Abstract
Ensemble-decision aliquot ranking (eDAR) is a sensitive and high-throughput method to analyze circulating tumor cells (CTCs) from peripheral blood. Here, we report the next generation of eDAR, where we designed and optimized a new hydrodynamic switching scheme for the active sorting step in eDAR, which provided fast cell sorting with an improved reproducibility and stability. The microfluidic chip was also simplified by incorporating a functional area for subsequent purification using microslits fabricated by standard lithography method. Using the reported second generation of eDAR, we were able to analyze 1-mL of whole-blood samples in 12.5 min, with a 95% recovery and a zero false positive rate (n=15).
Introduction
A number of analytical methods for studying circulating tumor cells (CTCs) have been developed because of the expectation that they will simultaneously facilitate more effective, less invasive cancer treatments and elucidate the process of cancer metastasis.1–3 These methods come with three major technological challenges.4 First, CTCs are usually scarce in peripheral blood samples (<100 per billions of blood cells), so all CTC-related technologies need to have an accurate and high-throughput enumeration. So far, the majority of the clinical applications of CTCs are still focused on enumerating these cells from patient blood and correlating their counts with the clinical progress. Enumeration and correlation of CTCs with disease progress has been verified by many studies involving breast,5 lung,6 colorectal,7 and prostate8 cancers.
The second technological challenge comes from the heterogeneity of cancer cells, whose physical and biological attributes can vary significantly within CTC populations and over time. Most of the current methods are positive selections based on one biological parameter, such as the expression of the widely used surface antigen, the epithelial cell adhesion molecule (EpCAM),9 or a physical property, such as the size or density of the cells.10 An ideal CTC-technology should be flexible in using different markers to enrich the tumor cells from blood samples so different subpopulations of CTCs are not lost during the analysis.
The final challenge lies in the need for downstream analyses of the isolated CTCs, providing cellular and molecular information at the single-cell level, which may be more important than simple enumeration. These analyses have been successfully performed by researchers investigating various cancer biomarkers, including the human epidermal growth factor receptor 2 (HER-2) in breast cancer,11 epidermal growth factor receptor (EGFR) in lung cancer,12 and TMPRSS2-ERG in prostate cancer.13 Enrichment ratio and purity of the captured CTCs are crucial to the downstream analyses, because they will determine the throughput and accuracy of these measurements. Imaging quality of the isolated CTCs is also important if downstream analyses are performed on the same device that captures CTCs, because many bioassays may be performed based on detailed labeling and imaging studies of individual CTCs.
Many CTC-analysis systems utilize microfluidic components to overcome these challenges as well as increase their sensitivity and improve their throughput.14 Various microfluidic systems have been fabricated, including the line-confocal flow-detection platform,15 the flow-counting method based on micro-Hall effects16, and the conductometric detection system.17 Other types of microfluidic CTC systems include those which select and isolate target cells based on: (i) binding to a cell surface marker, analogous to affinity chromatography type of methods,18–20 (ii) size via micro-filtration,21 (iii) size, density, or permittivity via field flow fractionation,10 (iv) morphology via high-speed photography,22 and (v) density based on the use of Dean flow.23 Of course, there are many CTC-analysis systems that do not involve microfluidics and instead rely on methods such as fiber-optic-array scanning24 and immunomagnetic separation.25–26 In fact, the only FDA-approved CTC analysis system, CellSearch, does not have microfluidic components, but rather selects and manipulates target cells via magnetic nanoparticles.25
We recently reported a CTC analysis method called ensemble-decision aliquot ranking, or eDAR,27 which combined the following components: multi-color line-confocal fluorescence detection with a high sensitivity, a hydrodynamic switching mechanism, a cell trapping and subsequent purification process, and an identification and downstream analysis section. It had a high throughput, analyzing 1 mL of whole blood in 20 minutes, with a 93% recovery ratio and a zero false positive rate. CTCs were captured onto a very small area (1 mm2) with a high enrichment ratio.27 Although eDAR was more sensitive than the CellSearch method in detecting CTCs from metastatic breast cancer patients,27 the original version of eDAR still has several factors that may limit its clinical application. The original microchip had six layers, which constrained the yield and efficiency of the chip production. Although the throughput of fluorescence imaging was greatly improved because of the very small area on which CTCs were trapped, the imaging quality could be adversely affected by the track-etched filter, generating a non-uniform background.
This paper describes the second generation of eDAR platform, which included an integrated filtration area fabricated by standard lithography methods. This microchip had only two layers of feature on the silicon master and could be fabricated with one-step replica molding into polydimethylsiloxane (PDMS) and then bonding to a glass substrate. The entire system was also simplified with a new active sorting scheme. We carefully evaluated several different designs of the microfluidic chip and hydrodynamic switching mechanisms, and optimized the analytical performance of the device. The recovery efficiency was 95% with a zero false positive rate (n=15), and the highest throughput tested was 4.8 mL of whole blood per hour. We successfully applied this method to whole-blood samples taken from metastatic pancreatic cancer patients.
Materials and Methods
Microfluidic Chips
The microfluidic chip had two functional areas integrated in the same design, the eDAR sorting area and the filtration unit based on slit structures. The main channel in the sorting unit, which introduced the blood into the sorting junction, had a height of 50 µm and width of 150 µm; all the other 4 channels were 50-µm tall and 200-µm wide. The slit-filters were 5-µm tall and 5-µm width. The maximum number of microslits we tested was 20,000.
