Abstract
Introduction
Tonic immobility (TI) is fear-induced freezing that animals may undergo when confronted by a threat. It is principally observed in prey species as defence mechanisms. In our preliminary research, we detected large inter-individual variations in the frequency and duration of freezing behavior among newly hatched domestic chicks (Gallus gallus). In this study we aim to identify the copy number variations (CNVs) in the genome of chicks as genetic candidates that underlie the behavioral plasticity to fearful stimuli.
Methods
A total of 110 domestic chicks were used for an association study between TI responses and copy number polymorphisms. Array comparative genomic hybridization (aCGH) was conducted between chicks with high and low TI scores using an Agilent 4×180 custom microarray. We specifically focused on 3 genomic regions (>60 Mb) of chromosome 1 where previous quantitative trait loci (QTL) analysis showed significant F-values for fearful responses.
Results
ACGH successfully detected short CNVs within the regions overlapping 3 QTL peaks. Eleven of these identified loci were validated by real-time quantitative polymerase chain reaction (qPCR) as copy number polymorphisms. Although there wkas no significant p value in the correlation analysis between TI scores and the relative copy number within each breed, several CNV loci showed significant differences in the relative copy number between 2 breeds of chicken (White Leghorn and Nagoya) which had different quantitative characteristics of fear-induced responses.
Conclusion
Our data shows the potential CNVs that may be responsible for innate fear response in domestic chicks.
Introduction
Tonic immobility (TI) is an unlearned defensive behavior characterized by temporal paralysis [1], and is widely used to measure the extent of fear responses in chickens [2], [3] and other animals [4]–[6]. An individual with fewer induction attempts and longer TI responses is generally considered to be more fearful than those with many induction attempts and shorter TI responses [7]. Indeed, previous studies have demonstrated that the Red Junglefowl (RJF) and domesticated White Leghorn (WL) can be discriminated by different quantitative distributions of TI indices [8], [9]. Our previous study also detected significant differences in the TI responses between WL and Nagoya breeds (NG) in newly hatched chicks (days 1–2 after hatching) [10]. These significant levels of interbreed heterogeneity may be attributed to the artificial selection of response insensitivity to human handling during the process of chicken domestication.
Considerable efforts have been taken to understand the molecular basis of anxiety and fear-based responses based on the hypothesis that genetic linkage or pleiotropic gene effects could explain different reactions to fearful stimuli. In chickens, there are 2 major quantitative trait loci (QTL) for individual growth on chromosome 1 (Growth1 and Growth2) [11], and surprisingly, Growth1 QTL contains several genes which, together, affect personality [8]. Moreover, an important finding has been made regarding genetic links between fear responses and major growth QTLs in an RJF × WL intercross [7]. These findings raise the possibility that the growth QTL may contain genes or genetic regions that influence the extent of fear-related behavior in chickens with far-reaching effects at the molecular and cellular levels.
Another effective and reliable approach for identifying genes or genomic regions responsible for normal behaviors is to perform genome-wide searches for copy number gains and losses. Copy number variation (CNV) is defined as genomic duplications or deletions in relatively long elements (1 kb to several Mb in size). With increasing resolution in the detection of smaller CNVs, this definition has expanded to include short structural variants less than 1 kb, known as short CNVs (sCNVs) [12]. In humans, CNVs have been linked to various behaviors including brain-related disorders [13]. In non-human vertebrates, including chickens, a growing number of studies have focused on the associations between CNVs and observed phenotypic heterogeneities [14], and thus CNV has been recently recognized as an important source of genetic variability that may affect phenotypes because of the rearrangement of the genes or regulatory elements.
The main goal of this study was to identify novel sCNVs between chicks with high and low TI scores by using an array comparative genomic hybridization (aCGH) approach. We targeted 3 different QTL in chromosome 1, for which significant F values had been detected for TI responses in chickens. Our approach provides an efficient way to narrow the number of plausible factors that account for differences in fear-induced behaviors by focusing on the regions containing interesting QTL.
Materials and Methods
Animals Used in this Study
We used 3 breeds/strains of chicken with different selection histories (NG5 [n = 32], NG7 [n = 39], and WL [n = 39]). NG was chosen as the target chicken breed in this study because of the following reason: NG chicks occasionally panic and are hurt when they are frightened by sounds or small stimuli. It is especially important for future management of economically significant breeds to uncover their genetic basis of fear-related behaviors.
