Abstract
A picture is now starting to emerge regarding the liver-bile acid-microbiome axis. Increasing levels of the primary bile acid cholic acid (CA) causes a dramatic shift toward the Firmicutes, particularly Clostridium cluster XIVa and increasing production of the harmful secondary bile acid deoxycholic acid (DCA). During progression of cirrhosis, the microbiome, both through their metabolism, cell wall components (LPS) and translocation lead to inflammation. Inflammation suppresses synthesis of bile acids in the liver leading to a positive-feedback mechanism. Decrease in bile acids entering the intestines appears to favor overgrowth of pathogenic and pro-inflammatory members of the microbiome including Porphyromonadaceae and Enterobacteriaceae. Decreasing bile acid concentration in the colon in cirrhosis is also associated with decreases in Clostridium cluster XIVa, which includes bile acid 7α-dehydroxylating bacteria which produce DCA. Rifaximin treatment appears to act by suppressing DCA production, reducing endotoxemia and harmful metabolites without significantly altering microbiome structure. Taken together, the bile acid pool size and composition appear to be a major regulator of microbiome structure, which in turn appears to be an important regulator of bile acid pool size and composition. The balance between this equilibrium is critical for human health and disease.
Keywords: 7α-dehydroxylation, Clostridium cluster XIVa, MELD score, autochthonous, bile acids, cirrhosis, lachnospiraceae, rifaximin, ruminoccoccaceae
Alteration in gut microbiota or “dysbiosis” has been demonstrated in several chronic diseases, including chronic liver disease and cirrhosis. This dysbiosis is characterized by alteration in the Bacteroides/Firmicutes ratio and increase in potentially pathogenic taxa such as Enterobacteriaceae in diseased states.1,2 In cirrhosis, these changes have been associated with complications such as hepatic encephalopathy (HE).1 Therefore an understanding of the multiple factors that influence these changes in gut microbiota may increase insight into the progression of liver disease. One critical component of the intestinal milieu are bile acids which have downstream effects on processes as varied as GI motility, nutrition, carcinogenesis and intestinal permeability. A picture is emerging in which the bile acid pool size and composition modulates the size and composition of the gut microbiome and vice versa.
The study of the interaction between bile acids and gut microbiota in the context of liver disease is essential because the human liver is the only organ in the body that produces all 14 enzymes required for de novo synthesis of the primary bile acids.3 The “classical” or “neutral” pathway of bile acid synthesis begins with cholesterol 7α-hydroxylase (CYP7A1), which produces both the dihydroxy bile acid chenodeoxycholic acid (CDCA; 3α, 7α) and the trihydroxy bile acid cholic acid (CA; 3α, 7α, 12α). The “acidic” pathway produces mostly CDCA and is initiated by mitochondrial sterol 27 hydroxylase (CYP27A1) catalyzed side-chain oxidation, which is followed by cleavage of a three-carbon side chain resulting in the C-24 bile acids. CA synthesis requires sterol 12α-hydroxylase (CYP8B1) followed by side-chain oxidation and cleavage.4 The classical pathway produces the majority of the two primary bile acids in healthy humans. In the diseased liver, the classical pathway is down-regulated and the acidic pathway produces most of the primary bile acids.5 Bile acids are conjugated to glycine or taurine before being secreted from the liver and stored in the gallbladder. Eating stimulates gallbladder contraction and emptying of the contents into the small intestine. Bile salts function to solubilize fats and fat-soluble vitamins before they are actively transported across the ileum, entering the portal circulation, and return to the liver in what is termed the entero-hepatic circulation (EHC). The EHC is 95% efficient, with roughly 600–800 mg bile salts escaping into the large bowel daily.6