The silicon master was fabricated using two photolithography processes (Figure 1S). The features were designed using AutoCAD (Autodesk, San Rafael, CA), and written on a chrome mask (TRICR Corporation, SF, CA). Positive resist lithography and deep reactive ion etching (DRIE) were chosen for forming the first layer, the micro-filter feature. AZ 1512 was used as a positive photoresist, which was provided by Micromanufacturing Facility (MMF) at the University of Washington. DRIE process was optimized to achieve a depth in the range of 4.5–5 µm. The second layer of the eDAR feature was fabricated using the SU-8-3050 as a negative photoresist (MicroChem, Newton, MA), and the height of the feature was controlled to be 50 µm. After the master was silanized using tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (Sigma-Aldrich, St. Louis, MO), uncured PDMS was poured onto the silicon wafer and baked for 2 hours at 70°C. The piece of PDMS with the desired micro-feature was peeled off the silicon master, and then bonded with a piece of cover glass using the standard process of plasma oxidation.
Biological and clinical samples
Three breast cancer cell lines, SKBr-3, MCF-7, and MDA-MB-231 cells (American Type Culture Collection (ATCC), Manassas, VA) were used to characterize and optimize the eDAR system. SKBr-3 cells were cultured in McCoy’s 5, MCF-7 was cultured in Eagle's Minimum Essential Medium (EMEM), and MDA-MB-231 was cultured in Dulbecco's Modified Eagle's Medium (DMEM) (ATCC, Manassas, VA). All cell culture media also contained 2 mM L-glutamine, 10% fetal bovine serum (FBS) (ATCC, Manassas, VA), and 50 µg/mL penicillin/streptomycin (ATCC, Manassas, VA). Incubations were done at 37 °C with 5% CO2 in a humidified environment. The MDA-MB-231-GFP cell line was provided by Prof. Gail Sonenshein at Tuffs University and cultured in the DMEM medium with 10% FBS and 1 µg/mL puromycin (Life Technologies, Carlsbad, CA). Control blood from healthy donors was purchased from Plasma International Lab (Everett, WA); the first tube of the blood draw was discarded to prevent any possible contamination from skin cells. Whole blood samples were drawn from patients with metastatic pancreatic cancer based on a protocol approved by Fred Hutchinson Cancer Research Center’s institutional review board. Patient samples were collected at the Seattle Cancer Care Alliance (SCCA) using Vacutainer tubes (BD, Franklin Lakes, NJ) containing EDTA as an anti-coagulant, stored at 4 °C, and analyzed within 4 hours.
Sample preparation and eDAR analysis
Isoton (Beckman Coulter Inc., Chino, CA) was used as the buffer for all the experiments unless otherwise specified. For a typical eDAR experiment, 1 mL of whole blood samples was labeled with anti-EpCAM conjugated with phycoerythrin (PE) (Lot No. 515776, Abnova, Walnut, CA) for 30 minutes at room temperature in dark. All the labeling parameters have been optimized based on our previous work.27 The labeled samples were diluted to 14 mL and then centrifuged to remove the free antibodies. The final volume was adjusted to be the same as the initial volume. The prepared sample was next injected to the microfluidic chip using a syringe pump. Traces from fiber-coupled avalanche photodiodes (APDs) (Excelitas Technologies, Waltham, MA) were collected by a PCI data acquisition card (PCI 6602, National Instruments, Austin, TX). The sorting process of eDAR was automatically controlled using a home-written LabVIEW (National Instruments, Austin, TX) script and a field-programmable-gate-array (FPGA) device built in house. The sorting threshold was set at a signal-to-noise ratio that was 7 times the standard deviation of the noise level in each channel to rank the aliquots. The hydrodynamic switching, which collected the sorted aliquots, was controlled by a solenoid (INKA1226212H) purchased from the Lee Company (Westbrook, CT).
After all the positive aliquots were collected onto the filtration area, isoton was used to quickly wash the filtration area in less than 1 minute. After this, three microchannels were closed by turning off the in-line valves on the tubing connected with them, so the microchip only had one inlet, the buffer channel on the left, and one outlet, the waste collection channel after the filtration area. As a result, any further fixation, peameabilization and staining reagents would only flow through that area. If any cytoplasmic markers were used for the secondary labeling, 4% of paraformaldehyde (PFA) was loaded into the filtration area to fix the cells. Surfynol® 465 (Air product, Allentown, PA) was used to permeabilize the fixed cells. Anti-EpCAM-PE and anti-Cytokeratin-APC (Lot No. MAB5131, Abnova, Walnut, CA) and anti-CD45-fluorescein isothiocyanate (FITC) (Lot No. B116314, BioLegend San Diego, CA,) were typically used as the antibodies for the secondary labeling. Hoechst (Life Technologies, Carlsbad, CA) was also used as the nuclear stain to verify the labeled target was actually nucleated cells. Detailed procedures and experimental parameters were provided in our previous work.28
Results and Discussion
In a typical eDAR experiment, whole blood samples were labeled with antibodies conjugated with fluorophores. The operation of eDAR has been described in details previously,27 but briefly, blood sample was injected into the chip and divided into aliquots. A line-confocal detection scheme was then applied to rank the aliquots as “positive” or “negative”, which was determined by the labeling scheme. For example, in many applications, we labeled blood with anti-EpCAM-PE so only an aliquot that had a fluorescent signal from the particular dye conjugated to that antibody was ranked as a positive event. An automatic feedback scheme was applied to generate a switch of the direction of blood flow, which permitted the positive aliquot to be collected quickly. Due to the very low concentration of CTCs, more than 99.999% of the aliquots were discarded because of the absence of the desired fluorescence signal. Those aliquots that gave the positive fluorescence signal were transferred to another area on the microfluidic chip to be further purified and then counted. A series of downstream analyses can be performed on the trapped cells, such as a secondary immunostaining step, a more complicated staining/bleaching process,28 or the manipulation and culture at the single-cell level.