The NG breed was originated from a cross between a local chicken from the City of Nagoya and the Chinese Buff Cochin in the early 1880 s. In 1905, this breed was recognized as the first practical breed for poultry farming in Japan, and the NG was formally established in 1919 [15]. Of the various strains, NG5 and NG7 have distinct histories of selection either as a layer-type strain (NG5) or as a meat-type strain (NG7). The details of husbandry of chicks have been described elsewhere [10].
Tonic Immobility Test
A TI test was conducted using male chicks on days 1 and 2 after hatching. We measured TI responses 6 times for each chick (3 times on each of the days 1 and 2), regardless of the success rate of TI induction. We employed the same method used for assessing TI scores in adult chickens [1]; each chick was placed on its back in a V-shaped cradle and was kept there with light pressure on its breast for 5 s. After removing the pressure, chicks were not considered to be in TI status if the bird jumped up or righted itself within 5 s. We carried out the procedure 3 times in succession on each individual. The operator recorded the number of induction attempts required to induce a chick into the TI status as well as the time until righting in each successful TI induction (hereafter expressed as TIind and TIdur, respectively). If a bird did not enter TI status for all 6 attempts, TIind was assigned a score of 7. When 10 min had passed since the bird entered TI, the chick was forced to stand up, and TIdur was scored as 600 s. A fixed video camera was also used to ensure that environmental factors such as unexpected noise and changes in light intensity had no influence on chicks’ fear-relevant behaviors.
Microarray Design
Blood samples were collected from the 3 chicken breeds/strains of chickens and stored at −20°C until DNA extraction. We isolated genomic DNA from 110 chicks by using either the PUREGENE® DNA Purification Kit (Gentra Systems, Minneapolis, USA) or DNeasy® Blood and Tissue Kit (Qiagen, Tokyo, Japan). DNA concentration of each extract was measured using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). Four individuals with the highest average TIdur were chosen from NG (IDs: NG933 [TIind = 1; average TIdur = 248.8 s], NG4692 [TIind = 1; average TIdur = 172.3 s], and NG3557 [TIind = 1; average TIdur = 255.0 s]) and WL (WL3597 [TIind = 1; average TIdur = 199.3 s]) strains as samples compared in aCGH analysis. One NG chick (NG999 [TIind = 7; TIdur = 0]) that had not been induced the TI status in all 6 attempts was chosen as the reference sample. We used a chicken CGH Microarray 4×180K (Agilent Technologies, Tokyo, Japan), containing 180,000 custom probes of 60-mer, because Agilent’s 60-mer offers the highest sensitivity and reproducibility among the currently available commercial platforms [16]–[18]. We designed these probes by using the eArray software (Available: https://earray.chem.agilent.com/earray/Accessed 2011 May 19). Note that these probes covered a total of 60 Mb in exonic, intronic, and intergenic regions of chromosome 1, where significant F values were detected by previous QTL analysis for fear-related behaviors [7]. Information on QTL for TI attempts (trait ID: 2123) and duration (2124) in the chicken genome was obtained from the QTL database (Available: http://www.genome.iastate.edu/cgi-bin/QTLdb/GG/index. Accessed 2011 Oct 11). The mean probe spacing was 1,029 bp, and the median probe spacing was 264 bp. Our strategy was somewhat analogous to that employed by a previous study [19], which targeted for restricted chromosomal regions in the porcine genome. There were several reasons for targeting sCNVs as a candidate for TI response variability. Although no clear pattern for CNV effect versus CNV-gene distance has been observed, smaller variants less than 1 kb have been found to be more likely to regulate gene transcription than larger variants [12]. Moreover, a recent study suggested that sCNVs tend to originate from the presence of a variable number of tandem repeats, which could provide a source of genetic variability for modifying normal and abnormal human behaviors [20]. All hybridizations were performed using 2 dyes for labeling reference (Cy3) and sample DNA (Cy5). The hybridization and initial data analysis were performed by MACS® Molecular genomic service (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany).
Detection of Copy Number Variations
Statistical analysis for CNV detection was performed using Agilent GENOMIC WORKBENCH Standard Edition 6.5 software (Agilent Technologies). The minimum number of probes present in an aberrant region was 4, and aberrant segments were identified for a CNV locus when the average log2 ratio was greater than |±0.4|. In addition, a less stringent filter was used to infer aberrant regions under the condition that the minimum number of probes present in an aberrant region was 2. In both cases, the statistical analysis of aberrant regions was based on the aberrant detection algorithm ADM-2. The full data set and designs from the oligo aCGH experiments have been submitted to the GEO database [21] under the accession ID GSE38434.