The large bowel is the most densely populated natural environment known, containing roughly 1014 bacterial cells.7 Diversity in this environment is almost entirely at lower taxonomic levels (genus, species, strain).8 Phylum level diversity in the gut is remarkably low, with two major divisions, the Bacteroidetes and Firmicutes predominating, followed by Actinoacteria. Greater than 99% of the functional genes associated with the human body are microbial, and most of these are found in bacteria colonizing the large bowel.9 The lumen of the large bowel is a highly anaerobic environment and microbes that occupy this ecological niche must carry out fermentative metabolism in order to generate ATP and grow. The main endogenous substrate used by the large bowel microbiota consists of sloughed intestinal epithelial cells (100–200 g/day) and bile components, whereas, exogenous substrates include resistant starch, plant polysaccharides and proteins. From these substrates gut bacteria produce mostly short chain fatty acids (acetate, propionate and butyrate). During EHC bile acids enter the colon where they are metabolized by the large bowel microbiota. CA and CDCA are 7α-dehydroxylated exclusively by a small population of anaerobic bacteria to form deoxycholic acid (DCA) and lithocholic acid (LCA), respectively.10 Human gut bacteria carrying out bile acid 7α-dehydroxylation have been shown to belong to the genus Clostridium, which are gram-positive anaerobic spore forming members of the Firmicutes.11-13
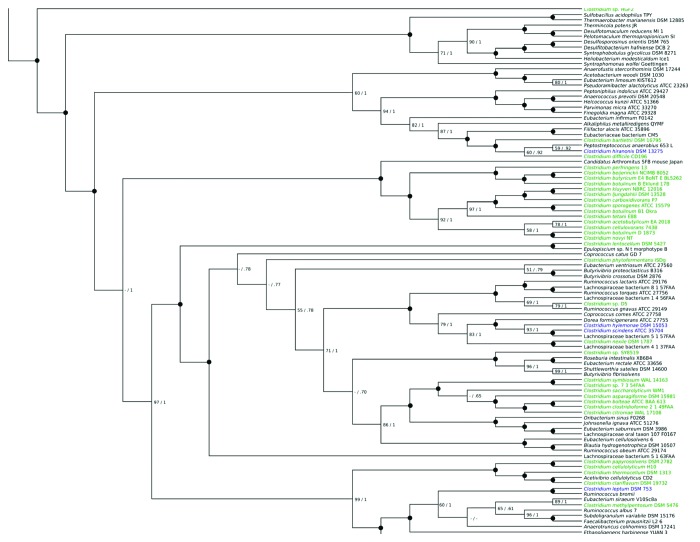
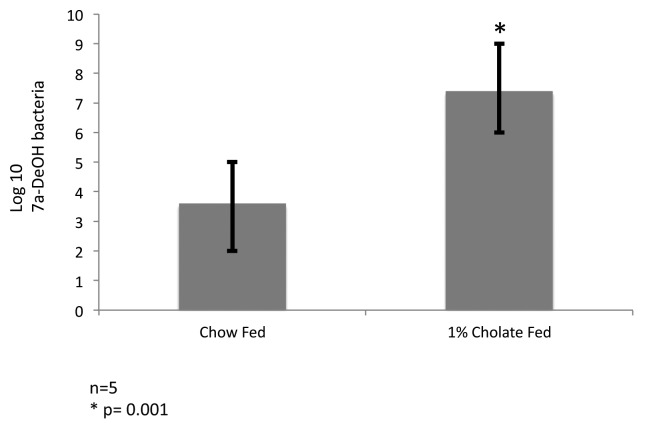
Animal model studies demonstrate that increased bile acid levels in the colon select against the Bacteroidetes and Actinobacteria and favor the Firmicutes including bile acid 7α-dehydroxylating bacteria in vivo.14 Indeed, Islam et al. (2011) reported a marked increase in Firmicutes (control 54%; CA feeding 95%) with greatest increase in members of clostridial cluster XIVa following CA feeding in rats and significant increase in DCA with higher input of CA.14 We have detected bile acid 7α-dehydroxylation activity in Clostridium scindens, C. hiranonis, C. hylemonae and C. sordellii, which are each nested within a phylogenetic tree containing members of Blautia, Ruminococcaceae and Lachnospiraceae all of which are in clostridial rRNA cluster XIVa (Fig. 1). We have determined the levels of bile acid 7α-dehydroxylating bacteria in control C57BL/6 mice fed a normal chow diet (NC) and in mice fed a NC diet plus 1% w/w cholic acid. After 2 weeks of feeding, the ceca were removed and immediately serially diluted in anaerobic brain heart infusion broth containing [24-14C]-cholic acid. We observed a 1000-fold increase in the levels of bile acid 7α-dehydroxylating bacteria (p = 0.001) with cholic acid feeding consistent with the hypothesis that increased primary bile acid substrate supports a larger population of these bacteria in the intestines (Fig. 2).
Figure 1. Reference species tree of the clostridia and related Clostridiales bacteria. Phylogenetic tree obtained from analysis. Maximum likelihood and Bayesian analyses of 20 concatenates single-copy proteins from 99 genomes, showing showing Clostridium scindens as distantly related to most other Clostridium species and being deeply embedded in a group of bacteria mainly from the Lachnospiraceae and Ruminococcaceae families. Streptococcus sanguinis SK36 was included as an outgroup. Numbers on nodes indicate support from bootstrap support and posterior probability (numbers below 50 or 0.5 not shown, indicated by a dash); black circles indicate support of 100 and 1, with all other values explicitly written.