There are two key factors that determined the feature and performance of an eDAR platform: an efficient and active sorting scheme and a subsequent efficient purification scheme. Here, we designed the second generation of eDAR-platform to optimize these two components.
Redesigned Hydrodynamic Sorting Scheme
In the first generation of eDAR, we designed a mechanical valve to control the active sorting step, which was fast (about 2 ms response time) and robust compared to other reported switching mechanisms, such as the electroosmotic flow29 or the sol-gel transformation.30 Although promising, some design factors of this mechanical valve scheme may constrain the potential application of eDAR. To form the mechanical valve on the chip, 3 individual structural layers were required—the solenoid, its PDMS thread, and the microchannels on a 150-µm PDMS film. This would make the chip preparation complicated and time-consuming. Another shortcoming is the direct contact between the captured blood aliquots and the mechanical valve, which might increase the risk of the loss or damage of CTCs. In this work, we replaced the solenoid with an off-chip model, which is normally closed but can be opened in 2 to 3 ms when a 5V DC voltage is applied. Because this in-line solenoid was not a part of the microchip, the preparation of the microfluidic device was significantly simplified. The solenoids could be easily connected with any microchannels, so we could test many possible hydrodynamic sorting schemes. We designed and tested 8 different schemes (Figure 2S) to drive the fluidic switch. Because of the structure of this type of solenoid and the elastic nature of PDMS, the fluidic performance varied a lot (Table 1S), and we had to characterize each scheme to choose the one that offered the best performance.
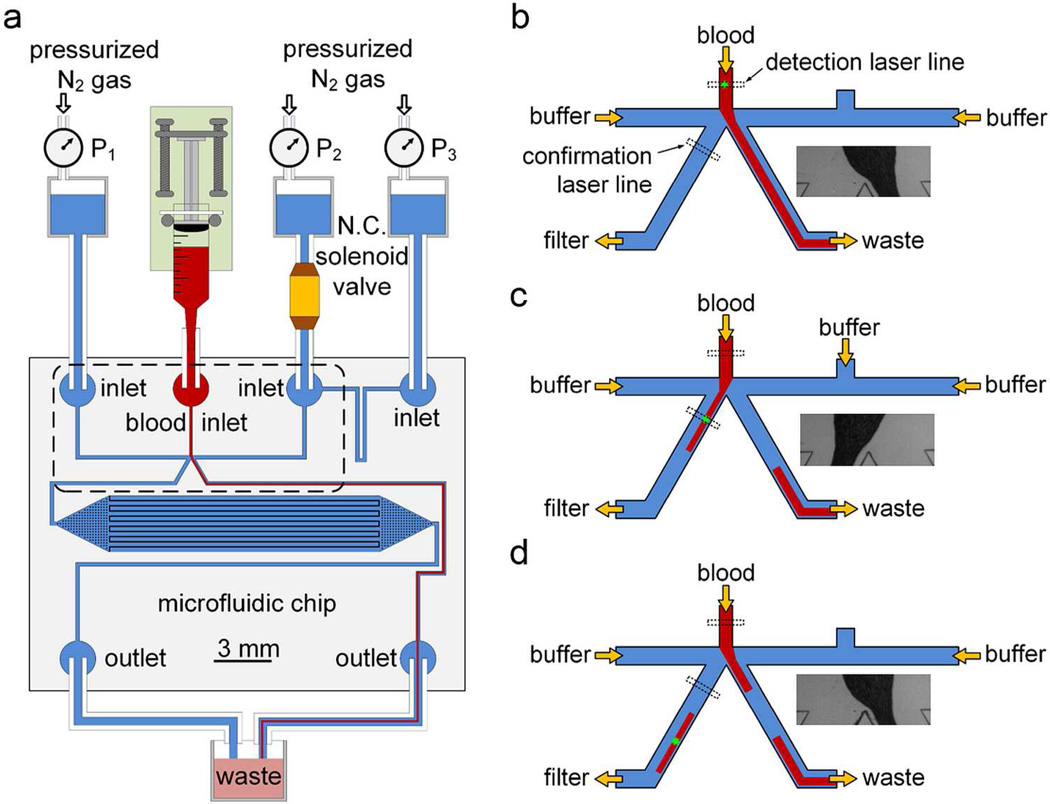
After characterization and optimization, the structure of the platform and the corresponding scheme of the hydrodynamic sorting were chosen for the reported second generation of eDAR (Figure 1). The labeled blood sample was injected into the top channel of the chip using a syringe pump (Figure 1a). Two side channels, where buffer flowed through, were used to control the active sorting step. There were two ports placed on the right-side channel, and both of them were connected to a pressurized buffer source. The normally closed solenoid was connected to the port near the sorting junction to control the hydrodynamic switch. There were two channels after the sorting junction. The one on the left was used to collect positive aliquots and deliver them to the filtration and collection area for further purification; the one on the right was the waste collection channel where all the negative aliquots flowed through.
