Abstract
Background
Toll-like receptor 4 plays a role in pathogen recognition, and common polymorphisms may alter host susceptibility to infectious diseases.
Purpose
To review the association of two common polymorphisms (TLR4 896A>G and TLR4 1196C>T) with infectious diseases.
Data Sources
We searched PubMed and EMBASE up to March 2013 for pertinent literature in English, and complemented search with references lists of eligible studies.
Study Selection
We included all studies that: reported an infectious outcome; had a case-control design and reported the TLR4 896A>G and/or TLR4 1196C>T genotype frequencies; 59 studies fulfilled these criteria and were analyzed.
Data Extraction
Two authors independently extracted study data.
Data Synthesis
The generalized odds ratio metric (ORG) was used to quantify the impact of TLR4 variants on disease susceptibility. A meta-analysis was undertaken for outcomes reported in >1 study. Eleven of 37 distinct outcomes were significant. TLR4 896 A>G increased risk for all parasitic infections (ORG 1.59; 95%CI 1.05-2.42), malaria (1.31; 95%CI 1.04-1.66), brucellosis (2.66; 95%CI 1.66-4.27), cutaneous leishmaniasis (7.22; 95%CI 1.91-27.29), neurocysticercosis (4.39; 95%CI 2.53-7.61), Streptococcus pyogenes tonsillar disease (2.93; 95%CI 1.24-6.93) , typhoid fever (2.51; 95%CI 1.18-5.34) and adult urinary tract infections (1.98; 95%CI 1.04-3.98), but was protective for leprosy (0.36; 95%CI 0.22-0.60). TLR4 1196 C>T effects were similar to TLR4 896 A>G for brucellosis, cutaneous leishmaniasis, leprosy, typhoid fever and S. pyogenes tonsillar disease, and was protective for bacterial vaginosis in pregnancy (0.55; 95%CI 0.31-0.98) and Haemophilus influenzae tonsillar disease (0.42; 95%CI 0.17-1.00). The majority of significant associations were among predominantly Asian populations and significant associations were rare among European populations.
Conclusions
Depending on the type of infection and population, TLR4 polymorphisms are associated with increased, decreased or no difference in infectious disease. This may be due to differential functional expression of TLR4, the co-segregation of TLR4 variants or a favorable inflammatory response.
Introduction
Toll-like receptors (TLRs) are a class of highly conserved membrane bound pattern recognition receptors (PRRs) that play an integral role in the regulation of the immune system through the recognition of pathogen-associated molecular patterns (PAMPs) and the activation of immune response genes [1,2]. Toll-like receptor 4 (TLR4), is a well-studied TLR, specifically recognizing lipopolysaccharide from Gram-negative bacteria [3,4] and initiating intracellular signal cascades, that involve the adaptor protein encoded by the myeloid differentiation primary response gene 88 (MyD88), which ultimately activates nuclear factor kappa B [5] and leads to interferon production [6]. TLR4 has also been shown to recognize mannans of fungal pathogens [7], Mycobacterium tuberculosis [8], and the fusion protein of respiratory syncytial virus [9].
Two single nucleotide polymorphisms (SNPs), TLR4 896 A>G (corresponding to an Asp299Gly substitution mutation ; SNP ID: rs 4986790) and TLR4 1196 C>T (corresponding to a Thr399Ile substitution mutation; SNP ID: rs 4986791), have been shown to be associated with LPS hyporesponsiveness [10,11]. In whites, the two SNPs are in linkage disequilibrium (D=1 and r2=0.791, HapMap accessible at: http://hapmap.ncbi.nlm.nih.gov/). Structurally, these mutations are found outside of the ligand binding domain of TLR4 and crystal structures have shown that these mutations have no effect on LPS binding. Instead, they do cause local conformational changes around the area of the mutation that may affect folding efficiency, cell surface expression, protein stability, as well as interaction with downstream messenger proteins [12]. At the molecular level, it has been shown that the TLR4 896 A>G mutation interferes with TLR4 interaction with MyD88 and other downstream messengers [13]. These mutations also appear to affect the levels of functional TLR4 expression, leading to a 2-fold reduction [14]. This reduction is further amplified to 10-fold in the absence of myeloid differentiation factor 2 (MD-2) which forms a complex with TLR4 and LPS [14,15].
There has been great interest regarding the association of the TLR4 SNPs TLR4 896 A>G and TLR4 1196 C>T to susceptibility for infection and other non-infectious disease states. Clinical studies associating these SNPs to infectious disease susceptibility have produced mixed results [16-19]. The present study aims to reassess the association of TLR4 896 A>G and TLR4 1196 C>T with infectious disease susceptibility using the Generalized Odds Ratio (ORG), which can elucidate the magnitude and association of individual genotypes with susceptibility to disease [20].
Materials and Methods
Study Selection
We conducted searches on Pubmed and EMBASE up to March, 2013 (last access on March 3, 2013). The search terms included: “(toll AND like AND receptor AND 4 AND polymorphism) OR (TLR4 AND polymorphism) OR Asp299Gly OR D299G OR Thr399Ile OR T399I” for PubMED; “('tlr4'/exp OR 'tlr4') AND ('receptor'/exp OR 'receptor') AND (polymorphism OR asp299gly OR d299g OR thr399ile OR t399i)” for EMBASE. The titles and abstracts of the studies were reviewed; titles that included TLR4 polymorphisms and risk for infectious disease were included for more detailed evaluation. Studies that reviewed TLR4 polymorphisms and their association with non-infectious disease were excluded, as were studies that were not published in English. An eligible study fulfilled all of the following three criteria: (i) the study reported an infectious disease outcome, (ii) the study was performed using a case-control design, where “cases” refer to subjects with a disease outcome and controls refer to a healthy population (without the disease outcome), and, (iii) the study reported genotype frequencies for TLR4 896 A>C, TLR4 1196C>T, or both.
Data Extraction
Two authors (PDZ and MLP) independently extracted data from the final included articles. Any discrepancies were reviewed and resolved by consensus. The information extracted included name of first author, origin of population being studied, number of cases and controls being studied subdivided by genotype frequencies (homozygous wild-type, heterozygous, and homozygous mutant), the disease being studied, and the conclusions reportedly drawn from each study.
Data Synthesis
We used the generalized odds ratio (ORG) along with its 95% Confidence Interval (95% CI) to address the association of TLR4 896 A>C and TLR4 1196 C>T polymorphisms with outcomes of interest (disease susceptibility). The ORG provides a model-free approach of estimating the genetic risk in genetic association studies (GAS) and meta-analysis of GAS, depending on the mutational load [20]. The ORG is defined as follows: for any two subjects, one diseased (case) and one non-diseased (control), the ORG estimates the odds of being diseased relative to the odds of being non-diseased when the diseased subject has higher mutational load than the non-diseased subject, i.e. the risk of disease is proportional to the increased genetic exposure. Alternatively, the ORG shows how many diseased-healthy pairs exist in the study for which the diseased have the larger mutational load, relative to the number of pairs for which the non-diseased have the larger mutational load [20][21]. The ORG estimates the overall genetic risk effect by utilizing the complete genotype distribution whereas the OR of conventional genetic models (additive, dominant, recessive, co-dominant) is calculated by merging genotypes. In addition, the conventional genetic models are not independent and thus, the interpretation of results is difficult when more than one model is significant [22]. In the meta-analysis of GAS, heterogeneity was quantified using the Cochran’s Q and I2 metric [20]. The existence of the differential magnitude of effect in large versus small studies was checked using the Harbord’s test [23] for meta-analysis involving at least four studies. Also, the Hardy-Weinberg equilibrium (HWE) was used as a quality criterion for control populations. HWE deviations may result in biased estimations as they can influence type-I error in single study effects, and may alter statistical significance in meta-analysis of gene-disease associations [24,25]. The HWE deviations amongst the control populations were screened using the chi-square test [26]. For single studies deviating from HWE, a sensitivity analysis was performed after correction of control group with the expected genotype frequencies [22,27]. ORG was calculated using the ORGGASMA application available at http://biomath.med.uth.gr [20]. This study complies with the PRISMA guidelines for reporting reviews and meta-analyses (Checklist S1) [28].
Results
A total of 962 studies from PubMed and 1615 from EMBASE were initially retrieved, comprising a total of 2,197 non-duplicate studies (Table S1-Flow diagram). After reading the title and the abstract, 117 studies were found to be suitable for further evaluation. Of the 117 articles reviewed in detail, 58 studies were excluded (18 studies did not publish genotypic frequencies, 13 had no healthy controls in their experimental design, 5 focused on in vitro functional studies, 8 did not study the desired polymorphisms, 6 were either reviews or a meta-analysis, and 8 studies had non extractable data for other reasons). A total of 59 case-control studies [29-87] were included in the analysis, reporting 37 different disease outcomes (Tables 1 and 2). The origin of studies was in descending order Europe (28 studies), Asia (12 studies), South America (7 studies), Africa (6 studies), North America (5 studies), Australia (1 studies).