Figure 2. Cholic acid feeding stimulates growth of bile acid 7α-dehydroxylating bacteria in mice. Twelve week old C57Bl/6 mice were fed normal chow (n = 5), or 1% w/w cholic acid (n = 5) for 2 weeks. Cecal contents were then serially diluted (10-fold) in anaerobic brain heart infusion broth containing 10 μM cholic acid and 0.01 μCi [24-14C] cholic acid. Bile acids were extracted from the medium after 48 h growth, separated by thin layer chromatography (solvent system toluene:dioxane:glacial acetic acid 75:20:2 v:v:v). Semi-quantitative estimates of bile acid 7α-dehydroxylating bacteria were made based on the last dilution tube in which [24-14C] deoxycholic acid was detected by audoradiography on Kodak Biomax MS film after 48 h exposure. *p = 0.001, T-test.
The current publication by Kakiyama et al. (2013) provides interesting data in which dysbiosis is occurring in patients with cirrhosis in part due to low bile acid input into the gut.15 This data suggests that in the absence of bile acids, the bile acid 7α-dehydroxylating bacterial population collapses. Two observations point to this conclusion. First, total bile acids in feces of patients with advanced cirrhosis decreased roughly 5-fold and the ratios of DCA/CA and LCA/CDCA decreased significantly. Second, there is a significant positive correlation between the presence of members of the Clostridium cluster XIVa and DCA and LCA concentration. Members of Clostridium cluster XIVa, which includes the bile acid 7α-dehydroxylating bacteria, decrease in the intestines as cirrhosis severity advanced. Taken together, these data show a direct relationship between the bile acid pool size and the relative abundance of Clostridium cluster XIVa. We have previously elucidated a multi-step bile acid 7α-dehydroxylating pathway in these bacteria that allow them to use primary bile acids as an electron acceptor allowing for increased ATP formation and growth.10,16 However, most members of Clostridium cluster XIVa lack the bai operon and thus do not convert CA to DCA, so for most of these microbes, energetic considerations can be ignored. Energetics may be ancillary to production of and resistance to DCA, a potent antimicrobial agent that reduces competition for growth substrates. Is the expansion of the bile acid 7α-dehydroxylating bacteria population due mainly to selection for bile tolerance coupled with reduced competition for growth substrates, or does metabolism of bile acid determine population size of members with the bai operon? We know that growth of these microbes in vitro is not dependent on the presence of bile acids. However, competition for resources in vivo is fierce in the colon, and their low levels (0.0001% of the microbiota) indicate a specialized niche.10 Inhibition of the bile acid 7α-dehydroxylating pathway without inhibition of the organism itself either pharmacologically or through genetic-knockout of the bile acid 7α-dehydroxylating pathway will be necessary to determine the in vivo role of the bile acid 7α-dehydroxylating pathway, particularly in the presence of exogenously added DCA.
At least in rodents, bile acid 7α-dehydroxylating bacteria are capable of regulating bile acid synthesis in the liver by removing an FXR-antagonist, tauro-β-muricholic acid, in the ileum.17 In humans, other members of the microbiome are capable of shrinking the bile acid pool through inhibition of bile acid synthesis in the liver by inflammation.18 As cirrhosis progresses, expression of the ileal bile acid transporter increases, resulting in less bile acids reaching the large bowel, likely due to decreased concentration of bile salts.19 Bacterial overgrowth occurs in the small bowel, often by gram-negative members of the oral and gut microbiota such as Enterobacteriaceae, Veillonellaceae, Alcaligeneaceae and Porphyromonadaceae.1,20 Indeed, we have previously shown a positive correlation between levels of Alcaligeneaceae and Porphyromonadaceae and cognitive impairment in cirrhotic patients who develop hepatic encephalopathy.1 Kakiyama et al. (2013) observed a positive correlation between fecal levels of CDCA and members of the Enterobacteriaceae. Previously, Bajaj et al. (2012) found a positive linkage between Enterobacteriaceae, endotoxemia and inflammation, which was also positively correlated with development of HE.1