Figure 1.
Microfluidic chip and hydrodynamic switching scheme of eDAR. a) General structure of the microfluidic chip and the configuration of the eDAR platform. The bottom left channel was to collect sorted aliquots and transfer them to the subsequent purification area, which had 20,000 microslits. The area marked with a dashed box is further explained in b-d. b) The flow condition when no positive aliquot was ranked. c) The blood flow was switched to the CTC collection channel, and the sorted aliquot was confirmed by the second APD. d) The blood flow was switched back after the aliquot was sorted.
When those aliquots were ranked as “negative” (Figure 1b), there was no voltage applied on the solenoid so it was closed. An initial pressure drop was set between the No.1 and 3 buffer sources in figure 1a, so the blood could only flow into the channel that collected the waste, which is also shown in the bright field image in figure 1b. When a positive event was detected by the first detection window, a 5V DC voltage was immediately applied on the solenoid to open the buffer flow from the No.2 buffer reservoir. This decreased the flow resistance of the buffer channel on the right side and generated a higher flow rate there. The blood flow was pushed from the right side to the left to collect the positive aliquot (Figure 1c). After this aliquot was collected and confirmed by the second detection window, the solenoid was closed to switch the blood flow back to the waste collection channel (Figure 1d). The time required for the switch-over and back was determined to be 2 to 3 ms for each (Figure 3S). This process was stable enough for eDAR even after more than 105 on-off cycles that we tested. The in-line solenoid was placed on the buffer line so blood could not come into contact with the solenoid, which eliminated the possibility of the blood-coagulation and cross-contamination. Moreover, in this scheme, there was a constant flow of buffer in the CTC collection channel during the eDAR process. This improved the efficiency of the subsequent purification step and prevented the formation of aggregates of cells.
Design and optimization of the further purification mechanism
In the first generation of eDAR, we used a piece of track-etched polycarbonate filter to retain and purify the captured CTCs. However, it required two additional layers in the micro-chip, as well as a complicated procedure to bond the filter to the PDMS. Here, we developed a new scheme of on-chip filtration based micro-slit structures, which were made of PDMS and did not require additional layers.
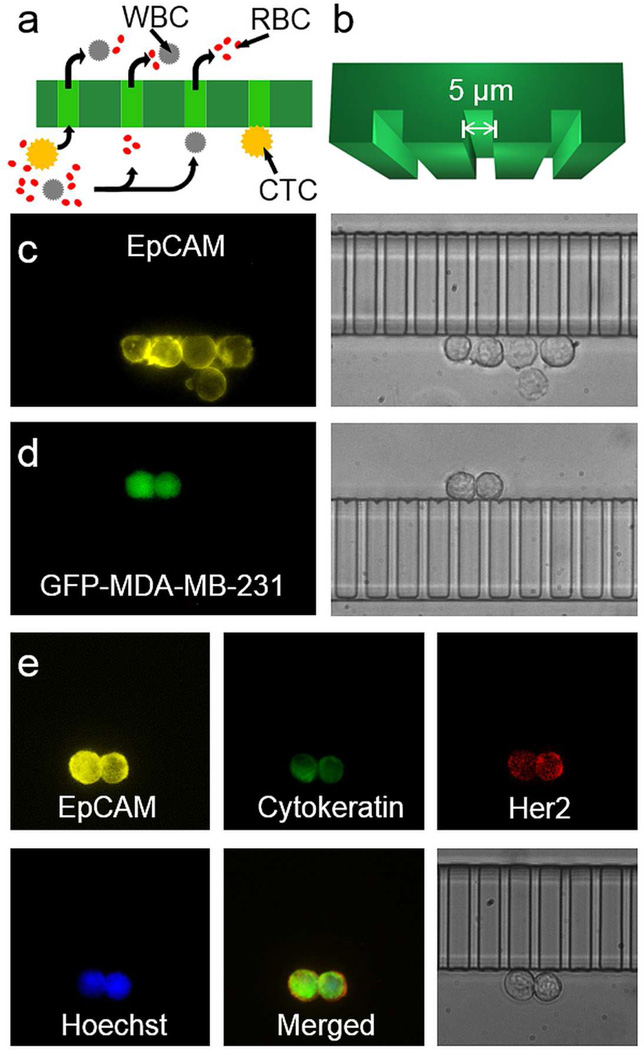
Figure 2a shows the basic structure of these microslits, which captured the CTCs without retaining any red blood cells (RBCs). The size of the slit was optimized to be 5-µm tall and 5-µm wide (Figure 2b), which is smaller than the ones used in most of the CTC methods based on filtration. With this size of microslits, we minimized the risk of losing small CTCs, while still allowing many WBCs to deform and pass through the filter. The purity of trapped CTCs was similar to the first generation of eDAR. Because the micro-filter was made of PDMS and bonded with a piece of coverslip, the imaging quality was improved significantly (Figure 2c and 2d) compared to the polycarbonate filter, which is not fully transparent and may generate the scattering and aberration. Moreover, because the cells could only be trapped along the array of slits, they could be easily referenced and tracked; in many other methods, the cells are distributed randomly on the surface. This trapping along the slits made the imaging procedure faster and the results of enumeration more accurate. The slits also made it faster and more efficient to perform the secondary labeling on the trapped CTCs. Figure 2e shows that two cancer cells labeled with anti-EpCAM-PE were trapped on the microslit. We fixed, permeabilzed, and labeled them using anti-Cytokeratin-Alexa488, anti-Her2-Alexa647 and Hoechst. Fluorescence images showed the expression of these markers on these two cells clearly, and the bright field image also confirmed their morphology. We also labeled these two cells with anti-CD45-Alexa700 as a negative control marker, and did not find any signal from the color channel that corresponded to this tag.