Table 1. Genotypic frequencies reported for the TLR4 896 A>G SNP and association with disease outcome; significant effects are in bold; outcomes that have been studied more than once have been grouped together in the table, with the overall effect described in the shaded area† genotypic frequencies of controls that did not satisfy Hardy Weinberg Equilibrium, [effects in brackets after correction of HWE deviations].
| Control Genotype |
Case Genotype |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Population | A/A | A/G | G/G | A/A | A/G | G/G | Disease Outcome | Conclusion Reported | ORG (95% CI) |
| Carvalho et al [29] | England | 70 | 10 | 0 | 58 | 18 | 0 | Aspergillosis | Overall susceptibility not studied | 2.10 (0.92-4.81) |
| Rezazadeh et al [30] | Iran | 65 | 46 | 0 | 68 | 127 | 3 | Brucellosis | Increased risk† | 2.66(1.66-4.27) [2.69 (1.67-4.33)] |
| Doorduyn et al [31] | Netherlands | 608 | 72 | 3 | 405 | 49 | 1 | Campylobacter | No association | 1.00 (0.68-1.46) |
| Plantinga et al [32] | Tanzania | 99 | 9 | 0 | 107 | 10 | 0 | Oropharyngeal candidiasis in HIV | No association | 1.02(0.41-2.55) |
| Laisk et al [33] | Estonia | 287 | 35 | 1 | 61 | 9 | 0 | C.trachomatis(women) | No association | 1.24 (0.58-2.67) |
| Szebeni et al [34] | Hungary | 108 | 10 | 0 | 37 | 4 | 0 | NecEnterocolitis in LBW infants | No association | 1.26 (0.40-4.00) |
| Lee, et al [35] | United States | 431 | 11 | 21 | 103 | 2 | 3 | Gram –ve infections in liver transplant | No association† | 0.66 (0.26-1.70) [0.42(0.16-1.66)] |
| Ajdary, et al [36] | Iran | 73 | 2 | 0 | 102 | 26 | 0 | Leishmaniasis (Cutaneous) | Increased risk | 7.22 (1.91-27.29) |
| Rasouli et al [37] | Iran | 137 | 18 | 0 | 110 | 11 | 1 | Leishmaniasis (Visceral) | No asscociation | 0.81 (0.38-1.75) |
| Bochud et al [38] | East Africa | 155 | 37 | 2 | 375 | 32 | 2 | Leprosy | Protective | 0.36 (0.22-0.60) |
| West, et al [39] | Thailand | 1377 | 20 | 1 | 484 | 5 | 0 | Meliodosis | No association† | 0.74(0.29-1.92) [0.70(0.27-1.78)] |
| Verma, et al [40] | India | 127 | 22 | 1 | 77 | 61 | 2 | Neurocysticercosis | Increased risk | 4.39(2.53-7.61) |
| Montes et al [41] | Spain | 135 | 20 | 0 | 65 | 12 | 3 | Osteomyelitis | Increased risk | 1.55 (0.76-3.20) |
| Emonts et al [42] | Netherlands | 374 | 58 | 1 | 293 | 42 | 2 | Otitis media (acute) | Overall susceptibility not studied | 0.96 (0.63-1.45) |
| Moens et al [43] | Belgium | 161 | 16 | 1 | 84 | 13 | 2 | Invasive pneumococcal infection | No association | 1.69 (0.81-3.54) |
| Mrazek et al [44] | Czechoslovakia | 217 | 34 | 1 | 89 | 9 | 0 | Prosthetic joint infection | No association | 0.66 (0.31-1.42) |
| Doorduyn et al [31] | Netherlands | 608 | 72 | 3 | 173 | 20 | 0 | Salmonella gastroenteritis | No association | 0.96 (0.57-1.60) |
| Yuan et al [45] | Australia | 364 | 44 | 1 | 82 | 3 | 0 | S. pneumoniae | Protective | 0.35 (0.12-1.07) |
| Liadaki, et al [46] | Greece | 195 | 27 | 0 | 99 | 6 | 0 | Tonsillar Disease (H.influenzae) | No association | 0.47 (0.19-1.14) |
| Liadaki, et al [46] | Greece | 264 | 25 | 0 | 30 | 8 | 0 | Tonsillar Disease (S.pyogenes) | Increased risk | 2.93 (1.24-6.93) |
| Bhuvanendran, et al [47] | Malaysia | 241 | 9 | 0 | 277 | 27 | 0 | Typhoid Fever | Increased Risk | 2.51 (1.18-5.34) |
| Yin, et al [48] | China | 227 | 21 | 0 | 109 | 20 | 0 | UTI (Adults) | Increased risk | 1.98 (1.04-3.98) |
| Hawn et al [49] | United States | 274 | 33 | 6 | 585 | 65 | 2 | UTI (Women) | No association | 0.79 (0.52-1.20) |
| Chagas Disease | 1.06 (0.53-2.14) | |||||||||
| Weitzel, et al [50] | Northern Chile | 42 | 3 | 0 | 114 | 11 | 0 | Chagas Disease | No association | 1.20 (0.35-4.14) |
| Zafra et al [51] | Colombia | 191 | 9 | 0 | 262 | 10 | 3 | Chagas Disease | No association | 1.00 (0.43-2.36) |
| H. pylori | 0.91 (0.61-1.36) | |||||||||
| Achyut et al [52] | India | 168 | 32 | 0 | 110 | 20 | 0 | H. pylori | No association | 0.97 (0.53-1.76) |
| Moura et al [53] | Brazil | 222 | 28 | 4 | 206 | 25 | 1 | H. pylori | No association† | 0.87(0.50-1.50) [0.81(0.47-1.40)] |
| Malaria | 1.31 (1.04-1.66) | |||||||||
| Esposito, et al [54] | Burundi | 300 | 36 | 1 | 528 | 72 | 2 | Malaria (children) | No association | 1.13 (0.74-1.73) |
| Zakeri, et al [55] | Iran | 287 | 33 | 0 | 276 | 39 | 5 | Malaria (all ages) | No association | 1.38 (0.86-2.22) |
| Mockenhaupt et al [56] | Ghana | 239 | 47 | 4 | 444 | 129 | 7 | Malaria (pregnancy) | Overall susceptibility not studied | 1.42 (0.99-2.02) |
| Meningococcal disease | 1.10 (0.90-1.34) | |||||||||
| Biebl et al [57] | Austria | 678 | 88 | 3 | 167 | 18 | 0 | Meningococcal disease (all ages) | No association | 0.82 (0.49-1.40) |
| Read et al [58] | England | 787 | 81 | 11 | 924 | 110 | 13 | Meningococcal disease (all ages) | No association† | 1.13 (0.86-1.51) [1.05(0.79-1.38)] |
| Faber et al [59] | Europe | 190 | 23 | 1 | 165 | 27 | 5 | Meningococcal disease (infants) | Increased risk | 1.55(0.89-2.72) |
| Allen et al [60] | Gambia | 198 | 51 | 2 | 198 | 51 | 3 | Meningococcal meningitis (children) | No association | 1.02(0.67-1.56) |
| Periodontitis (aggressive) | 1.04 (0.53-2.04) | |||||||||
| Brett et al [61] | England | 90 | 7 | 0 | 37 | 8 | 0 | Aggressive periodontitis | No association | 2.73 (0.96-7.76) |
| Emingil et al [62] | West Europe | 147 | 7 | 1 | 86 | 4 | 0 | Aggressive periodontitis | No association† | 0.96 (0.30-3.12) [0.81(0.26-2.54)] |
| James et al [63] | West Europe | 103 | 20 | 0 | 69 | 4 | 0 | Aggressive periodontitis | No association | 0.33 (0.12-0.97) |
| Noack et al [64] | Germany | 71 | 9 | 0 | 100 | 11 | 0 | Aggressive periodontitis | No association | 0.86 (0.35-2.13) |
| Schulz et al [65] | Germany | 73 | 7 | 0 | 52 | 8 | 0 | Aggressive periodontitis | No association | 1.58 (0.56-4.47) |
| Periodontitis (chronic) | 0.94 (0.75-1.18) | |||||||||
| Garlet, et al [66] | Brazil | 131 | 74 | 12 | 135 | 56 | 6 | Chronic periodontitis | No association | 0.70 (0.47-1.03) |
| Noack et al [67] | Germany | 68 | 8 | 0 | 96 | 12 | 0 | Chronic periodontitis | No association | 1.04 (0.42-2.61) |
| Sahingur et al [68] | United States | 59 | 17 | 1 | 95 | 19 | 0 | Chronic periodontitis | No association | 0.67 (0.33-1.37) |
| Schulz et al [65] | Germany | 73 | 7 | 0 | 66 | 7 | 0 | Chronic periodontitis | No association | 1.10 (0.38-3.19) |
| Izakovicova Holla et al [69] | Czechoslovakia | 195 | 23 | 0 | 147 | 24 | 0 | Chronic periodontitis | No association | 1.38 (0.76-2.53) |
| Berdeli et al [70] | Turkey | 100 | 6 | 0 | 79 | 4 | 0 | Chronic periodontitis | No association | 0.88 (0.26-3.01) |
| James et al [63] | West Europe | 78 | 16 | 0 | 77 | 17 | 1 | Chronic periodontitis | No association | 1.11 (0.53-2.31) |
| Brett et al [61] | England | 90 | 7 | 0 | 47 | 6 | 0 | Chronic periodontitis | No association | 1.66 (0.55-4.97) |
| Laine et al [71] | Netherlands | 90 | 8 | 1 | 90 | 10 | 0 | Chronic periodontitis | No association | 1.16 (0.46-2.93) |
| Folwaczny et al [72] | Germany | 236 | 8 | 0 | 234 | 10 | 0 | Chronic periodontitis | No association | 1.24 (0.50-3.12) |
| Respiratory Syncytial Virus | 1.02 (0.72-1.44) | |||||||||
| Lofgren, et al [73] | Finland | 290 | 59 | 7 | 251 | 55 | 6 | Respiratory Syncytial Virus | No association | 1.06 (0.73-1.66) |
| Paulus et al [74] | Canada | 97 | 9 | 0 | 218 | 17 | 1 | Respiratory Syncytial Virus | No association | 0.84(0.37-1.91) |
| Sepsis | 0.81 (0.42-1.56) | |||||||||
| Ahmad-Nejad et al [75] | Germany | 99 | 12 | 1 | 31 | 6 | 1 | Sepsis (ICU) | No association | 1.72 (0.64-4.63) |
| Carregaro et al [76] | Brazil | 178 | 26 | 1 | 88 | 9 | 0 | Sepsis (ICU) | No association | 0.71 (0.33-1.56) |
| Feterowski et al [77] | Germany | 135 | 19 | 0 | 143 | 10 | 0 | Sepsis (ICU) | No association | 0.51 (0.23-1.19) |
| Tuberculosis | 1.18 (0.80-1.73) | |||||||||
| Najmi et al [78] | India | 206 | 44 | 0 | 95 | 34 | 6 | Tuberculosis | Increased association | 2.00 (1.23-3.25) |
| Newport et al [79] | Gambia | 235 | 58 | 5 | 241 | 62 | 4 | Tuberculosis | No association | 1.01(0.69-1.49) |
| Sanchez, et al [80] | Colombia | 270 | 29 | 1 | 429 | 36 | 1 | Tuberculosis | No association | 0.78 (0.47-1.28) |
| Selvaraj et al [81] | South India | 151 | 53 | 3 | 153 | 47 | 4 | Tuberculosis | No association | 0.91 (0.59-1.40) |
| Rosas-Taraco et al [82] | Mexico | 110 | 4 | 0 | 94 | 10 | 0 | Tuberculosis | No association | 2.70 (0.87-8.39) |
| UTI | 1.41 (0.70-2.84) | |||||||||
| Akil, et al [83] | Turkey | 79 | 14 | 0 | 97 | 14 | 1 | UTI-children | No association | 0.85 (0.39-1.84) |
| Ertan, et al [84] | Turkey | 29 | 1 | 0 | 28 | 2 | 0 | UTI-children | No association | 1.70 (0.22-13.37) |
| Karoly et al [85] | Hungary | 218 | 17 | 0 | 88 | 15 | 0 | UTI-children | Increased risk | 2.18 (1.06-4.52) |
Table 2. Genotypic frequencies reported for the TLR4 1196 C>TSNP and association with disease outcome; significant effects are in bold; outcomes that have been studied more than once have been grouped together in the table, with the overall effect described in the shaded area.