Lipopolysaccharide (LPS) or endotoxin, a component of the cell wall of gram-negative bacteria varies between species.21 This structural variance leads to differing degrees of inflammation in the host. Quantitatively, species of Bacteroides are among the most predominate genera in the human GI tract, while genera within the Enterobacteriaceae, Porphyromonadaceae, and Alcaligeneaceae are only minor members of the gut microbiome of healthy humans, while several members of these families are human pathogens.8 However, LPS from members of the Enterobacteriaceae, for instance, show potency on the order of 4–50-fold that of LPS from members of the Bacteroidetes in TNF-α assays.22 Indeed, animal models of non-alcoholic fatty liver disease (NAFLD) found a positive correlation between Porphyromonadaceae and exacerbation of hepatic steatosis and inflammation through TLR4 and TLR9 activation of TNF-α.23 Porphyromonas gingivalis LPS has been shown to induce TNF-α through TLR-2 and TLR-4.24 Therefore, the data presented by Kakiyama et al. (2013) confirm previous observations that increase in specific gram-negative taxa, particularly Enterobacteriaceae and Porphyromonadaceae could lead to inflammation, which contributes to cirrhosis and its complications. Interestingly, we have previously shown that while the stool microbiome between HE and non-HE cirrhotic patients were not significantly different, the mucosal microbiome differed markedly and toward pathogenic genera in HE.25 Indeed in the current study, and in recently published studies by our group and others, it was shown that rifaximin treatment reduced endotoxemia,26,27 reduced levels of secondary bile acids,15 and reduced harmful metabolites. These metabolites derived from aromatic amino acid metabolism, ammonia metabolism and oxidative stress indicators, which were positively correlated with Bacteroidaceae, Enterobacteriaceae, Porphyromonadaceae without significantly altering microbiome community structure.26 Recent studies also suggest that DCA is involved in barrier dysfunction.28,29 While it is unclear if DCA exacerbates barrier dysfunction in cirrhotic patients, our observation of a combination of reduced DCA/CA ratio, reduction in toxic metabolites from key microbial taxa, improvement in cognitive function and reduction in endotoxemia, warrant further investigation. LCA is likely to play a lesser role due to its insolubility in fecal water. Indeed, fecal levels of LCA increase in early cirrhosis, as bile acid synthesis through the “acidic” pathway is favored; however, LCA was not detected in serum in our study.15
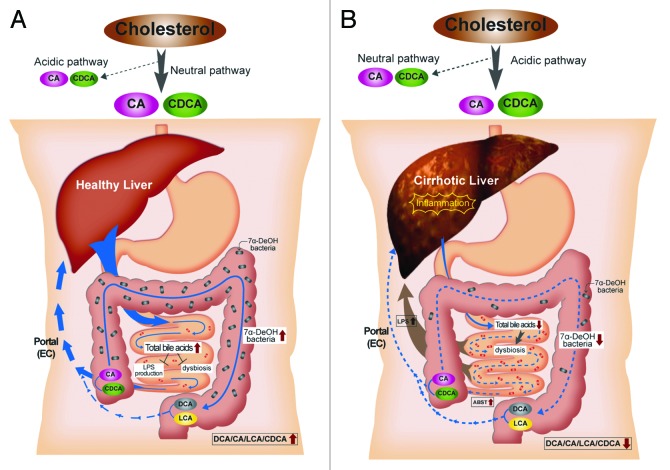
We hypothesize that a shrinking bile acid pool leads to increases in microbes with potent pro-inflammatory molecules coupled with production of harmful metabolites, which in turn lead to further down-regulation of bile acid synthesis in a positive-feedback mechanism via inflammation. Inhibition of CYP7A1 through inflammation leads to decreased total bile acids18,30 and a shift toward CDCA production through the alternative pathway.3,4 This explains the current observation of 1) decreased total bile acid synthesis, 2) CDCA synthesis predominating over CA. As the input of CA into the colon decreases so do the levels of DCA. DCA is by far the most potent antimicrobial among bile acids.31 Key changes in the microbiome may thus serve as a marker for changes in the bile acid pool composition and vice versa. Figure 3 summarizes a model that may explain the relationship between bile acids, key elements in the microbiome and cirrhosis.
Figure 3. A model for the relationship between bile acids, the microbiome and cirrhosis. In healthy individuals, cholesterol is primarily converted to CA and CDCA by the neutral bile acid biosynthetic pathway. Sufficient quantities of bile salts enter the small intestine to prevent dysbiosis and the release of inflammatory markers (i.e., LPS). Bile acid 7α-dehydroxylating bacteria are found in normal range (103–105 cells per gram wet weight), and the ratio of secondary to primary bile acids in stool is high. In cirrhosis, the neutral pathway is repressed due to downregulation of CYP7A1 by proinflammatory cytokines, and the acidic pathway is the primary pathway for bile acid synthesis. Dysbiosis occurs due to lower concentration of bile salts entering the small bowel. This dybiosis is characterized by inflammation due to an increase in organisms with potent LPS such as members of the Enterobacteriaceae. The population of 7α-dehydroxylating bacteria in the colon is hypothesized to decrease due to lower levels of primary bile acids which are thought to serve the as an energy source. Consequently, ratio of secondary/primary bile acids is low in cirrhosis.
