Abstract
Poly(lactic acid) (PLA) is a versatile synthetic polyester. We noted that this depsipeptide analog of polyalanine has a helical structure that resembles a polyproline II helix. Using natural bond orbital analysis, we find that n→π* interactions between sequential ester carbonyl groups contribute 0.44 kcal/mol per monomer to the conformational stability of PLA helices. We conclude that analogous n→π* interactions could direct the folding of a polypeptide chain into a polyproline II helix prior to the formation of hydrogen bonds between backbone amides.
Polyesters have found widespread utility due to their inexpensive preparation and structural integrity.1 Poly(lactic acid) (PLA) is a recyclable polyester that can be prepared by condensing lactic acid, a renewable resource (Fig. 1).2-4 The thermal and structural properties of PLA can be adjusted by varying the ratio of l- and d-lactic acid monomers, or by altering the polymer processing conditions. These ensuing materials have received significant attention amongst macromolecular scientists, especially for use in biocompatible and biodegradable devices.2-4,5
Fig. 1.
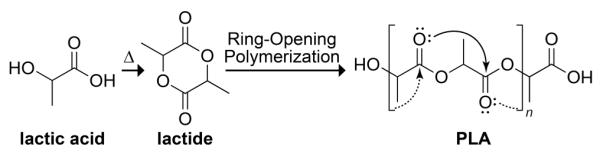
Synthetic route to PLA. Curved arrows indicate putative n→π* interactions.
Interest in tuning properties of PLA has motivated the determination of its structure in atomic detail. These analyses have revealed the existence of conformational isomers, which arise from different preparation conditions.6-7 The α form of PLA has received the most attention, due to its high stability.
Fibre diffraction of α-PLA has proven challenging, and the ensuing structure has been revised several times since its first report.8-12 Recently, data from neutron diffraction and NMR spectroscopy have been used to complement X-ray diffraction data.13-14 Though refinement of the structural model continues, the overall topology appears to be consistent between studies (Fig. 2A).
Fig. 2.
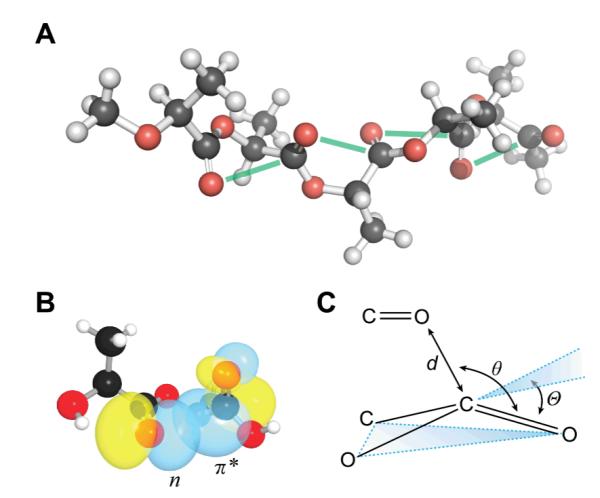
n→π* Interactions in PLA. (A) Five-residue segment of α-PLA from l-lactic acid (i.e., isotactic PLLA).12 Green lines show putative n→π* interactions. (B) Overlap of the n and π* orbitals in di(l-lactic acid) (CCDC Refcode: DUZMER). (C) Structural parameters describing the n→π* interaction and the resulting pyramidalization of the acceptor carbonyl.