| Control Genotype |
Case Genotype |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Population | C/C | C/T | T/T | C/C | C/T | T/T | Disease Outcome | Conclusion Reported | ORG (95% CI) | ||
| Goepfert et al [86] | United States | 316 | 28 | 0 | 435 | 21 | 0 | Bacterial Vaginosis in Pregnant | Protective | 0.55 (0.31-0.98) | ||
| Laisk et al [33] | Estonia | 287 | 35 | 1 | 61 | 9 | 0 | C. trachomatis(women) | No association | 1.24 (0.58-2.67) | ||
| Szebeni et al [34] | Hungary | 108 | 10 | 0 | 37 | 4 | 0 | NecEnterocolitis in LBW infants | No association | 1.26 (0.39-4.00) | ||
| Lee, et al [35] | United States | 395 | 64 | 4 | 89 | 18 | 1 | Gram –ve infections in liver transplant | No association | 1.23 (0.71-2.15) | ||
| Achyut et al [52] | India | 188 | 11 | 1 | 115 | 9 | 6 | H pylori | No association | 2.08 (0.95-4.54) | ||
| Ajdary, et al [36] | North Iran | 74 | 1 | 0 | 105 | 21 | 2 | Leishmaniasis (Cutaneous) | Increased risk of infection | 10.14 (1.90-54.16) | ||
| Rasouli et al [37] | Iran | 137 | 18 | 0 | 112 | 9 | 1 | Leishmaniasis (Visceral) | No association | 0.67 (0.30-1.49) | ||
| Bochud et al [38] | East Africa | 179 | 15 | 1 | 407 | 8 | 0 | Leprosy | Protective | 0.23 (0.10-0.55) | ||
| West, et al [39] | Thailand | 1379 | 22 | 1 | 486 | 3 | 0 | Meliodosis | No association † | 0.43 (0.14-1.33) [0.41(0.13-1.25)] | ||
| Verma, et al [40] | India | 140 | 9 | 1 | 114 | 25 | 1 | Neurocysticercosis | Increased risk | 3.13 (1.46-6.73) | ||
| Montes et al [41] | Spain | 133 | 22 | 0 | 67 | 10 | 3 | Osteomyelitis | Increased risk | 1.19 (0.57-2.47) | ||
| Mrazek et al [44] | Czechoslovakia | 219 | 33 | 0 | 88 | 10 | 0 | Prosthetic joint infection | No association | 0.78 (0.38-1.63) | ||
| Ahmad-Nejad et al [75] | Germany | 98 | 13 | 1 | 31 | 6 | 1 | Sepsis (ICU) | No association | 1.58 (0.60-4.23) | ||
| Yuan et al [45] | Australia | 365 | 43 | 1 | 82 | 3 | 0 | S. pneumoniae | Protective | 0.36 (0.12-1.09) | ||
| Liadaki, et al [46] | Greece | 192 | 30 | 0 | 99 | 6 | 0 | Tonsillar Disease (H.influenzae) | Protective | 0.42 (0.17-1.00) | ||
| Liadaki, et al [46] | Greece | 262 | 27 | 0 | 29 | 9 | 0 | Tonsillar Disease (S.pyogenes) | Increased risk | 3.12 (1.36-7.13) | ||
| Bhuvanendran, et al [47] | Malaysia | 242 | 8 | 0 | 282 | 22 | 0 | Typhoid Fever | Increased Risk | 2.26 (1.01-5.07) | ||
| Hawn et al [49] | United States | 277 | 35 | 4 | 589 | 69 | 0 | UTI - Women | No association | 0.83 (0.55-1.26) | ||
| Chagas Disease | 1.03 (0.49-2.18) | |||||||||||
| Weitzel, et al [50] | Northern Chile | 42 | 3 | 0 | 114 | 11 | 0 | Chagas Disease | No association | 1.19 (0.35-4.14) | ||
| Zafra et al [51] | Colombia | 282 | 9 | 0 | 267 | 8 | 0 | Chagas disease | No association | 0.95 (0.37-2.42) | ||
| Malaria | 1.30 (0.64-2.65) | |||||||||||
| Zakeri, et al [55] | Iran | 270 | 50 | 0 | 271 | 49 | 0 | Malaria (all ages) | No association | 0.98(0.64-1.50) | ||
| Mockenhaupt et al [56] | Ghana | 283 | 7 | 0 | 550 | 28 | 2 | Malaria (pregnancy) | Overall susceptibility not studied | 2.05 (0.91-4.62) | ||
| Periodontitis (aggressive) | 0.78(0.42-1.65) | |||||||||||
| Brett et al [61] | England | 78 | 17 | 0 | 46 | 3 | 0 | Aggressive periodontitis | No association | 0.35 (0.11-1.16) | ||
| Emingil et al [62] | Turkey | 148 | 7 | 0 | 88 | 2 | 0 | Aggressive periodontitis | No association | 0.57 (0.13-2.41) | ||
| Noack et al [64] | Germany | 71 | 9 | 0 | 100 | 11 | 0 | Aggressive periodontitis | No association | 0.86 (0.35-2.13) | ||
| Schulz et al [65] | Germany | 73 | 7 | 0 | 52 | 8 | 0 | Aggressive periodontitis | No association | 1.58 (0.56-4.47) | ||
| Periodontitis (chronic) | 1.12 (0.83-1.52) | |||||||||||
| Brett et al [61] | England | 78 | 17 | 0 | 50 | 4 | 0 | Chronic periodontitis | No association | 0.41 (0.14-1.22) | ||
| Reddy et al [87] | South India | 59 | 1 | 0 | 56 | 3 | 1 | Chronic periodontitis | No association | 2.77 (0.42-18.48) | ||
| Schulz et al [65] | Germany | 73 | 7 | 0 | 67 | 7 | 0 | Chronic periodontitis | No association | 1.09 (0.38-3.14) | ||
| IzakovicaHolla et al [69] | Czechoslovakia | 196 | 22 | 0 | 147 | 24 | 0 | Chronic periodontitis | No association | 1.45 (0.79-2.67) | ||
| Berdeli et al [70] | Turkey | 101 | 5 | 0 | 80 | 3 | 0 | Chronic periodontitis | No association | 0.81(0.20-3.16) | ||
| James et al [63] | West Europe | 74 | 18 | 0 | 73 | 20 | 1 | Chronic periodontitis | No association | 1.16 (0.58-2.32) | ||
| Noack et al [67] | Germany | 68 | 8 | 0 | 96 | 12 | 0 | Chronic periodontitis | No association | 1.04 (0.42-2.61) | ||
| Laine et al [71] | Netherlands | 90 | 8 | 1 | 90 | 10 | 0 | Chronic periodontitis | No association | 1.15 (0.46-2.93) | ||
| Folwaczny et al [72] | Germany | 235 | 9 | 0 | 233 | 11 | 0 | Chronic periodontitis | No association | 1.22(0.51-2.93) | ||
| Tuberculosis | 1.07 (0.81-1.42) | |||||||||||
| Najmi et al [56] | India | 206 | 43 | 1 | 105 | 26 | 4 | Tuberculosis | No association | 1.37 (0.82-2.28) | ||
| Sanchez, et al [80] | Colombia | 272 | 26 | 1 | 429 | 36 | 1 | Tuberculosis | No association | 0.87 (0.52-1.46) | ||
| Selvaraj et al [81] | South India | 152 | 46 | 5 | 150 | 49 | 4 | Tuberculosis | No association | 1.04 (0.68-1.61) | ||
genotypic frequencies of controls that did not satisfy Hardy Weinberg Equilibrium, [effects in brackets after correction of HWE deviations].