Figure 2.
Microslits and multicolor fluorescence imaging of captured CTCs. a) The sorted aliquots were further purified through the array of microslits. Objects in yellow represent CTCs; red and grey objects represent RBCs and WBCs, respectively. The curved arrows show the flow paths across the microslits. b) The 3D model of the microslits with a 5-µm height and a 5-µm width. c) Fluorescence (left) and bright field (right) images of five MCF-7 cells captured via eDAR. d) Fluorescence (left) and bright field (right) images of two MDA-MB-231-GFP cells captured based on their GFP signal without any prelabeling. E) Two SKBr-3 cells were captured by eDAR, and further labeled with additional markers.
To optimize the performance, we prepared and tested microchips with 1000, 5000, and 20,000 microslits. These chips helped us to determine the flow resistance across the filtration area, which could affect the hydrodynamic switching and the stress on the trapped cells. The eDAR-chip with 20,000 slits required a low pressure (< 4 psi) on the two side-buffer channels to balance the hydrodynamic switching process. The pressure drop across the microfilter was also lower, which would minimize the stress and deformation of the cell.
Characterization and analytical performance of eDAR
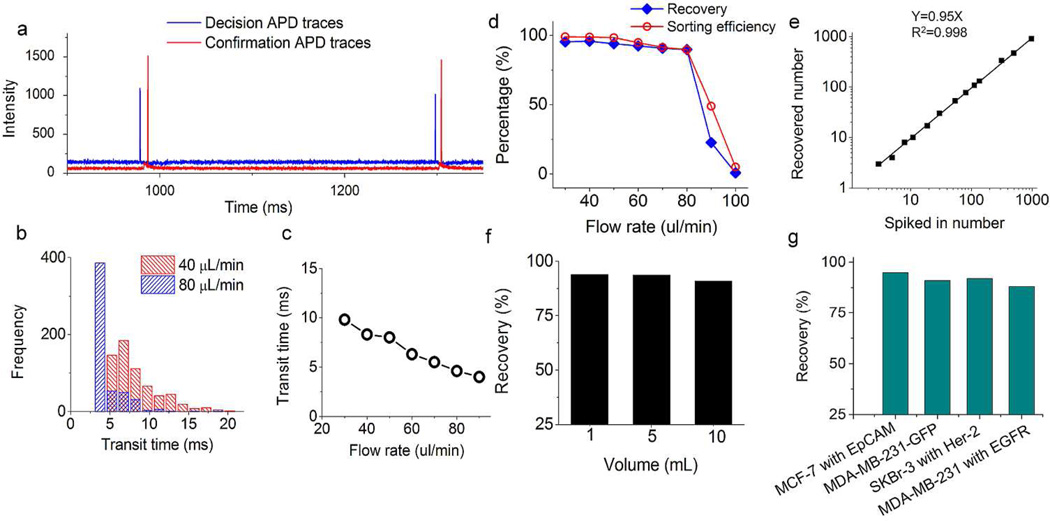
The efficiency of the active sorting step was monitored in real time. Figure 3a shows a small portion of the APD data from a pancreatic cancer patient sample. The signals in blue were from the first detection window that ranked the aliquots and controlled the sorting. The two peaks at 978 and 1298 ms represented two CTCs labeled with anti-EpCAM-PE that triggered the aliquot sorting. The two peaks in red show that two cancer cells flowed through the second detection window located on the collection channel, confirming that the two positive aliquots were actually sorted. It is worthwhile to point out that the background change from the second detector (Figure 3a) also confirmed that only a small portion of blood was collected by eDAR, contributing to the high enrichment ratio of CTCs ( up to a million fold for a typical clinical sample).
Figure 3.
Characterization and analytical performances of eDAR. a) The segment of the APD data from a pancreatic cancer sample that shows two events triggered the sorting which then were confirmed by the second detection window. b) The distribution of transit time at flow rates of 40 and 80 µL/min, respectively. c) A plot shows the fastest average transit time was about 4 ms when the flow rate was 90 µL/min. d) The recovery and sorting efficiency value versus different flow rate. e) The recovery ratio of MCF-7 cells spiked into whole blood. f) The recovery ratio of 300 MCF-7 cells spiked into 1, 5 and 10 mL of whole blood aliquots. g) The recovery ratio of 4 selection schemes of 4 breast cancer cell lines spiked into whole blood.