We became interested in PLA due to its similarity to polyalanine. Indeed, PLA is the depsipeptide counterpart to polyalanine wherein each amide linkage is replaced by an ester. The amide-to-ester modification has proven useful for revealing the contribution of hydrogen bonds to the structure and stability of peptides and proteins because incorporation of the ester linkage deletes a backbone hydrogen-bond donor and reduces the strength of a hydrogen bond with the carbonyl oxygen.15-20
By examining the structure of a peptide-like polymer that is incapable of forming intramolecular hydrogen bonds, we sought to isolate other interactions that bias the conformation of peptide chains. In particular, we wished to determine the role of the n→π* interaction in dictating the conformational geometry of PLA. In an n→π* interaction, the filled p-type lone pair (n) of a carbonyl oxygen overlaps with the empty π* antibonding orbital of a nearby carbonyl group (Fig. 1 and 2B). This overlap allows for orbital mixing and the subsequent release of energy. Such an interaction occurs when the donor oxygen contacts the acceptor carbonyl carbon within the sum of their van der Waals radii (rO + rC = 3.22 Å), and along the Bürgi–Dunitz trajectory for nucleophilic addition (∠O⋯C=O = ~109°).21 We have estimated that n→π* interactions between amides likely contribute 0.27 kcal/mol of stabilization energy per interaction,22 and we have shown that these interactions are present in protein structures, especially helices.23 The question remains, however, does the n→π* interaction bias a peptide toward a particular conformation, or is the interaction an artefact of a particular structural motif? By examining the structure of a polymer devoid of hydrogen bonds, we hoped to ascertain the relevance of the n→π* interaction to macromolecular conformation.
Upon inspection of the structure of α-PLA,12 we observed that its backbone torsion angles (Table 1) bear striking similarity to those of the polyproline II helix, which has backbone torsion angles of and , and the strands of a collagen triple helix.24 We showed previously that these torsion angles allow for effective n→π* interactions.23 Indeed, the average O⋯C distance in the α-PLA structure is 2.98 Å, which is 0.24 Å less than the sum of the van der Waals radii; moreover, the average ∠O⋯C=O is 94°, which is consistent with an n→π* interaction.23
Table 1.
Structural parameters and n→Π* energies of lactic acid polymers
| Polymer | ϕ (°) | ψ (°) | d (Å)d | θ (°)d |
En→π* (kcal/mol)e |
Θ (°)d |
|---|---|---|---|---|---|---|
| α-PLAa | −63.7 | 154.4 | 2.98 | 93.6 | 0.44 | ND |
| Di(l-lactic acid)b | −69.2 | 148.0 | 2.90 | 102.0 | 0.67 | 2.62 |
| Tri(l-lactic acid)c | −69.2 | 163.5 | 2.98 | 92.7 | 0.41 | 3.40 |
Values are the mean over five consecutive l-lactic acid residues.12 For an image, see: Fig. 2A. ND, not determined.
Values are the mean from two molecules in the unit cell of CCDC Refcode DUZMER. For an image, see: Fig. 2B.
Values are the mean from three residues in CCDC Refcode DUZMIV.
For definitions, see: Fig. 2C.
Values are from second-order perturbation theory.
To evaluate whether or not an n→π* interaction is operative in the structure of α-PLA, we conducted natural bond orbital (NBO) analysis of its crystalline structure at the B3LYP/6-311+G(2d,p) level of theory.25-27 We observed an average n→π* energy of 0.44 kcal/mol per interaction. This value is consistent with a strong n→π* interaction between the carbonyl groups of adjacent backbone esters in α-PLA.
To establish further the presence of an n→π* interaction, we searched for a structural signature. As the n→π* interaction populates the π* orbital of the acceptor carbonyl, it induces a pyramidalization of the carbonyl group from planar sp2 geometry (Fig. 2C), which can be detected in high-resolution crystal structures.28-31,22 Unfortunately, the initial α-PLA structure-determination assumed planarity of the ester bond,12 thereby obscuring the most definitive signature of an n→π* interaction. Later structures do not provide enough resolution to determine pyramidalization accurately.13 Accordingly, we sought high-resolution structures of lactic-acid oligomers.
We analysed structures of di(l-lactic acid) and tri(l-lactic acid), which were obtained from the Cambridge Structural Database.32 To ensure that these short oligomers are appropriate models for the structure of α-PLA, we compared their backbone torsion angles to those observed in α-PLA and found gratifying agreement (Table 1). We also employed DFT calculations and NBO analysis to estimate the energy of the n→π* interaction in these molecules and found that the n→π* energies are consistent with those observed in the polymer. Confident that these structures are an accurate reflection of the structure of α-PLA, we then determined the distortion of the backbone esters from planarity, as measured by the angle Θ. In both structures, we observed substantial pyramidalization of the putative n→π* acceptor toward the putative donor. In the absence of an attractive interaction, one would expect distortion to occur in the opposite direction, so as to reduce unfavorable Pauli repulsion.33 Accordingly, the observed pyramidalization is strong evidence of an attractive n→π* interaction between the monomeric units in α-PLA.
