage Non-Small Cell. Lung Cancer - Cancer Res 73: 4801-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP et al. (2009) MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 50: 113-121. doi: 10.1016/S0168-8278(09)60290-7. PubMed: 19441017. [DOI] [PubMed] [Google Scholar]
- 32. Liu Q, Fu H, Sun F, Zhang H, Tie Y et al. (2008) miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391-5404. doi: 10.1093/nar/gkn522. PubMed: 18701644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J et al. (2010) Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg 251: 499-505. doi: 10.1097/SLA.0b013e3181cc939f. PubMed: 20134314. [DOI] [PubMed] [Google Scholar]
- 34. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P (2005) Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309: 1577-1581. doi: 10.1126/science.1113329. PubMed: 16141076. [DOI] [PubMed] [Google Scholar]
- 35. Esau C, Davis S, Murray SF, Yu XX, Pandey SK et al. (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3: 87-98. doi: 10.1016/j.cmet.2006.01.005. PubMed: 16459310. [DOI] [PubMed] [Google Scholar]
- 36. Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O et al. (2009) Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev 23: 1313-1326. doi: 10.1101/gad.1781009. PubMed: 19487572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S et al. (2007) Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 67: 6092-6099. doi: 10.1158/0008-5472.CAN-06-4607. PubMed: 17616664. [DOI] [PubMed] [Google Scholar]
- 38. Bai S, Nasser MW, Wang B, Hsu SH, Datta J et al. (2009) MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem 284: 32015-32027. doi: 10.1074/jbc.M109.016774. PubMed: 19726678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW et al. (2009) MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology 49: 1571-1582. doi: 10.1002/hep.22806. PubMed: 19296470. [DOI] [PubMed] [Google Scholar]
- 40. Zeng C, Wang R, Li D, Lin XJ, Wei QK et al. (2010) A novel GSK-3 beta-C/EBP alpha-miR-122-insulin-like growth factor 1 receptor regulatory circuitry in human hepatocellular carcinoma. Hepatology 52: 1702-1712. doi: 10.1002/hep.23875. PubMed: 21038412. [DOI] [PubMed] [Google Scholar]
- 41. Lan Q, Hsiung CA, Matsuo K, Hong YC, Seow A et al. (2012) Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 44: 1330-1335. doi: 10.1038/ng.2456. PubMed: 23143601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EGFR Exon18-21 PCR Primers. Table S1shown the EGFR primers. DNA for EGFR sequencing test was extracted from the surgical specimen using the pheno-chloroform method. PCR was performed on 100 ng DNA samples to identify mutations in the EGFR exons 18-21.
(DOCX)
Assay IDs for the microRNA assays (Applied Biosystems, Foster City, CA). Each sample was tested in triplicate using the TaqMan microRNA assays, the assay IDs were shown in Table S2 along with the TaqMan probes and primers (Applied Biosystems, Foster City, CA).
(DOCX)