Because the labeled CTC had to flow from the first detection window to the second one, we could observe a time difference between the decision APD peak and its confirmation signal. This time difference was defined as the transit time of the sorted CTCs, which might vary because the CTCs can have different linear flow rates due to the nature of laminar flow in the microchannel. Figure 3b shows the distribution histogram of the transit time at flow rates of 40 and 80 µL/min. Generally, a higher volumetric flow rate of the blood resulted in a shortened transit time of the sorted CTCs (Figure 3c). When the flow rate was 90 µL/min, the average transit time was lowered to 4 ms, very close to the switching time of the sorting scheme (2 to 3 ms), which implies that this is the limit for the throughput for this design of eDAR.
If the transit time for a sorted CTC was shorter than the hydrodynamic switching time, the cell could not be sorted on this platform. The sorting efficiency was thus defined as the number of collected events versus the total number of events that triggered the sorting. Figure 3d shows the values of sorting efficiency at the flow rate of 30 to 100 µL/min. When the flow rate was 30 µL/min, the sorting efficiency was almost 100% because the average transit time at that flow rate was around 10 ms (Figure 3c). This transit time was long enough for the active sorting step to collect the CTCs. The sorting efficiency decreased to 90% at the flow rate of 80 µL/min, and then dropped to 49% when the flow rate was 90 µL/min. Figure 3d also shows the recovery efficiency of eDAR at different flow rates, which had a similar trend compared to the sorting efficiency. However, the recovery efficiency was defined as the number of spiked-in cells versus the number of recovered cells counted using multicolor fluorescence imaging on the eDAR chip. This performance is a combination of many factors, including the antibody-labeling efficiency, the line-confocal detection efficiency and the sorting efficiency. This explains the difference between the recovery and sorting efficiency at the same flow rate. As a result, for this second generation of eDAR, the upper limit of the throughput was 80 µL/min (12.5 min for 1 mL of blood) with an 88% recovery ratio. Although this throughput is higher than most CTC technologies for the analysis of whole blood, it can be further improved by designing a wider blood inlet channel or moving the first detection beam farther up.
Three to 975 MCF-7 cells were spiked into 1 mL of healthy blood to analyze the recovery efficiency at the flow rate of 50 µL/min. To ensure the accuracy of the cell numbers at the low end, we used a capillary counting method31 to precisely spike in cultured cells when the concentration was lower than 100 cells/mL. The average recovery ratio was 95% with an R2 value of 0.998 (Figure 3e), which is a little higher than the first generation of eDAR (93%).27 Because the concentration of CTCs is usually very low, the enumeration results were affected by the Poisson distribution. In this case, the ability to analyze a larger volume of whole blood sample with an acceptable throughput and recovery ratio was very important. In the first generation of eDAR, the capacity was limited by the number of pores on the polycarbonate filter, which was around 1000. In this reported second-generation eDAR, we had 20,000 mircoslits, which resulted in significantly increased capacity. We spiked the same number of MCF-7 cells into 1, 5 and 10 mL of healthy blood, and then analyzed these 3 samples at the flow rate of 50 µL/min. There was no significant change in their recovery ratio (Figure 3f), which shows that our method is capable of running a large amount of whole blood with high efficiency and throughput.
Although EpCAM was used in most of the CTC studies to select tumor cells, increasingly more studies have reported that CTCs with a low EpCAM expression have more mesenchymal characteristics and are more aggressive.32 The latest eDAR platform is sufficiently flexible to use any labeling scheme to select rare cells so we can capture tumor cells using biomarkers other than EpCAM. We designed three schemes to select different cultured breast cancer cell lines (Figure 3g). EpCAM was used to select MCF-7 cells, Her-2 was used to select SKBr-3 cells, and EGFR was used to select MDA-MB-231 cells. All these three schemes isolated and trapped the targeted cells with a recovery ratio higher than 88%. Another unique and important feature of eDAR is the independence of where the marker is located. For example, other technologies, such as the surface capture methods or immunomagnetic methods, only can capture the antigens on the cell surface. But our method could select cells with an intracellular marker, such as GFP (Figure 2d). The recovery ratio of the MDA-MB-231-GFP cells spiked into whole human blood was 91% (Figure 3g). Since fluorescent proteins are widely used in animal models to study the progression and mechanisms of metastasis,33 eDAR could be an ideal tool to select CTCs in these models without any immunostaining steps.