These observations have broad implications. First, they imply a new means to modulate the structure of organic polymers. We found previously that the nucleophilicity of sulfur in thioamides can be exploited to increase the strength of an n→π* interaction28,22 and that surrogate alkenes and fluoroalkenes can be used to attenuate an n→π* interaction.33 These isosteres20 could be used to produce polymeric materials with tailored structural and thermal properties. Secondly, as the n→π* interaction is likely to reduce the electrophilicity of the acceptor carbonyl by contributing additional electron density,34-35 it could contribute to the observed hydrolytic stability of PLA. Thirdly, because n→π* interactions are extant in PLA even without the potential for intramolecular hydrogen bonding, we conclude that the n→π* interaction can operate independently of the geometric constraints imposed by hydrogen-bonding patterns. Finally, the observation of polyproline-like structure in PLA is itself significant, given that this structural motif is prevalent in the unfolded state of proteins.36-40 During their folding, polypeptide chains are likely to sample highly local interactions sooner than less local ones. Operating between adjacent residues (that is, i→i+1), the n→π* interaction is considerably more local than common hydrogen-bonding patterns such as that in the α-helix (i→i+4). Thus, the presence of n→π* interactions in the structure of PLA suggests that—before hydrogen bonds can form—the conformation of polypeptide chains can be guided by n→π* interactions.41
Acknowledgments
We thank M. K. Mahanthappa and M. R. Aronoff for contributive discussions. R.W.N. was supported by NIH Biotechnology Training Grant T32 GM008349. This work was supported by grants R01 AR04476 (NIH) and CHE-1124944 (NSF). High-performance computing was supported by grant CHE-0840494 (NSF).
Notes and references
- 1.Scheirs J, Long TE, editors. Modern Polyesters: Chemistry and Technology of Polyesters and Copolyesters. John Wiley & Sons; West Sussex, England: 2003. [Google Scholar]
- 2.Garlotta D. J. Polym. Environ. 2001;9:63–84. [Google Scholar]
- 3.Piemonte V, editor. Polylactic Acid: Synthesis, Properties and Applications. Nova Science Publishers; Hauppauge, NY: 2011. [Google Scholar]
- 4.Sin LT, Rahmat AR, Rahman WA. Polylactic Acid: PLA Biopolymer Technology and Applications. Elsevier; Oxford, UK: 2012. W. A. [Google Scholar]
- 5.Ikada Y, Tsuji H. Macromol. Rapid Commun. 1999;21:117–132. [Google Scholar]
- 6.Cartier L, Okihara T, Ikada Y, Tsuji H, Puiggali J, Lotz B. Polymer. 2000;41:8909–8919. [Google Scholar]
- 7.Puiggali J, Ikada Y, Tsuji H, Cartier L, Okihara T, Lotz B. Polymer. 2000;41:8921–8930. [Google Scholar]
- 8.de Santis P, Kovacs AJ. Biopolymers. 1968;6:299–306. doi: 10.1002/bip.1968.360060305. [DOI] [PubMed] [Google Scholar]
- 9.Hoogsteen W, Postema AR, Pennings AJ, ten Brinke G. Macromolecules. 1990;23:634–642. [Google Scholar]
- 10.Kobayashi J, Asahi T, Ichiki M, Oikawa A, Suzuki H, Watanabe T, Fukada E, Shikinami Y. J. App. Phys. 1995;77:2957–2973. [Google Scholar]
- 11.Aleman C, Lotz B, Puiggali J. Macromolecules. 2001;34:4795–4801. [Google Scholar]
- 12.Sasaki S, Asakura T. Macromolecules. 2003;36:8385–8390. [Google Scholar]
- 13.Wasanasuk K, Tashiro K, Hanesaka M, Ohhara T, Kurihara K, Kuroki R, Tamada T, Ozeki T, Kanamoto T. Macromolecules. 2011;44:6441–6452. [Google Scholar]
- 14.Pawlak T, Jaworska M, Potrzebowski MJ. Phys. Chem. Chem. Phys. 2013;15:3137–3145. doi: 10.1039/c2cp43174b. [DOI] [PubMed] [Google Scholar]