Copy Number Validation by Quantitative Polymerase Chain Reaction
To validate representative aCGH results, quantitative polymerase chain reaction (qPCR) was performed using the chicken β-actin gene (ACTB) as a reference for normalization of the real-time PCR experiments. On the basis of the aCGH aberration data, 52 sets of PCR primers for candidate sites were designed using Primer3 software [22]. Each set of primer pair was expected to yield PCR products, ranging from 150 to 200 bp based on the sequence in the chicken genome assembly build 3.1. The primer sequences are listed in Table S1. Prior to real-time PCR, each product was electrophoresed on a 2.0% agarose gel in order to verify the expected product size. Amplification curve and Ct values were generated with the Thermal Cycler Dice® Real Time System (Takara Bio, Shiga, Japan). Each qPCR reaction (15 µL) contained 10 ng of genomic DNA, 2.0 µL of 5 µM forward and reverse primers, 0.3 µL of ROX reference dye, and 7.5 µL of the 2× SYBR® Premix Ex Taq™ II (Takara Bio). The PCR cycle consisted of initial denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 31 s. Each sample was run 3 times to obtain accurate qPCR results. For relative amount quantification, Ct value differences were used to quantify the relationship between relative copy number and β-actin. This was calculated as follows: relative copy number = log2ΔCt, where ΔCt = Ctβ-actin – Cttarget. The Ct values obtained were in agreement within and between runs.
Statistical Analysis
Since almost all test data showed non-normal frequency distributions that could not be transformed to meet the requirements of parametric statistical analysis, the Kruskal-Wallis test was carried out to examine the difference in TIind and TIdur scores between breeds/strains. In each sCNV locus, we compared TI scores with the distribution of relative copy number, calculated as log2ΔCt. To compare relative copy number between chicks with high and low TI scores, each chicken cohort was classified into high (TIind = 1; TIdur ≥60 s) and low (TIind = 2∼7; TIdur <60 s) groups. Biased relationships between copy number and each TI score were examined using F-statistics under the null hypothesis of no association between copy number on the target locus and TI responses.
Ethical Note
All aspects of the study were performed according to the guidelines established by the Ministry of Education, Culture, Sports, Science and Technology in Japan (Notice No. 71). This study fulfilled ethical guidelines of the International Society of Applied Ethology [23]. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Wildlife Research Center of Kyoto University (Permit No. WRC-2012-EC001).
Results
A total of 180,000 unique probes was designed in chicken chromosome 1 targeting 3 identical QTL for fear traits (Figure 1). Table 1 shows the main results from the TI experiment in each cohort of newly hatched chicks. WL chicks were easily induced into TI status according to the number of successful TI inductions to total attempts and TIind as compared with those of NG (F 1,107 = 16.18; p<0.001), whereas TIdur in WL was significantly shorter than that of NG (F 1,107 = 4.56; p<0.05). Data from NG5 and NG7 were combined for statistical analysis, since we did not find any difference in their TI scores (p>0.05).
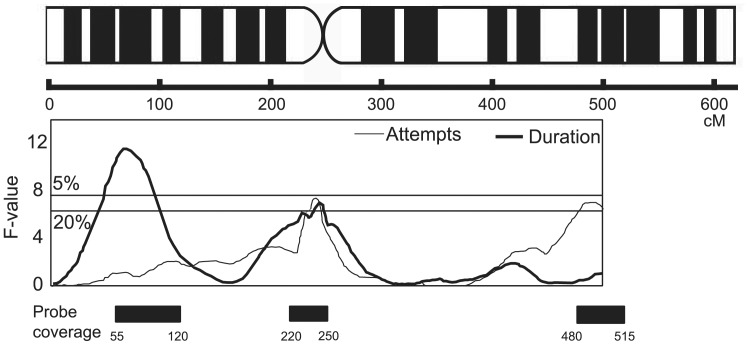
Figure 1. Probe coverage on chicken chromosome 1 for array comparative genomic hybridization.
Probes are designed in 3 regions (>60 Mb) where significant F-values have been identified by previous quantitative trait loci analysis. Genome-wide F-values for tonic immobility duration (thick line) and induction attempts (thin line) are quoted from [7].
Table 1. The induction and duration of TI response in each chicken breed/strain.
| Strains | n | Induction/attempts | Induction | TI duration | ||
| NG5 | 32 | 51/192 (0.27) | 4.4 | 99.4 | ± | 15.8 |
| NG7 | 39 | 45/234 (0.19) | 4.6 | 114.2 | ± | 22.5 |
| WL | 39 | 82/234 (0.35) | 2.7 | 74.8 | ± | 10.2 |
Note: Standard error of the mean (SEM) are shown with the time until righting (sec).