TLR4 896 A>G and disease susceptibility
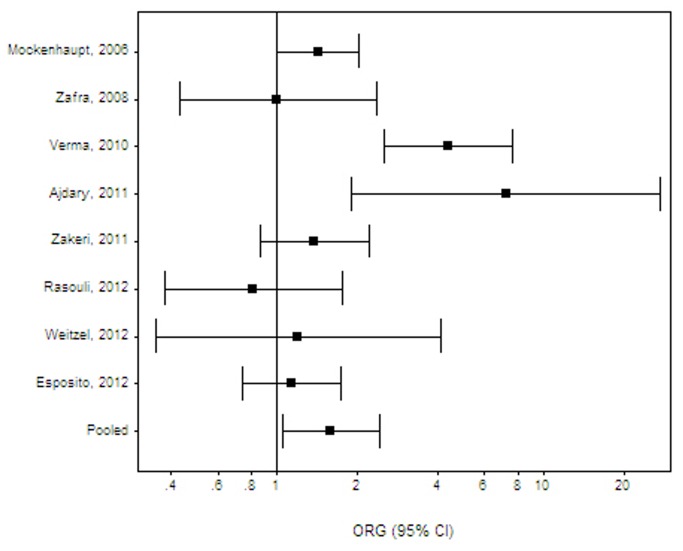
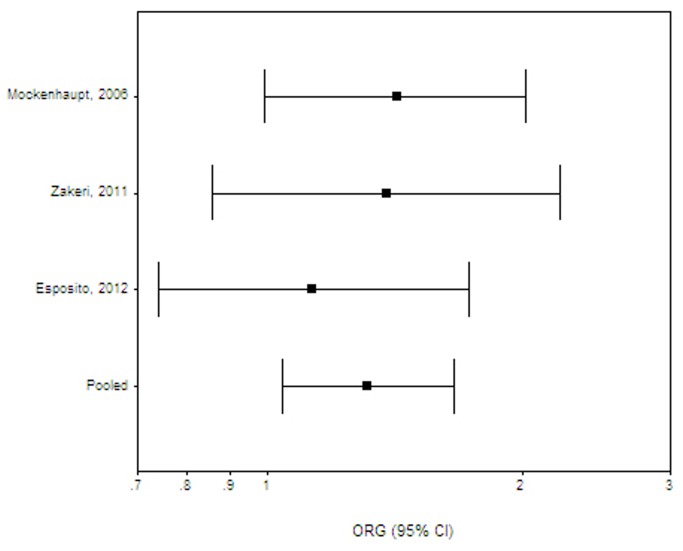
For outcomes with more than 1 available study, a meta-analysis was performed for chronic periodontitis (10 studies) [61,63,65-72], Helicobacter pylori infection (2 studies) [52,53], malaria (3 studies) [54-56], meningococcal disease (4 studies) [57-60], sepsis (3 studies) [75-77], respiratory syncytial virus (2 studies) [73,74], tuberculosis (5 studies) [78-82] and urinary tract infections in children (3 studies) [83-85]. Combined effects were also calculated for all Gram negative infections [30,31,33,35,39,46,47,52,53,57-60], all Gram positive infections [43,45,46] and all parasitic infections [36,37,40,50,51,54-56] (table 3). A significant risk was found for all parasitic infections combined (ORG 1.59; 95% CI 1.05-2.42, effect derived from Asian, African and South American populations; Figure 1) and malaria (ORG 1.31; 95% CI 1.04-1.66, a combined effect for African and Asian studies; Figure 2) . The effect on malaria was of marginal significance across African studies [54,56] (ORG 1.29; 95% CI 0.99-1.69). All other effects were insignificant, namely all Gram negative infections (ORG 1.10; 95% CI 0.90-1.38), all Gram positive infections (ORG 1.28; 95% CI 0.43-3.81), Chagas disease (ORG 1.06; 95% CI 0.53-2.14) , H. pylori (ORG 0.91; 95% CI 0.61-1.36), meningococcal disease (ORG 1.10; 95% CI 0.90-1.34), aggressive or chronic periodontitis (ORG 1.04; 95% CI 0.53-2.04 and ORG 0.94; 95% CI 0.75-1.18, respectively), respiratory syncytial virus (ORG1.02; 95% CI 0.72-1.44), sepsis (ORG 0.81; 95% CI 0.41-1.56) and tuberculosis (ORG 1.18; 95% CI 0.80-1.73). The meta-analysis results are summarized in Table 3. Statistical heterogeneity varied from absent to moderate. The Harbord’s test indicated that there is no differential magnitude of effect in large versus small studies for all outcomes (p≥0.05). Across populations of European ancestry, the risk of meningococcal disease [57-59] (ORG 1.12; 95% CI 0.85-1.49) and chronic periodontitis (excluding the two non-European studies [66,68]; ORG 1.06; 95% CI 0.53-2.14) remained insignificant. The effects on meningococcal disease and aggressive periodontitis did not alter after removing from analysis the two studies not in HWE equilibrium (data not shown) [58,62]. Effects on tuberculosis remained insignificant across Indian [78,81] (ORG 1.34; 95% CI 0.62-2.90) or S. American [80,82] populations (ORG 1.30; 95% CI 0.39-4.33). For outcomes with a single available study, a significant risk was present for brucellosis (ORG 2.66; 95% CI 1.66-4.27) [30], cutaneous leishmaniasis (ORG 7.22; 95% CI 1.91-27.29) [36], neurocysticercosis (ORG 4.39; 95% CI 2.53-7.61) [40], and typhoid fever (ORG 2.51; 95% CI 1.18-5.34) [47]. All the significant single-study effects are summarized in Table 4.
Table 3. Summary of disease associations derived from meta-analysis of case-control studies.
| Disease Outcome | Studies | Polymorphism | Effect (ORG ; 95% CI) | PQ | I2 | PH |
|---|---|---|---|---|---|---|
| All Gram - infections | 13 | TLR4 896 A>G | 1.10 (0.90-1.38) | 0.01 | 52% | 0.32 |
| 6 | TLR4 1196 C>T | 1.11 (0.66-1.87) | 0.02 | 61% | 0.59 | |
| Helicobacter pylori | 2 | TLR4 896 A>G | 0.91 (0.61-1.36) | 0.79 | - | - |
| Meningococcal Disease | 4 | TLR4 896 A>G | 1.10 (0.90-1.34) | 0.43 | 0 | 0.93 |
| All Gram + infections | 3 | TLR4 896 A>G | 1.28 (0.43-3.81) | 0.01 | 77% | - |
| 2 | TLR4 1196 C>T | 1.09(0.13-9.09) | 0.002 | - | - | |
| All parasitic infections | 8 | TLR4 896 A>G | 1.59 (1.05-2.42) | <0.001 | 72% | 0.72 |
| 7 | TLR4 1196 C>T | 1.50 (0.88-2.56) | 0.01 | 64% | 0.5 | |
| Chagas Disease | 2 | TLR4 896 A>G | 1.06 (0.53-2.14) | 0.82 | - | - |
| 2 | TLR4 1196 C>T | 1.03 (0.49-2.18) | 0.76 | - | - | |
| Malaria | 3 | TLR4 896 A>G | 1.31 (1.04-1.66) | 0.71 | 0 | - |
| 2 | TLR4 1196 C>T | 1.30 (0.64-2.65) | 0.11 | - | - | |
| Periodontitis(Aggressive) | 5 | TLR4 896 A>G | 1.04 (0.53-2.04) | 0.07 | 52% | 0.16 |
| 4 | TLR4 1196 C>T | 0.78 (0.42-1.65) | 0.29 | 20% | 0.92 | |
| Periodontitis (Chronic) | 10 | TLR4 896 A>G | 0.94 (0.75-1.18) | 0.68 | 0 | 0.74 |
| 9 | TLR4 1196 C>T | 1.12 (0.83-1.52) | 0.74 | 0 | 0.93 | |
| RSV | 2 | TLR4 896 A>G | 1.02 (0.72-1.44) | 0.61 | - | - |
| Sepsis | 3 | TLR4 896 A>G | 0.81 (0.41-1.56) | 0.16 | 45% | - |
| Tuberculosis | 5 | TLR4 896 A>G | 1.18(0.80-1.73) | 0.03 | 63% | 0.43 |
| 3 | TLR4 1196 C>T | 1.07 (0.81-1.42) | 0.47 | 0 | - | |
| UTI (Children) | 3 | TLR4 896 A>G | 1.41 (0.70-2.84) | 0.21 | 35% | - |
PQ= p value for Q homogeneity test; PH= p value for Harbord’s small study effects test, -=not applicable
Figure 1. All parasitic infections: Random effects (RE) generalized odds ratio (ORG) estimates with the corresponding 95% confidence interval (CI) for the variant TLR4 896 A>G.