The study of secondary BA formation as a potential marker for disease progression in human cirrhosis is important because while the 7α-dehydroxylation can be construed as a “functional test” for microbiota, the production of these secondary BAs does not provide an energy advantage to the human host, but to the bacteria that achieve this conversion. Due to their membrane-destabilizing properties and potential for worsening intestinal permeability, the decrease in secondary BAs in advancing cirrhosis, actually may be protective. This hypothesis is also supported by the reduction in the secondary/primary BA ratio after rifaximin in early cirrhotic patients. Therefore this initial study of BAs in the modulation of the fecal microbiome in worsening cirrhosis raises several important questions that need to be answered in future studies in order to delineate this complex liver-bile acid-microbiome interaction within the gut milieu.
Acknowledgments
This work was supported by RO1AA020203 from the National Institute on Alcohol Abuse and Alcoholism, grant RO1DK087913 from the National Institute of Diabetes and Digestive and Kidney Diseases (both to JSB), VA Merit Grant BX001328 to PBH. The authors would like to thank Mikyung Kang for graphic assistance with figures.
Submitted
04/22/2013
Revised
6/10/2013
Accepted
07/11/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/25723
References
- 1.Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168–75. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–72. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 3.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–66. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlahcevic ZR, Heuman DM, Hylemon PB. Regulation of bile acid synthesis. Hepatology. 1991;13:590–600. doi: 10.1002/hep.1840130331. [DOI] [PubMed] [Google Scholar]
- 5.Axelson M, Sjövall J. Potential bile acid precursors in plasma--possible indicators of biosynthetic pathways to cholic and chenodeoxycholic acids in man. J Steroid Biochem. 1990;36:631–40. doi: 10.1016/0022-4731(90)90182-R. [DOI] [PubMed] [Google Scholar]
- 6.Vlahcevic ZR, Heuman DM, Hylemon PB. 1996. Physiology and pathophysiology of enterohepatic circulation of bile acids. In Hepatology: A Textbook of Liver Disease. 3rd edition. Vol. 1. D. Zakim and T. Boyer, editors. Saunders, Philadelphia, PA. 376–417. [Google Scholar]
- 7.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 8.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–59. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Mallonee DH, White WB, Hylemon PB. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J Bacteriol. 1990;172:7011–9. doi: 10.1128/jb.172.12.7011-7019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wells JE, Hylemon PB. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol. 2000;66:1107–13. doi: 10.1128/AEM.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridlon JM, Kang DJ, Hylemon PB. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe. 2010;16:137–46. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–81. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 15.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–55. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J Lipid Res. 2012;53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–35. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Dikopoulos N, Weidenbach H, Adler G, Schmid RM. Lipopolysaccharide represses cholesterol 7-alpha hydroxylase and induces binding activity to the bile acid response element II. Eur J Clin Invest. 2003;33:58–64. doi: 10.1046/j.1365-2362.2003.01079.x. [DOI] [PubMed] [Google Scholar]
- 19.Neimark E, Chen F, Li X, Shneider BL. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 2004;40:149–56. doi: 10.1002/hep.20295. [DOI] [PubMed] [Google Scholar]
- 20.Quigley EM, Quera R. Small intestinal bacterial overgrowth: roles of antibiotics, prebiotics, and probiotics. Gastroenterology. 2006;130(Suppl 1):S78–90. doi: 10.1053/j.gastro.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 21.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delahooke DM, Barclay GR, Poxton IR. A re-appraisal of the biological activity of bacteroides LPS. J Med Microbiol. 1995;42:102–12. doi: 10.1099/00222615-42-2-102. [DOI] [PubMed] [Google Scholar]
- 23.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, et al. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. 2004;72:5041–51. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–85. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8:e60042. doi: 10.1371/journal.pone.0060042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalambokis GN, Mouzaki A, Rodi M, Pappas K, Fotopoulos A, Xourgia X, et al. Rifaximin improves systemic hemodynamics and renal function in patients with alcohol-related cirrhosis and ascites. Clin Gastroenterol Hepatol. 2012;10:815–8. doi: 10.1016/j.cgh.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 28.Stenman LK, Holma R, Korpela R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World J Gastroenterol. 2012;18:923–9. doi: 10.3748/wjg.v18.i9.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol Gastrointest Liver Physiol. 2013;304:G227–34. doi: 10.1152/ajpgi.00267.2012. [DOI] [PubMed] [Google Scholar]
- 30.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–22. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 31.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–51. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]