A large number of aberrant loci were detected in chicken chromosome1 based on the aCGH analysis between chicks characterized with high and low TI scores. The total number of aberrant segments identified in the 4 comparisons was 202 (average 50.5) in a stringent setting (4 probes) and 477 (average 119.3) in a less stringent setting (2 probes). Of these segments, 288 showed loss variation, and the remaining 391 segments showed gain variation. The duplicated segments (gain) occupied 57.6% in total length aberration. The average length of gain or loss segments was estimated at 3,552 bp (4 probes) and 1,833 bp (2 probes). The 477 CNVs found under the less stringent criteria encompassed 874.4 kb, which accounted for 1.46% of the total target region (60 Mb) in this study. This ratio was similar to the value suggested by whole genome analysis, indicating that CNVs occupied 1.34% of the entire chicken genome [24]. CNVs were not equally distributed throughout the target regions; the distal QTL peak (480–515 cM) of chromosome 1 contained a greater number of sCNVs than the other 2 QTL regions (Fisher’s exact test; p<0.01). Since a previous study also detected a large number of CNVs in this region [24], this chromosomal region can be regarded as a “CNV hotspot” in which mutations leading to copy number differences between individuals occur more frequently than expected. We further screened these aberrant segments under the following two conditions: the segments were commonly detected in all genomic comparisons with log2 ratio |±0.4|, and the segments were detected in at least 2 comparisons with log2 ratio |±1.2|. From the whole data set (4 & 2 probes), we extracted 52 loci that satisfied either of these conditions.
Real-time qPCR was performed to validate the aCGH data for 52 candidate loci. For preliminary screening procedure of qPCR validation, 12 DNA samples from NG and WL strains, including samples used for aCGH analysis, were employed as templates. The qPCR analysis displayed different patterns of quantitative variations that could be classified into 5 categories: (1) the same level of ΔCt values was detected in all samples except for the reference sample, whose PCR product was completely absent (described as “deletion” in Table 2); (2) an apparent variation of the ΔCt values was observed in 12 samples including the reference sample (described as “CNV” in Table 2) (3) almost the same level of ΔCt values was observed in all samples including a reference sample, i.e., monomorphic loci; (4) no PCR product was detected in several samples including or excluding the reference sample; and (5) failed PCR amplification in all specimens, probably due to primer mismatch. We found quantitative variations in the following percentage: (1) 11.5%, (2) 9.6%, (3) 63.5%, (4) 3.8%, and (5) 11.5%. We identified 11 loci, belonging to either category (1) or (2), as candidate sCNVs, and sCNVs in the other category were briefly summarized in Table S2.
Table 2. Candidate short Copy Number Variations identified by array Comparative Genomic Hybridization and subsequent qPCR validation.
| locus ID | probes | Start | Stop | bp | Gene | qPCR |
| TIC_03 | 2 | 25773437 | 25773746 | 309 | Non-coding | CNV |
| TIC_04 | 2 | 28079752 | 28080201 | 449 | Non-coding | CNV |
| TIC_05 | 2 | 28432127 | 28432740 | 613 | Non-coding | deletion |
| TIC_15 | 2,4 | 87064880 | 87065977 | 1097 | NME7 | deletion |
| TIC_16 | 2 | 170375957 | 170376395 | 438 | KIAA0564 | CNV |
| TIC_18 | 2,4 | 176058363 | 176059210 | 847 | Non-coding | CNV |
| TIC_42 | 2 | 177404311 | 177404572 | 261 | NBEA | CNV |
| TIC_44 | 2 | 180954362 | 180954612 | 250 | CDK8 | CNV |
| TIC_19 | 2 | 181380041 | 181380759 | 718 | NUPL1 | CNV |
| TIC_20 | 2,4 | 186702464 | 186703251 | 787 | Non-coding | CNV |
| TIC_21 | 2 | 188457807 | 188458430 | 623 | Non-coding | CNV |
Note: Only loci displaying quantitative difference in qPCR validation are shown here.