High-throughput analysis of samples from patients with pancreatic cancer
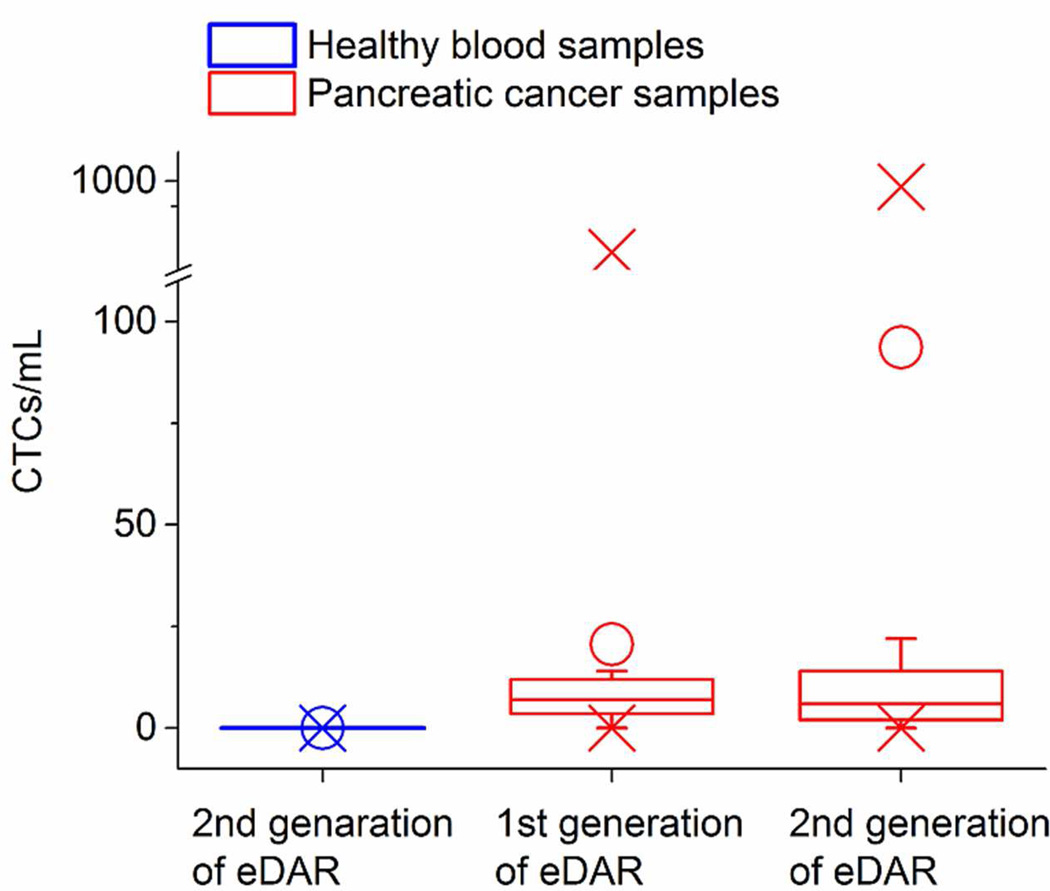
Blood samples from 15 healthy donors were used to evaluate the false positive ratio of this method; no CTCs were found in any of them. We collected 26 blood samples from the patients with pancreatic cancer. Sixteen of them were analyzed using the first generation of eDAR and the other 10 samples were analyzed using the newer eDAR platform. Figure 4 shows the distribution of the three data sets: the control blood analyzed by the current method, pancreatic cancer samples analyzed by the first generation of eDAR, and the pancreatic cancer samples analyzed by the current method. The raw data of those clinical samples are in Table 2S. With our current method, we detected CTCs in 80% (8 of 10) of the samples ranging from 2 to 872 cells/mL. CTC clusters, reported by previous studies,34 were also observed in the patient blood samples. It is interesting to point out that many of the clusters observed in our experiments had low EpCAM expression. Figure 4S shows a cluster of CTCs which had a high expression of cytokeratin but a low expression of EpCAM. We also compared the clinical performance of the two generations of eDAR using the analysis of variance (ANOVA) method, which showed that the two data sets were not significantly different (p=0.30, α=0.05).
Figure 4.
The distribution of 15 control samples and 10 pancreatic cancer samples analyzed by the method reported here, as well as the distribution of 16 pancreatic cancer samples analyzed by the first generation of eDAR. O shows the average values for each data set; X shows the minimum and maximum values we found in each data set.
Conclusion
eDAR has proven to be an “all-in-one” platform for analyzing rare cells in complicated biological matrices, such as CTCs in peripheral blood. Here, we redesigned the platform by incorporating a new active sorting scheme and a microfabricated filter based on PDMS slits. The structure of the microchip was much simplified, resulting in higher yield and better quality control in chip preparation. Previously, the production cycle of a batch of microfluidic chips was almost two days with ~ 50% yield; in the reported method, it only took us 2 hours to prepare a batch of microchips with an almost 100% yield. The imaging quality to enumerate and analyze CTCs was also improved. The analytical performance was optimized so that we were able to analyze 1 mL of whole blood in 12.5 min with an average recovery ratio at 95% and a zero false positive rate (n=15). Biomarkers other than EpCAM were also successfully applied to select different subpopulations of CTCs, including intracellular markers. We also validated this method by analyzing pancreatic cancer samples taken from patients. We believe the second generation of the eDAR platform offers greatly simplified chip preparation and is more robust and flexible in analyzing CTCs from patient blood while offering higher throughput. It has the potential to benefit the analysis of other types of rare cells as well.
Supplementary Material
Acknowledgement
We wish to thank all the patients who participated in the study. We thank Prof. Gail Sonenshein at Tuffs University for providing the cultured cancer cells. R. K. A. is supported by a T32 grant from the NIH NCI (T32CA138312). We are grateful to the Department of Defense CDMRP Program (BC100510), NIH (R21CA147831) and Life Science Discovery Fund for support of this work.
Footnotes
Conflict of interest
The authors declare the following competing financial interest(s): D.T.C. and P.G.S. have financial interest in MiCareo, which has licensed the described technology from the University of Washington.