- 15.Koh JT, Cornish VW, Schultz PG. Biochemistry. 1997;36:11314–11322. doi: 10.1021/bi9707685. [DOI] [PubMed] [Google Scholar]
- 16.Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 17.Deechongkit S, Dawson PE, Kelly JW. J. Am. Chem. Soc. 2004;126:16762–16771. doi: 10.1021/ja045934s. [DOI] [PubMed] [Google Scholar]
- 18.Fu Y, Gao J, Bieschke J, Dendle MA, Kelly JW. J. Am. Chem. Soc. 2006;128:15948–15949. doi: 10.1021/ja065303t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Kelly JW. Protein Sci. 2008;17:1096–1011. doi: 10.1110/ps.083439708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary A, Raines RT. ChemBioChem. 2011;12:1801–1807. doi: 10.1002/cbic.201100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bürgi HD, Dunitz JD, Shefter E. Acta Crystallogr. 1974;B30:1517–1527. [Google Scholar]
- 22.Newberry RW, VanVeller B, Guzei IA, Raines RT. J. Am. Chem. Soc. 2013;135:7843–7846. doi: 10.1021/ja4033583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartlett GJ, Choudhary A, Raines RT, Woolfson DN. Nat. Chem. Biol. 2010;6:615–620. doi: 10.1038/nchembio.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoulders MD, Raines RT. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed AE, Curtiss LA, Weinhold F. Chem. Rev. 1988;88:899–926. [Google Scholar]
- 26.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr., Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- 27.Glendening ED, Badenhoop JK, Reed AE, Carpenter JE, Bohmann JA, Morales CM, Weinhold F. Theoretical Chemistry Institute, University of Wisconsin–Madison; Madison, WI: 2012. [Google Scholar]
- 28.Choudhary A, Gandla D, Krow GR, Raines RT. J. Am. Chem. Soc. 2009;131:7244–7246. doi: 10.1021/ja901188y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhary A, Pua KH, Raines RT. Amino Acids. 2011;41:181–186. doi: 10.1007/s00726-010-0504-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choudhary A, Raines RT. Protein Sci. 2011;20:1077–1081. doi: 10.1002/pro.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhary A, Kamer KJ, Raines RT. J. Org. Chem. 2011;76:7933–7937. doi: 10.1021/jo201389d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen FH. Acta Crystallogr. 2002;B58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- 33.Jakobsche CE, Choudhary A, Miller SJ, Raines RT. J. Am. Chem. Soc. 2010;132:6651–6653. doi: 10.1021/ja100931y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhary A, Kamer KJ, Powner MW, Sutherland JD, Raines RT. ACS Chem. Biol. 2010;5:655–657. doi: 10.1021/cb100093g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollock SB, Kent SBH. Chem. Commun. 2011;47:2342–2344. doi: 10.1039/c0cc04120c. [DOI] [PubMed] [Google Scholar]
- 36.Shi Z, Olson CA, Rose GD, Baldwin RL, Kallenbach NR. Proc. Natl. Acad. Sci. USA. 2002;99:9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mezei M, Fleming PJ, Srinivasan R, Rose GD. Proteins. 2004;55:502–507. doi: 10.1002/prot.20050. [DOI] [PubMed] [Google Scholar]
- 38.Hamburger JB, Ferreon JC, Whitten ST, Hilser VJ. Biochemistry. 2004;43:9790–9799. doi: 10.1021/bi049352z. [DOI] [PubMed] [Google Scholar]
- 39.Whittington SJ, Chellgren BW, Hermann VM, Creamer TP. Biochemistry. 2005;44:6269–6275. doi: 10.1021/bi050124u. [DOI] [PubMed] [Google Scholar]
- 40.Grdadolnik J, Mohacek-Grosev V, Baldwin RL, Avbelj F. Proc. Natl. Acad. Sci. USA. 2011;108:1794–1798. doi: 10.1073/pnas.1017317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Z, Kallenbach NR. Proc. Natl. Acad. Sci. USA. 2011;108:3–4. doi: 10.1073/pnas.1017021108. [DOI] [PMC free article] [PubMed] [Google Scholar]