For the 11 candidate loci, we examined the association between sCNVs and TIind or TIdur scores in 110 chicks. With regard to both TI indices, we detected significant differences in the copy number distributions between NG and WL in TIC_3 (F 1,107 = 4.10; p<0.05), TIC_18 (F 1,107 = 19.05; p<0.001), and TIC_42 (F 1,107 = 236.59; p<0.001; Figure 2). However, there was no difference in the relative copy number for all target CNVs between chicks with high and low TI scores (TIind & TIdur) in each NG and WL. Scatter plots of correlation analysis between TI scores and the relative copy number in each locus were shown in the supporting information (Figures S1 and S2).
Figure 2. Comparison of relative copy number between chicken breeds with different sensitivity to fear.
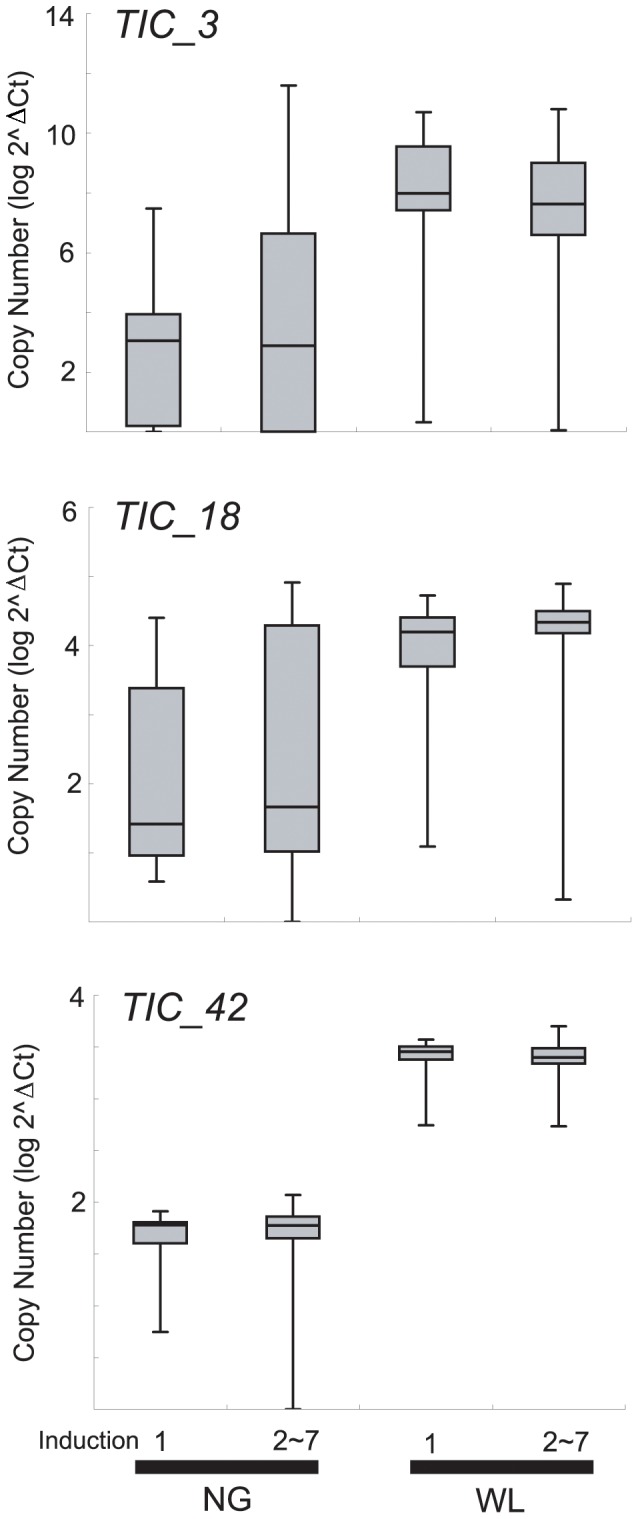
Relative copy number is calculated as log2ΔCt, where ΔCt = Ctβ-actin – Cttarget. NG and WL indicate Nagoya and White Leghorn, respectively. The number of samples in each group was; n = 13 (NG; TI induction 1), n = 58 (NG; TI induction 2∼7), n = 13 (WL; TI induction 1), and n = 26 (WL; TI induction 2∼7).
Discussion
Until now, most studies on chicken domestication have focused on the genetic and behavioral heterogeneities between RJF and WL in order to highlight the alternative histories of domestication. Additionally, our preliminary study [10] detected large differences in the quantitative traits of TI responses between WL and NG populations. WL chicks were prone to be induced into the TI status with fewer attempts (low TIind), whereas NG had longer TIdur in each successful TI induction. These empirical data strongly support the previous hypothesis that the TI behavior in chickens has a genetic basis with breed- or strain-specific behavioral characteristics [8], [25], [26].