The horizontal axis is plotted on a log scale.
Figure 2. Malaria: Random effects (RE) generalized odds ratio (ORG) estimates with the corresponding 95% confidence interval (CI) for the variant TLR4 896 A>G.
The horizontal axis is plotted on a log scale.
Table 4. Summary of significant associations with disease outcomes, derived from single case-control studies.
| Study | Population | Disease Outcome | Polymorphism | ORG (95% CI) |
|---|---|---|---|---|
| Goepfert [86] | USA | Bacterial vaginosis (pregnancy) | TLR4 1196 C>T | 0.55 (0.31-0.98) |
| Rezazadeh[30] | Iran | Brucellosis | TLR4 896 A>G | 2.66 (1.66-4.27) |
| Ajdary [36] | Iran | Cutaneous leishmaniasis | TLR4 896 A>G | 7.22 (1.91-27.29) |
| TLR4 1196 C>T | 10.14 (1.90-54.16) | |||
| Bochud [38] | East Africa | Leprosy | TLR4 896 A>G | 0.36 (0.22-0.60) |
| TLR4 1196 C>T | 0.23(0.10-0.55) | |||
| Verma [40] | India | Neurocysticercosis | TLR4 896 A>G | 4.39 (2.53-7.61) |
| TLR4 1196 C>T | 3.13 (1.46-6.73) | |||
| Liadaki [46] | Greece | H.influenzae (tonsillitis) | TLR4 1196 C>T | 0.42 (0.17-1.00) |
| Liadaki [46] | Greece | S.pyogenes (tonsillitis) | TLR4 896 A>G | 2.93 (1.24-6.93) |
| TLR4 1196 C>T | 3.12 (1.36-7.13) | |||
| Bhuvanedran [47] | Malaysia | Typhoid fever | TLR4 896 A>G | 2.51 (1.18-5.34) |
| TLR4 1196 C>T | 2.26 (1.01-5.07) | |||
| Yin [48] | China | UTI (Adults) | TLR4 896 A>G | 1.98 (1.04-3.98) |
Of note, all these effects were derived from Asian studies. Increased risk for tonsillar infection due to Streptococcus pyogenes (ORG 2.93; 95% CI 1.24-6.93) [46] was noted in the Greek pediatric population, as was an increased risk for urinary tract infections in adults (ORG 1.98; 95% CI 1.04-3.98) in a Chinese population [48]. Interestingly, not all outcomes were negative and the TLR4 896 A>G polymorphism was associated with significant protection against leprosy (ORG 0.36; 95% CI 0.22-0.60) in East Africa [38].
The use of the ORG metric resulted in more conservative estimates of associations, as two reportedly significant associations (1 reporting increased risk for Gram-negative osteomyelitis [41] and 1 reporting a protective effect for Streptococcus pneumoniae in children [45]) were downgraded to non-significant. Six control populations deviated for HWE equilibrium [30,35,39,53,58,62], and associations of TLR4 variants with disease were readdressed after correcting genotypes with their expected frequencies. These effects did not change (they appear in brackets in Tables 1,2). Specifically, the association of TLR4 896 A>G and brucellosis [30] remained significant after HWE correction (ORG 2.69; 95% CI 1.67-4.33).
TLR4 1196 C>T and disease susceptibility
A meta-analysis of GAS was performed for malaria (2 studies) [55,56], aggressive periodontitis (4 studies) [61,62,64,65], chronic periodontitis (9 studies) [61,63,65,67,69-72,87], and tuberculosis (3 studies) [78,80,81] and revealed no significant effects. Statistical heterogeneity varied from absent to moderate. Specifically, the combined effects were ORG 1.30 (95% CI 0.64-2.65) for malaria, ORG 0.78 (95% CI 0.42-1.65) for aggressive and ORG 1.12 (0.83-1.52) for chronic periodontitis, and ORG 1.07 (95% CI 0.81-1.42) for tuberculosis. Effects were also insignificant for all Gram negative infections combined [ORG 1.11 (95% CI 0.66-1.87)][33,35,39,46,47,52], all Gram positive infections combined [ORG 1.09 (95% CI 0.13-9.09)] [45,46] and all parasitic infections combined [ORG 1.50 (95% CI 0.88-2.56)] [36,37,40,50,51,55,56]. The meta-analysis results are summarized in Table 3. The Harbord’s test indicated that there is no differential magnitude of effect in large versus small studies for all outcomes (p≥0.05).
For outcomes with a single available study, a significant risk was present for cutaneous leishmaniasis in Iran (ORG 10.14; 95% CI 1.90-54.16) [36], neurocysticercosis in India (ORG 3.10; 95% CI 1.45-6.67) [40], S. pyogenes tonsillar disease in Greece (ORG 3.12; 95% CI 1.36-7.13) [46] and typhoid fever in Malaysia (ORG 2.26; 95% CI 1.01-5.07) [47]. A significant protection was conferred for bacterial vaginosis in pregnancy (ORG 0.55;95% CI 0.31-0.98) in the United States (notably, African Americans comprised 78% of the cases) [86], leprosy in East Africa (ORG 0.23; 95% CI 0.10-0.55) [38], and Haemophilus influenzae tonsillar disease in a Greek pediatric population (ORG 0.42; 95% CI 0.17-1.00) [46]. The significant results are summarized in Table 4. Only 1 control population deviated from HWE equilibrium that assessed the risk of meliodosis [39], a risk that did not change after correction with the expected genotype frequencies (Table 2). Two reportedly significant associations for Gram-negative osteomyelitis (increased risk) and S. pneumoniae (protection) were not confirmed in this analysis with the use of the ORG metric.
The significant effects were unidirectional and similar in magnitude when both TLR4 896 A>G and 1196 C>T were examined (Table 4), that is if TLR4 896 A>G was protective then 1196 C>T was also protective. When TLR4 896 A>G increased risk, then 1196 C>T increased risk. Specifically, the point estimates for 896 A>G and 1196 C>T variants were (respectively): 7.22 and 10.14 for cutaneous leishmaniasis, 4.39 and 3.13 for neurocysticercosis, 2.93 and 3.12 for S. pyogenes tonsillar disease, 2.51 and 2.26 for typhoid fever, 0.36 and 0.23 for leprosy. An exception to the rule was H. influenzae tonsillar disease, where the protective effect of TLR4 896 A>G did not reach statistical significance (ORG 0.47; 95% CI 0.19-1.14), while 1196 C>T showed significant association (ORG 0.42; 95% CI 0.17-1.00).
Discussion
We performed a systematic literature review to address the potential association of 2 common TLR4 single nucleotide polymorphisms (TLR4 896 A>G, TLR4 1196 C>T) with infectious diseases. An increased risk was documented for all parasitic infections combined, malaria [54-56], brucellosis [30], cutaneous leishmaniasis [36], typhoid fever [47], neurocysticercosis [40] and adult urinary tract infections [48]. Interestingly, all these effects were reported in populations of Asian descent, with the exception of parasitic infections and malaria where the effect was a combined effect from Asian, African and South American populations. This finding is more striking when we consider that European populations comprised the majority of GAS data (28 out of 59 studies, 48%) and a significant risk was found only for TLR4 polymorphisms and S. pyogenes tonsillitis among Greek children [46]. Another notable finding is that, for some infections, these single nucleotide polymorphisms were associated with lower infection rates. Overall, these effects sum to a total of 11 significant SNPs-disease associations that represent almost one third (30%) of all outcomes addressed in the eligible studies and there was consistency of effects (risk or protection) between 896 A>G and 1196 C>T variants when both associations were studied.
In this study we utilized the generalized odds ratio (ORG) metric to quantify the magnitude of associations. This metric provides a straightforward interpretation of the relative risk effect, based solely on genotype distribution [20]. The generalized odds ratio overcomes this problem by directly quantifying the magnitude of association of a gene with disease [20]. Implementing the ORG obviates the need for selecting, estimating and interpreting individual genotype contrasts (dominant, recessive and co-dominant) and their effect. ORG can also be used in meta-analysis of GAS to summarize effects and produce robust results, avoiding the shortcomings of multiple model testing, namely the lack of biologic justification and non-independency of effects [20,88,89]. For example, for TLR4 896 A>G association with malaria, the combined ORG showed that the probability of having malaria might be 31% higher for subjects having higher mutational load relative to those with lower mutational load (subjects who are homozygous for G allele have the highest mutational load, those homozygous for A allele have the lowest, and heterozygous have an intermediate level). The application of the ORG metric also resulted in a more conservative estimate of associations, given that associations for infections such as osteomyelitis (39) and S. pneumonia (43) were downgraded to insignificant. The associations derived from tuberculosis data were insignificant similar to those reported [90].