References
- 1.Chaffer CL, Weinberg RA. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Mocellin S, Keilholz U, Rossi CR, Nitti D. Trends Mol. Med. 2006;12:130–139. doi: 10.1016/j.molmed.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Pantel K, Alix-Panabieres C, Riethdorf S. Nat. Rev. Clin. Oncol. 2009;6:339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 4.Paterlini-Brechot P, Benali NL. Cancer Lett. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Andreopoulou E, Cristofanilli M. Expert Rev. Anticancer Ther. 2010;10:171–177. doi: 10.1586/era.09.105. [DOI] [PubMed] [Google Scholar]
- 6.Hou JM, Krebs M, Ward T, Sloane R, Priest L, Hughes A, Clack G, Ranson M, Blackhall F, Dive C. Am. J. Pathol. 2011;178:989–996. doi: 10.1016/j.ajpath.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen L, Meropol NJ. J. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 8.Danila DC, Fleisher M, Scher HI. Clin. Cancer Res. 2011;17:3903–3912. doi: 10.1158/1078-0432.CCR-10-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patriarca C, Macchi RM, Marschner AK, Mellstedt H. Cancer Treat. Rev. 2012;38:68–75. doi: 10.1016/j.ctrv.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HTK, Lee W, Amini H, Di Carlo D. Anal. Bioanal. Chem. 2010;397:3249–3267. doi: 10.1007/s00216-010-3721-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riethdorf S, Mueller V, Zhang L, Rau T, Loibl S, Komor M, Roller M, Huober J, Fehm T, Schrader I, Hilfrich J, Holms F, Tesch H, Eidtmann H, Untch M, von Minckwitz G, Pantel K. Clin. Cancer Res. 2010;16:2634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 12.Balasubramanian P, Yang L, Lang JC, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Mol. Pharm. 2009;6:1402–1408. doi: 10.1021/mp9000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby BJ, Jodari M, Loftus MS, Gakhar G, Pratt ED, Chanel-Vos C, Gleghorn JP, Santana SM, Liu H, Smith JP, Navarro VN, Tagawa ST, Bander NH, Nanus DM, Giannakakou P. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P, Stratton ZS, Dao M, Ritz J, Huang TJ. Lab Chip. 2013;13:602–609. doi: 10.1039/c2lc90148j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao M, Schiro PG, Kuo JS, Koehler KM, Sabath DE, Popov V, Feng Q, Chiu DT. Anal. Chem. 2013;85:2465–2471. doi: 10.1021/ac400193b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Issadore D, Chung J, Shao H, Liong M, Ghazani AA, Castro CM, Weissleder R, Lee H. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Goettert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ST, Liu K, Liu JA, Yu ZTF, Xu XW, Zhao LB, Lee T, Lee EK, Reiss J, Lee YK, Chung LWK, Huang JT, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng HR. Angew. Chem. Int. Ed. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng W, Chen T, Kamath R, Xiong X, Tan W, Fan ZH. Anal. Chem. 2012;84:4199–4206. doi: 10.1021/ac3005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai Y-C, Cote RJ. Clin. Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goda K, Ayazi A, Gossett DR, Sadasivam J, Lonappan CK, Sollier E, Fard AM, Hur SC, Adam J, Murray C, Wang C, Brackbill N, Di Carlo D, Jalali B. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11630–11635. doi: 10.1073/pnas.1204718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun JS, Li MM, Liu C, Zhang Y, Liu DB, Liu WW, Hu GQ, Jiang XY. Lab Chip. 2012;12:3952–3960. doi: 10.1039/c2lc40679a. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh HB, Marrinucci D, Bethel K, Curry DN, Humphrey M, Krivacic RT, Kroener J, Kroener L, Ladanyi A, Lazarus N, Kuhn P, Bruce RH, Nieva J. Biosens. Bioelectron. 2006;21:1893–1899. doi: 10.1016/j.bios.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 25.Riethdorf S, Fritsche H, Mueller V, Rau T, Schindibeck C, Rack B, Janni W, Coith C, Beck K, Jaenicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Clin. Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang XJ. Lab Chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiro PG, Zhao M, Kuo JS, Koehler KM, Sabath DE, Chiu DT. Angew. Chem. Int. Ed. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao M, Wei B, Chiu DT. Methods. 2013 doi: 10.1016/j.ymeth.2013.08.006. (In press), http://dx.doi.org/10.1016/j.ymeth.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Schiro PG, Gadd JC, Yen GS, Chiu DT. J. Phys. Chem. B. 2012;116:10490–10495. doi: 10.1021/jp3019233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirasaki Y, Tanaka J, Makazu H, Tashiro K, Shoji S, Tsukita S, Funatsu T. Anal. Chem. 2006;78:695–701. doi: 10.1021/ac0511041. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Schiro PG, Kuo J, Ng L, Chiu DT. Anal. Chem. 2008;81:1285–1290. doi: 10.1021/ac802250d. [DOI] [PubMed] [Google Scholar]
- 32.Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, von Ahsen O. BMC Cancer. 2012;12 doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yagublu V, Ahmadova Z, Hafner M, Keese M. In Vivo. 2012;26:599–607. [PubMed] [Google Scholar]
- 34.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.