However, we did not detect a difference in the relative copy number between high and low TI groups within each chicken breed. There are several reasons for this, one of which is that the number of chicks used in this study may be insufficient to detect an association between sCNVs and variations in the chicks’ sensitivity to fearful stimuli. Considering that a TI response is not simple but has a complicated contextual framework involving cognitive and neural processes, there may be multiple genetic determinants associated with the quantitative traits of TI responses. Therefore, to detect an effect of CNVs on TI, sample sizes would need to be increased more than 20-fold over the current study design, and other genetic and epigenetic factors such as single nucleotide polymorphisms (SNPs) and methylation patterns should be included as possible candidates for affecting fear-induced behaviors in future research. Another explanation for the lack of significant results in this study is the inconsistency in chicken breeds between the reference genomic DNA sequence derived from NG and the genome shotgun sequence of RJF [27] used for primer design in qPCR validation. Based on the patterns of qPCR amplification, more than half (63.5%) of candidate CNV loci showed monomorphism in amplification plots. This is probably due to considerable sequence diversity between NG/WL and RJF, which might lead to an apparent loss of copy number polymorphisms in the validation phase. According to archaeological findings [28], the divergence time of domestic chickens from RJF is estimated to be nearly 8,000 years. Comparison of DNA sequences from 30 introns at 25 nuclear loci revealed that the extent of nucleotide divergence after the split of RJF from their chicken ancestor is as small as 0.01% [29]. However, a more recent study indicated that NG lines were genetically distinct from commercial gene pools, thus making it a unique genetic resource [30]. A way to avoid this complication would be to change the strategy by including target chicken breeds in TI measurements. Finally, we cannot exclude the possibility that innate and learned fear responses are modulated differently by independent neural networks and mechanisms. It should be emphasized that in previous QTL analysis, TI tests were conducted when chickens were 29–30 weeks of age [7], whereas newly hatched chicks were used here for TI testing to preclude secondary social and environmental effects on TI responses. Therefore, if TI scores would fluctuate during the growing stages of chicks and juveniles, as has been suggested by previous studies [31], [32], these conflicts may have some impact on the outcome of genome-wide association studies (GWAS) between genetic variants and quantitative TI scores. We therefore consider our aCGH approach for analyzing inter-individual variations in freezing behavior, a useful preliminary analysis capable of generating further hypotheses for future evaluation.
Among the candidate sCNVs, which were excluded by aCGH screening and subsequent qPCR, we detected significant differences in the relative copy number between NG and WL in TIC_3, TIC_18, and TIC_42. The TIC_3 locus is embedded within Growth1 QTL, for which the highest p value was obtained in a QTL study for TI traits [7]. The fact that this locus was found in a relatively large non-coding region may imply that it affects gene expression through far-reaching cis- or trans-acting mechanisms. The TIC_18 locus is located upstream of the TRPC4 gene (ENSGALG00000017044), which plays a role in multiple processes, including neurotransmitter release and exocytosis [33]. A recent study suggested that TRPC4 play a pivotal role in regulating dopamine release, which may modulate emotional and cognitive responses in rats [34]. Thus, it is worthwhile investigating the expression of orthologous genes in the chicken brain and further examining the correlation between levels of expression and innate fear responsiveness. The other candidate sCNV (TIC_42) was found in the NBEA gene (ENSGALG00000017062), which has been identified as a putative regulator of membrane protein trafficking associated with the trans-Golgi network. [35]. NBEA-deficient mice died immediately after birth apparently from respiratory paralysis [36]. NBEA plays a complex role in the development and functioning of synapses and is believed to play a role in autism spectrum disorders by evoking an excitatory-inhibitory imbalance in synaptic activity [37]. Further information on differential gene expression of these candidate genes in the brain between various chicken selection lines, will provide opportunities to examine their role in shaping behavioral differences.
In conclusion, we identified 11 sCNVs that potentially account for the robust differences observed in TI responses among chicken breeds. None of the genes identified in this study have been directly implicated in TI responses; however, studies on several of these genes, such as TRPC4 and NBEA, have reported brain functions that might be involved in abnormal behaviors in humans and rodents. Future experiments involving gene expression assays and mutagenic approaches are needed to determine whether any of the novel candidate genes identified here play a role in modulating innate fear responses.
Supporting Information
Correlation analysis between the induction score of Tonic immobility (TIind; x-axis) and relative copy number (ΔCt; y-axis).