In our analysis, TLR4 polymorphisms were associated with susceptibility to a diverse spectrum of infections including Gram-negative, Gram-positive bacteria as well as parasitic infections, such as cutaneous leishmaniasis and neurocysticercosis. This wide spectrum of associations correlates with the spectrum of recognition molecules by TLR4. Indeed, TLR4 is involved in induction of cell-mediated immunity to Brucella abortus in mice [7] and TLR4 signaling also upregulates macrophage anti-leishmanial activity [91]. Similarly, binding of the Salmonella typhi porin OmpS1 to TLR4 leads to overexpression of MHCII and CD40 molecules and activation of dendritic cells [92]. TLR4 can recognize LPS of Gram-negative bacteria [3,4], glycans of the helminth Taenia solium [93] as well as the fusion protein of respiratory syncytial virus[9].
Interestingly, our analysis also confirmed that these polymorphisms are also protective for certain types of infection, such as leprosy. It is not clear why such polymorphisms confer increased susceptibility to some infection, but protect from others. It could be speculated that in some infections the immune response leads to an inflammatory response that is protective, whereas in others such response may be essential in the pathogenesis of the infectious process. An example is Mycobacterium leprae where the TLR4-mediated immune response to the pathogen may modulate inflammatory processes that influence disease manifestations but are not attributable to direct stimulation by M. leprae. Indeed, Bochud et al [38] found that the stimulation of monocytes with M. leprae inhibited their subsequent response to TLR4 stimulation with LPS.
Among Indo-European populations, 6-14% of the individuals are double heterozygous for both polymorphisms [94]. It is suggested that the double heterozygous TLR4 896 A>G/TLR4 1196 C>T haplotype does not functionally differ from wild type TLR4. Therefore, co-segregation may result in a functionally neutral phenotype and, as seen in European populations, lead to the lack of significant associations. Conversely, TLR4 896 A>G was frequently found (10-18%) among African populations, with only 2% having TLR4 1196 C>T co-segregation. Two studies (on typhoid fever and leprosy) indicated weak linkage disequilibrium in Malaysian [47] and East African populations [38]. These differences between Europeans (co-segregation) compared to Asian and African population (lack of co-segregation) may explain why the majority of significant associations were noted for endemic diseases of Asia and Africa.
Our analysis on the impact of these polymorphisms in periodontitis illustrates the different impact of polymorphisms based on the population. More specifically, despite the bulk of studies on aggressive and chronic periodontitis, TLR4 variants did not show any significant association, even though TLR4 has been shown to be overexpressed in gingival epithelial cells and gingival fibroblasts [95-97] in association with periodontal inflammation involving pathogens related to periodontitis, such as Porphyromonas gingivalis, Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans [98-101]. One possible explanation is that this finding was because all relevant studies were almost exclusively confined to European ancestry populations and the lack of susceptibility may be related to the strong linkage disequilibrium, that is the non-random association between 896 A>G and 1196 C>T in Europeans [94].
Importantly, our analysis highlights the need to evaluate the impact of these polymorphisms in different populations and various clinical conditions. Moreover, the absence of significant associations in meta-analysis data for periodontitis, tuberculosis, meningococcal disease and sepsis, signifies that the functional alterations related to polymorphic TLR4 variants may not be critical to produce the clinical phenotype. Lack of reproducibility stands as a barrier for conclusive evidence, and design, sample size and environmental and genetic heterogeneity between populations may affect results. Finally, the presence of a significant effect may rely on the magnitude of functional expression of TLR4. Protection or risk may be moderated by the level of TLR4 functional expression, which is modulated by TLR4 polymorphism and MD-2 presence [14,15]. Therefore, it is essential to explore whether MD-2 is important in the response to some infections, but not others, or that levels of TLR4 vary in one infection compared to another.
The heterogeneity of the populations studied along with multiple endpoints should also be considered as potential study limitation that may influence statistical power. Moreover, different populations mount diverse immunologic responses and the clinical relevance of polymorphisms is not always straightforward. The lack of association for a disease phenotype highlights that gene-to-gene interactions and gene-environment interactions may be influential parameters of disease association. Case-control design of individual GAS precludes adjusted analysis for gene-gene-environment interactions and may have reduced the efficiency of genetic risk estimates, though it is unlikely to inflate false-positive results [89].
Despite these limitations, genetic markers of immune response such as TLR4 variants, are valuable not only to classify high-risk patients based on disease susceptibility but also to predict disease severity and other sequelae. The associations of TLR4 896A>G with hearing loss in survivors of bacterial meningitis [102] and the increased risk of tympanostomy among toddlers with history of bronchiolitis [103] are indicative examples.
In conclusion, our analysis highlights the complex effect of TLR variants in susceptibility to infectious disease. Some of the effects, such as in malaria, are validated in a variety of studies, whereas single case-control studies should be cautiously interpreted until more information on the specific outcomes is added. Taken in their totality, our results indicate that depending on the infection and the population studied, the same polymorphism may be associated with risk, protection or have no effect. In this context, our analysis provides the rationale for understanding the protective or adverse effect of TLR4 polymorphisms and may provide a basis to explain the maintenance of these polymorphisms.
Supporting Information
PRISMA checklist.
(DOC)
Flow diagram of meta-analysis.
(DOCX)
Funding Statement
The Brown University Infectious Diseases Program in Outcomes Research is supported through funding from the Warren Alpert School of Brown University, the Department of Medicine and the Division of Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Creagh EM, O'Neill LA (2006) TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol 27: 352-357. doi: 10.1016/j.it.2006.06.003. PubMed: 16807108. [DOI] [PubMed] [Google Scholar]
- 2. Beutler B (2004) Innate immunity: an overview. Mol Immunol 40: 845-859. doi: 10.1016/j.molimm.2003.10.005. PubMed: 14698223. [DOI] [PubMed] [Google Scholar]
- 3. Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C et al. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282: 2085-2088. doi: 10.1126/science.282.5396.2085. PubMed: 9851930. [DOI] [PubMed] [Google Scholar]
- 4. Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T et al. (1999) Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol 162: 3749-3752. PubMed: 10201887. [PubMed] [Google Scholar]
- 5. Miggin SM, O'Neill LA (2006) New insights into the regulation of TLR signaling. J Leukoc Biol 80: 220-226. doi: 10.1189/jlb.1105672. PubMed: 16698941. [DOI] [PubMed] [Google Scholar]
- 6. Akira S (2003) Toll-like receptor signaling. J Biol Chem 278: 38105-38108. doi: 10.1074/jbc.R300028200. PubMed: 12893815. [DOI] [PubMed] [Google Scholar]
- 7. Campos MA, Rosinha GM, Almeida IC, Salgueiro XS, Jarvis BW et al. (2004) Role of Toll-like receptor 4 in induction of cell-mediated immunity and resistance to Brucella abortus infection in mice. Infect Immun 72: 176-186. doi: 10.1128/IAI.72.1.176-186.2004. PubMed: 14688095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sánchez D, Rojas M, Hernández I, Radzioch D, García LF et al. (2010) Role of TLR2- and TLR4-mediated signaling in Mycobacterium tuberculosis-induced macrophage death. Cell Immunol 260: 128-136. doi: 10.1016/j.cellimm.2009.10.007. PubMed: 19919859. [DOI] [PubMed] [Google Scholar]
- 9. Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP et al. (2000) Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol 1: 398-401. doi: 10.1038/80833. PubMed: 11062499. [DOI] [PubMed] [Google Scholar]