(PDF)
Correlation analysis between the duration score of Tonic immobility (TIdur; x-axis) and relative copy number (ΔCt; y-axis).
(PDF)
List of primers that show copy number polymorphisms in qPCR.
(PDF)
Another sets of candidate short Copy Number Variations examined in this study.
(PDF)
Acknowledgments
We thank Akihiro Nakamura, Junichi Ueda, and Hideshi Ohguchi (Aichi Agricultural Research Center) for their technical support. We also thank Azusa Hayano and Miyuki Sato (Kyoto University) for their valuable assistance with laboratory work.
Funding Statement
This study was supported financially by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) with a Grant-in-aid for Science Research (#21310150 and 25118005) and the Global Center of Excellence Program “Formation of a Strategie Base for Biodiversity and Evolutionary Research: from Genome to Ecosystem”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones RB (1986) The tonic immobility reaction of the domestic fowl: A review. World’s Poult Sci J 42: 82–96. [Google Scholar]
- 2. Maser JD, Gallup GG Jr (1974) Tonic immobility in the chicken: Catalepsy potentiation by uncontrollable shock and alleviation by imipramine. Psychosom Med 36: 199–205. [DOI] [PubMed] [Google Scholar]
- 3. Kujiyat SK, Craig JV, Dayton AD (1983) Duration of tonic immobility affected by housing environment in White Leghorn hens. Poult Sci 62: 2280–2282. [DOI] [PubMed] [Google Scholar]
- 4. Webster DG, Lanthorn TH, Dewsbury DA, Meyer ME (1981) Tonic immobility and the dorsal immobility response in twelve species of muroid rodents. Behav Neural Biol 31: 32–41. [DOI] [PubMed] [Google Scholar]
- 5. Cashner FM, Olson RD, Erickson DG, Olson GA (1981) Effects of MIF-I and sex differences on tonic immobility duration in the lizard, Anolis carolinensis. Peptides 2 Suppl 1161–165. [DOI] [PubMed] [Google Scholar]
- 6. Erhard HW, Mendl M, Christiansen SB (1999) Individual differences in tonic immobility may reflect behavioural strategies. Appl Anim Behav Sci 64: 31–46. [Google Scholar]
- 7. Schϋtz KE, Kerje S, Jacobsson L, Forkman B, Carlborg Ö, et al. (2004) Major growth QTLs in fowl are related to fearful behavior: possible genetic links between fear responses and production traits in a red junglefowl x white leghorn intercross. Behav Genet 34: 121–130. [DOI] [PubMed] [Google Scholar]
- 8. Wirén A, Jensen P (2010) A growth QTL on chicken chromosome 1 affects emotionality and sociality. Behav Genet 41: 303–311. [DOI] [PubMed] [Google Scholar]
- 9. Campler M, Jöngren M, Jensen P (2009) Fearfulness in red junglefowl and domesticated White Leghorn chickens. Behav Processes 81: 39–43. [DOI] [PubMed] [Google Scholar]
- 10. Abe H, Nagao K, Nakamura A, Inoue-Murayama M (2013) Differences in responses to repeated fear-relevant stimuli between Nagoya and White Leghorn chicks. Behav Process 99: 95–99. [DOI] [PubMed] [Google Scholar]
- 11. Kerje S, Carlborg O, Jacobsson L, Schütz K, Hartmann C, et al. (2003) The twofold difference in adult size between the red junglefowl and White Leghorn chickens is largely explained by a limited number of QTLs. Anim Genet 34: 264–274. [DOI] [PubMed] [Google Scholar]
- 12. Banerjee S, Oldridge D, Poptsova M, Hussain WM, Chakravarty D, et al. (2011) A computational framework discovers new copy number variants with functional importance. PLoS One 6: e17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wain LV, Armour JA, Tobin MD (2009) Genomic copy number variation, human health, and disease. Lancet 374: 340–350. [DOI] [PubMed] [Google Scholar]
- 14. Wright D, Boije H, Meadows JR, Bed’hom B, Gourichon D, et al. (2009) Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet 5: e1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura A, Noda K (2001) Breeding history and genetic characters of Nagoya breed, a Japanese poultry breed of Aichi prefecture. Surv Rep Anim Genet Resour 12: 77–97 (In Japanese).. [Google Scholar]
- 16. Greshock J, Feng B, Nogueira C, Ivanova E, Perna I, et al. (2007) A comparison of DNA copy number profiling platforms. Cancer Res 67: 10173–10180. [DOI] [PubMed] [Google Scholar]