- 10. Smirnova I, Mann N, Dols A, Derkx HH, Hibberd ML et al. (2003) Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc Natl Acad Sci U S A 100: 6075-6080. doi: 10.1073/pnas.1031605100. PubMed: 12730365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smirnova I, Poltorak A, Chan EK, McBride C, Beutler B (2000) Phylogenetic variation and polymorphism at the toll-like receptor 4 locus (TLR4). Genome Biol 1: RESEARCH002 PubMed: 11104518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohto U, Yamakawa N, Akashi-Takamura S, Miyake K, Shimizu T (2012) Structural analyses of human Toll-like receptor 4 polymorphisms D299G and T399I. J Biol Chem 287: 40611-40617. doi: 10.1074/jbc.M112.404608. PubMed: 23055527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Figueroa L, Xiong Y, Song C, Piao W, Vogel SN et al. (2012) The Asp299Gly polymorphism alters TLR4 signaling by interfering with recruitment of MyD88 and TRIF. J Immunol 188: 4506-4515. doi: 10.4049/jimmunol.1200202. PubMed: 22474023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prohinar P, Rallabhandi P, Weiss JP, Gioannini TL (2010) Expression of functional D299G.T399I polymorphic variant of TLR4 depends more on coexpression of MD-2 than does wild-type TLR4. J Immunol 184: 4362-4367. doi: 10.4049/jimmunol.0903142. PubMed: 20212095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park BS, Song DH, Kim HM, Choi BS, Lee H et al. (2009) The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458: 1191-1195. doi: 10.1038/nature07830. PubMed: 19252480. [DOI] [PubMed] [Google Scholar]
- 16. Zhu L, Li X, Miao C (2012) Lack of association between TLR4 Asp299Gly and Thr399Ile polymorphisms and sepsis susceptibility: a meta-analysis. Gene 501: 213-218. doi: 10.1016/j.gene.2012.04.027. PubMed: 22537674. [DOI] [PubMed] [Google Scholar]
- 17. Noreen M, Shah MA, Mall SM, Choudhary S, Hussain T et al. (2012) TLR4 polymorphisms and disease susceptibility. Inflamm Res 61: 177-188. doi: 10.1007/s00011-011-0427-1. PubMed: 22277994. [DOI] [PubMed] [Google Scholar]
- 18. Ferwerda B, McCall MB, Verheijen K, Kullberg BJ, van der Ven AJ et al. (2008) Functional consequences of toll-like receptor 4 polymorphisms. Mol Med 14: 346-352. PubMed: 18231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schröder NW, Schumann RR (2005) Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis 5: 156-164. doi: 10.1016/S1473-3099(05)70023-2. PubMed: 15766650. [DOI] [PubMed] [Google Scholar]
- 20. Zintzaras E (2010) The generalized odds ratio as a measure of genetic risk effect in the analysis and meta-analysis of association studies. Stat Appl Genet Mol Biol 9: Article21 [DOI] [PubMed] [Google Scholar]
- 21. Zintzaras E (2012) The power of generalized odds ratio in assessing association in genetic studies with known mode of inheritance. J Appl Stat 39: 2569-2581. doi: 10.1080/02664763.2012.722611. [DOI] [Google Scholar]
- 22. Zintzaras E, Lau J (2008) Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 61: 634-645. doi: 10.1016/j.jclinepi.2007.12.011. PubMed: 18538260. [DOI] [PubMed] [Google Scholar]
- 23. Harbord RM, Egger M, Sterne JA (2006) A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med 25: 3443-3457. doi: 10.1002/sim.2380. PubMed: 16345038. [DOI] [PubMed] [Google Scholar]
- 24. Schaid DJ, Jacobsen SJ (1999) Biased tests of association: comparisons of allele frequencies when departing from Hardy-Weinberg proportions. Am J Epidemiol 149: 706-711. doi: 10.1093/oxfordjournals.aje.a009878. PubMed: 10206619. [DOI] [PubMed] [Google Scholar]
- 25. Zintzaras E (2010) Impact of Hardy-Weinberg equilibrium deviation on allele-based risk effect of genetic association studies and meta-analysis. Eur J Epidemiol 25: 553-560. doi: 10.1007/s10654-010-9467-z. PubMed: 20526652. [DOI] [PubMed] [Google Scholar]
- 26. Rodriguez S, Gaunt TR, Day IN (2009) Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 169: 505-514. PubMed: 19126586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ziakas PD, Karsaliakos P, Prodromou ML, Mylonakis E (2013) Interleukin-6 polymorphisms and hematologic malignancy: a re-appraisal of evidence from genetic association studies. Biomarkers [e-pub ahead of print]. PubMed: 24059848. [DOI] [PubMed] [Google Scholar]
- 28. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339: b2535 doi:10.1136/bmj.b2535. PubMed: 19622551. [Google Scholar]
- 29. Carvalho A, Pasqualotto AC, Pitzurra L, Romani L, Denning DW et al. (2008) Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J Infect Dis 197: 618-621. doi: 10.1086/526500. PubMed: 18275280. [DOI] [PubMed] [Google Scholar]
- 30. Rezazadeh M, Hajilooi M, Rafiei A, Haidari M, Nikoopour E et al. (2006) TLR4 polymorphism in Iranian patients with brucellosis. J Infect 53: 206-210. doi: 10.1016/j.jinf.2005.10.018. PubMed: 16343635. [DOI] [PubMed] [Google Scholar]
- 31. Doorduyn Y, Van Pelt W, Siezen CL, Van Der Horst F, Van Duynhoven YT et al. (2008) Novel insight in the association between salmonellosis or campylobacteriosis and chronic illness, and the role of host genetics in susceptibility to these diseases. Epidemiol Infect 136: 1225-1234. PubMed: 18062835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Plantinga TS, Hamza OJ, Willment JA, Ferwerda B, van de Geer NM et al. (2010) Genetic variation of innate immune genes in HIV-infected african patients with or without oropharyngeal candidiasis. J Acquir Immune Defic Syndr 55: 87-94. doi: 10.1097/QAI.0b013e3181e53c64. PubMed: 20577092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laisk T, Peters M, Saare M, Haller-Kikkatalo K, Karro H et al. (2010) Association of CCR5, TLR2, TLR4 and MBL genetic variations with genital tract infections and tubal factor infertility. J Reprod Immunol 87: 74-81. doi: 10.1016/j.jri.2010.06.001. PubMed: 20598754. [DOI] [PubMed] [Google Scholar]
- 34. Szebeni B, Szekeres R, Rusai K, Vannay A, Veres G et al. (2006) Genetic polymorphisms of CD14, toll-like receptor 4, and caspase-recruitment domain 15 are not associated with necrotizing enterocolitis in very low birth weight infants. J Pediatr Gastroenterol Nutr 42: 27-31. doi: 10.1097/01.mpg.0000192246.47959.b2. PubMed: 16385250. [DOI] [PubMed] [Google Scholar]
- 35. Lee SO, Brown RA, Kang SH, Abdel Massih RC, Razonable RR (2011) Toll-like receptor 4 polymorphisms and the risk of gram-negative bacterial infections after liver transplantation. Transplantation 92: 690-696. doi: 10.1097/TP.0b013e31822b589f. PubMed: 21822168. [DOI] [PubMed] [Google Scholar]
- 36. Ajdary S, Ghamilouie MM, Alimohammadian MH, Riazi-Rad F, Pakzad SR (2011) Toll-like receptor 4 polymorphisms predispose to cutaneous leishmaniasis. Microbes Infect 13: 226-231. doi: 10.1016/j.micinf.2010.10.018. PubMed: 21056683. [DOI] [PubMed] [Google Scholar]
- 37. Rasouli M, Keshavarz M, Kalani M, Moravej A, Kiany S et al. (2012) Toll-like receptor 4 (TLR4) polymorphisms in Iranian patients with visceral leishmaniasis. Mol Biol Rep 39: 10795-10802. doi: 10.1007/s11033-012-1973-5. PubMed: 23053976. [DOI] [PubMed] [Google Scholar]
- 38. Bochud PY, Sinsimer D, Aderem A, Siddiqui MR, Saunderson P et al. (2009) Polymorphisms in Toll-like receptor 4 (TLR4) are associated with protection against leprosy. Eur J Clin Microbiol Infect Dis 28: 1055-1065. doi: 10.1007/s10096-009-0746-0. PubMed: 19430824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. West TE, Chierakul W, Chantratita N, Limmathurotsakul D, Wuthiekanun V et al. (2012) Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes Immun 13: 38-46. doi: 10.1038/gene.2011.49. PubMed: 21776015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verma A, Prasad KN, Gupta RK, Singh AK, Nyati KK et al. (2010) Toll-like receptor 4 polymorphism and its association with symptomatic neurocysticercosis. J Infect Dis 202: 1219-1225. doi: 10.1086/656395. PubMed: 20807077. [DOI] [PubMed] [Google Scholar]
- 41. Montes AH, Asensi V, Alvarez V, Valle E, Ocaña MG et al. (2006) The Toll-like receptor 4 (Asp299Gly) polymorphism is a risk factor for Gram-negative and haematogenous osteomyelitis. Clin Exp Immunol 143: 404-413. doi: 10.1111/j.1365-2249.2005.03002.x. PubMed: 16487238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emonts M, Veenhoven RH, Wiertsema SP, Houwing-Duistermaat JJ, Walraven V et al. (2007) Genetic polymorphisms in immunoresponse genes TNFA, IL6, IL10, and TLR4 are associated with recurrent acute otitis media. Pediatrics 120: 814-823. doi: 10.1542/peds.2007-0524. PubMed: 17908769. [DOI] [PubMed] [Google Scholar]
- 43. Moens L, Verhaegen J, Pierik M, Vermeire S, De Boeck K et al. (2007) Toll-like receptor 2 and Toll-like receptor 4 polymorphisms in invasive pneumococcal disease. Microbes Infect 9: 15-20. doi: 10.1016/j.micinf.2006.10.002. PubMed: 17196867. [DOI] [PubMed] [Google Scholar]
- 44. Mrazek F, Gallo J, Stahelova A, Petrek M (2013) Coding variants of TLR2 and TLR4 genes do not substantially contribute to prosthetic joint infection. Inflamm Res 62: 483-487. doi: 10.1007/s00011-013-0601-8. PubMed: 23417289. [DOI] [PubMed] [Google Scholar]