- 17. Baumbusch LO, Aaroe J, Johansen FE, Hicks J, Sun H, et al. (2008) Comparison of the Agilent, ROMA/NimbleGen and Illumina platforms for classification of copy number alterations in human breast tumors. BMC Genomics 9: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, et al. (2011) Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 29: 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fadista J, Nygaard M, Holm LE, Thomsen B, Bendixen C (2008) A snapshot of CNVs in the pig genome. PLoS One 3: e3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conrad DF, Bird C, Blackburne B, Lindsay S, Mamanova L, et al. (2010) Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat Genet 42: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, et al. (2009) NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 37: D885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 23. Sherwin CM, Christiansen SB, Duncan IJ, Erhard HW, Lay DC, et al. (2003) Guidelines for the ethical use of animals in applied ethology studies. Appl Anim Behav Sci 81: 291–305. [Google Scholar]
- 24. Wang X, Nahashon S, Feaster TK, Bohannon-Stewart A, Adefope N (2010) An initial map of chromosomal segmental copy number variations in the chicken. BMC Genomics 11: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Craig JV, Kujiyat SK, Dayton AD (1984) Tonic immobility responses of white leghorn hens affected by induction techniques and genetic stock differences. Poult Sci 63: 1–10. [DOI] [PubMed] [Google Scholar]
- 26. Albentosa MJ, Kjaer JB, Nicol CJ (2003) Strain and age differences in behaviour, fear response and pecking tendency in laying hens. Br Poult Sci 44: 333–344. [DOI] [PubMed] [Google Scholar]
- 27. Wallis JW, Aerts J, Groenen MA, Crooijmans RP, Layman D, et al. (2004) A physical map of the chicken genome. Nature 432: 761–764. [DOI] [PubMed] [Google Scholar]
- 28. Fumihito A, Miyake T, Sumi S, Takada M, Ohno S, et al. (1994) One subspecies of the red junglefowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proc Natl Acad Sci U S A 91: 12505–12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sawai H, Kim HL, Kuno K, Suzuki S, Gotoh H, et al. (2010) The origin and genetic variation of domestic chickens with special reference to junglefowls Gallus g. gallus and G. varius . PLoS One 5: e10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tadano R, Nakamura A, Kino K (2012) Analysis of genetic divergence between closely related lines of chickens. Poult Sci 91: 327–333. [DOI] [PubMed] [Google Scholar]
- 31. Heiblum R, Aizenstein O, Gvaryahu G, Voet H, Robinzon B, et al. (1998) Tonic immobility and open field responses in domestic fowl chicks during the first week of life. Appl Anim Behav Sci 60: 347–357. [Google Scholar]
- 32. Ghareeb K (2010) Presence of males within laying hens affects tonic immobility response and sociality. Int J Poult Sci 9: 1087–1091. [Google Scholar]
- 33. Obukhov AG, Nowycky MC (2002) TRPC4 can be activated by G-protein-coupled receptors and provides sufficient Ca(2+) to trigger exocytosis in neuroendocrine cells. J Biol Chem 277: 16172–16178. [DOI] [PubMed] [Google Scholar]
- 34.Illig KR, Rasmus KC, Varnell AL, Ostertag EM, Klipec WD, et al. (2011) TRPC4 ion channel protein is selectively expressed in a subpopulation of dopamine neurons in the ventral tegmental area. Available from Nature Precedings <http://dx.doi.org/10.1038/npre.2011.6577.1>.
- 35. Wang X, Herberg FW, Laue MM, Wullner C, Hu B, et al. (2000) Neurobeachin: A protein kinase A-anchoring, beige/Chediak-higashi proteinmolog implicated in neuronal membrane traffic. J Neurosci 20: 8551–8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Medrihan L, Rohlmann A, Fairless R, Andrae J, Döring M, et al. (2009) Neurobeachin, a protein implicated in membrane protein traffic and autism, is required for the formation and functioning of central synapses. J Physiol 587: 5095–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Persico AM, Bourgeron T (2006) Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci 29: 349–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation analysis between the induction score of Tonic immobility (TIind; x-axis) and relative copy number (ΔCt; y-axis).
(PDF)
Correlation analysis between the duration score of Tonic immobility (TIdur; x-axis) and relative copy number (ΔCt; y-axis).
(PDF)
List of primers that show copy number polymorphisms in qPCR.
(PDF)
Another sets of candidate short Copy Number Variations examined in this study.
(PDF)