- 45. Yuan FF, Marks K, Wong M, Watson S, de Leon E et al. (2008) Clinical relevance of TLR2, TLR4, CD14 and FcgammaRIIA gene polymorphisms in Streptococcus pneumoniae infection. Immunol Cell Biol 86: 268-270. doi: 10.1038/sj.icb.7100155. PubMed: 18180796. [DOI] [PubMed] [Google Scholar]
- 46. Liadaki K, Petinaki E, Skoulakis C, Tsirevelou P, Klapsa D et al. (2011) Toll-like receptor 4 gene (TLR4), but not TLR2, polymorphisms modify the risk of tonsillar disease due to Streptococcus pyogenes and Haemophilus influenzae. Clin Vaccine Immunol 18: 217-222. doi: 10.1128/CVI.00460-10. PubMed: 21159925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhuvanendran S, Hussin HM, Meran LP, Anthony AA, Zhang L et al. (2011) Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms and typhoid susceptibility in Asian Malay population in Malaysia. Microbes Infect 13: 844-851. doi: 10.1016/j.micinf.2011.04.007. PubMed: 21612766. [DOI] [PubMed] [Google Scholar]
- 48. Yin X, Hou T, Liu Y, Chen J, Yao Z et al. (2010) Association of Toll-like receptor 4 gene polymorphism and expression with urinary tract infection types in adults. PLOS ONE 5: e14223. doi: 10.1371/journal.pone.0014223. PubMed: 21151974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hawn TR, Scholes D, Li SS, Wang H, Yang Y et al. (2009) Toll-like receptor polymorphisms and susceptibility to urinary tract infections in adult women. PLOS ONE 4: e5990. doi: 10.1371/journal.pone.0005990. PubMed: 19543401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weitzel T, Zulantay I, Danquah I, Hamann L, Schumann RR et al. (2012) Mannose-binding lectin and Toll-like receptor polymorphisms and Chagas disease in Chile. Am J Trop Med Hyg 86: 229-232. doi: 10.4269/ajtmh.2012.11-0539. PubMed: 22302853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zafra G, Flórez O, Morillo CA, Echeverría LE, Martín J et al. (2008) Polymorphisms of toll-like receptor 2 and 4 genes in Chagas disease. Mem Inst Oswaldo Cruz 103: 27-30. doi: 10.1590/S0074-02762008000100004. PubMed: 18368233. [DOI] [PubMed] [Google Scholar]
- 52. Achyut BR, Ghoshal UC, Moorchung N, Mittal B (2007) Association of Toll-like receptor-4 (Asp299Gly and Thr399Ileu) gene polymorphisms with gastritis and precancerous lesions. Hum Immunol 68: 901-907. doi: 10.1016/j.humimm.2007.10.006. PubMed: 18082569. [DOI] [PubMed] [Google Scholar]
- 53. Moura SB, Almeida LR, Guerra JB, Rocha GA, Rocha et al Camargos. (2008) Toll-like receptor (TLR2, TLR4 and TLR5) gene polymorphisms and Helicobacter pylori infection in children with and without duodenal ulcer. Microbes Infect 10: 1477-1483. doi: 10.1016/j.micinf.2008.08.009. PubMed: 18809506. [DOI] [PubMed] [Google Scholar]
- 54. Esposito S, Molteni CG, Zampiero A, Baggi E, Lavizzari A et al. (2012) Role of polymorphisms of toll-like receptor (TLR) 4, TLR9, toll-interleukin 1 receptor domain containing adaptor protein (TIRAP) and FCGR2A genes in malaria susceptibility and severity in Burundian children. Malar J 11: 196. doi: 10.1186/1475-2875-11-196. PubMed: 22691414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zakeri S, Pirahmadi S, Mehrizi AA, Djadid ND (2011) Genetic variation of TLR-4, TLR-9 and TIRAP genes in Iranian malaria patients. Malar J 10: 77. doi: 10.1186/1475-2875-10-77. PubMed: 21457584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mockenhaupt FP, Cramer JP, Hamann L, Stegemann MS, Eckert J et al. (2006) Toll-like receptor (TLR) polymorphisms in African children: common TLR-4 variants predispose to severe malaria. J Commun Dis 38: 230-245. PubMed: 17373355. [PubMed] [Google Scholar]
- 57. Biebl A, Muendlein A, Kazakbaeva Z, Heuberger S, Sonderegger G et al. (2009) CD14 C-159T and toll-like receptor 4 Asp299Gly polymorphisms in surviving meningococcal disease patients. PLOS ONE 4: e7374. doi: 10.1371/journal.pone.0007374. PubMed: 19809507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Read RC, Pullin J, Gregory S, Borrow R, Kaczmarski EB et al. (2001) A functional polymorphism of toll-like receptor 4 is not associated with likelihood or severity of meningococcal disease. J Infect Dis 184: 640-642. doi: 10.1086/322798. PubMed: 11494169. [DOI] [PubMed] [Google Scholar]
- 59. Faber J, Meyer CU, Gemmer C, Russo A, Finn A et al. (2006) Human toll-like receptor 4 mutations are associated with susceptibility to invasive meningococcal disease in infancy. Pediatr Infect Dis J 25: 80-81. doi: 10.1097/01.inf.0000195595.22547.fe. PubMed: 16395111. [DOI] [PubMed] [Google Scholar]
- 60. Allen A, Obaro S, Bojang K, Awomoyi AA, Greenwood BM et al. (2003) Variation in Toll-like receptor 4 and susceptibility to group A meningococcal meningitis in Gambian children. Pediatr Infect Dis J 22: 1018-1019. doi: 10.1097/01.inf.0000095431.15606.68. PubMed: 14628773. [DOI] [PubMed] [Google Scholar]
- 61. Brett PM, Zygogianni P, Griffiths GS, Tomaz M, Parkar M et al. (2005) Functional gene polymorphisms in aggressive and chronic periodontitis. J Dent Res 84: 1149-1153. doi: 10.1177/154405910508401211. PubMed: 16304445. [DOI] [PubMed] [Google Scholar]
- 62. Emingil G, Berdeli A, Baylas H, Saygan BH, Gürkan A et al. (2007) Toll-like receptor 2 and 4 gene polymorphisms in generalized aggressive periodontitis. J Periodontol 78: 1968-1977. doi: 10.1902/jop.2007.060360. PubMed: 18062119. [DOI] [PubMed] [Google Scholar]
- 63. James JA, Poulton KV, Haworth SE, Payne D, McKay IJ et al. (2007) Polymorphisms of TLR4 but not CD14 are associated with a decreased risk of aggressive periodontitis. J Clin Periodontol 34: 111-117. PubMed: 17309585. [DOI] [PubMed] [Google Scholar]
- 64. Noack B, Görgens H, Lorenz K, Ziegler A, Hoffmann T et al. (2008) TLR4 and IL-18 gene variants in aggressive periodontitis. J Clin Periodontol 35: 1020-1026. doi: 10.1111/j.1600-051X.2008.01334.x. PubMed: 18983635. [DOI] [PubMed] [Google Scholar]
- 65. Schulz S, Zissler N, Altermann W, Klapproth J, Zimmermann U et al. (2008) Impact of genetic variants of CD14 and TLR4 on subgingival periodontopathogens. Int J Immunogenet 35: 457-464. doi: 10.1111/j.1744-313X.2008.00811.x. PubMed: 19046305. [DOI] [PubMed] [Google Scholar]
- 66. Garlet GP, Trombone AP, Menezes R, Letra A, Repeke CE et al. (2012) The use of chronic gingivitis as reference status increases the power and odds of periodontitis genetic studies: a proposal based in the exposure concept and clearer resistance and susceptibility phenotypes definition. J Clin Periodontol 39: 323-332. doi: 10.1111/j.1600-051X.2012.01859.x. PubMed: 22324464. [DOI] [PubMed] [Google Scholar]
- 67. Noack B, Görgens H, Lorenz K, Schackert HK, Hoffmann T (2009) TLR4 and IL-18 gene variants in chronic periodontitis: impact on disease susceptibility and severity. Immunol Invest 38: 297-310. doi: 10.1080/08820130902846290. PubMed: 19811440. [DOI] [PubMed] [Google Scholar]
- 68. Sahingur SE, Xia XJ, Gunsolley J, Schenkein HA, Genco RJ et al. (2011) Single nucleotide polymorphisms of pattern recognition receptors and chronic periodontitis. J Periodontal Res 46: 184-192. doi: 10.1111/j.1600-0765.2010.01327.x. PubMed: 21118416. [DOI] [PubMed] [Google Scholar]
- 69. Izakovicova Holla L, Buckova D, Fassmann A, Roubalikova L, Vanek J (2007) Lack of association between chronic periodontitis and the Toll-like receptor 4 gene polymorphisms in a Czech population. J Periodontal Res 42: 340-344. doi: 10.1111/j.1600-0765.2006.00954.x. PubMed: 17559631. [DOI] [PubMed] [Google Scholar]
- 70. Berdeli A, Emingil G, Han Saygan B, Gürkan A, Atilla G et al. (2007) TLR2 Arg753Gly, TLR4 Asp299Gly and Thr399Ile gene polymorphisms are not associated with chronic periodontitis in a Turkish population. J Clin Periodontol 34: 551-557. doi: 10.1111/j.1600-051X.2007.01092.x. PubMed: 17555409. [DOI] [PubMed] [Google Scholar]
- 71. Laine ML, Morré SA, Murillo LS, van Winkelhoff AJ, Peña AS (2005) CD14 and TLR4 gene polymorphisms in adult periodontitis. J Dent Res 84: 1042-1046. doi: 10.1177/154405910508401114. PubMed: 16246938. [DOI] [PubMed] [Google Scholar]
- 72. Folwaczny M, Glas J, Török HP, Limbersky O, Folwaczny C (2004) Toll-like receptor (TLR) 2 and 4 mutations in periodontal disease. Clin Exp Immunol 135: 330-335. doi: 10.1111/j.1365-2249.2004.02383.x. PubMed: 14738464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Löfgren J, Marttila R, Renko M, Rämet M, Hallman M (2010) Toll-like receptor 4 Asp299Gly polymorphism in respiratory syncytial virus epidemics. Pediatr Pulmonol 45: 687-692. doi: 10.1002/ppul.21248. PubMed: 20575099. [DOI] [PubMed] [Google Scholar]
- 74. Paulus SC, Hirschfeld AF, Victor RE, Brunstein J, Thomas E et al. (2007) Common human Toll-like receptor 4 polymorphisms--role in susceptibility to respiratory syncytial virus infection and functional immunological relevance. Clin Immunol 123: 252-257. doi: 10.1016/j.clim.2007.03.003. PubMed: 17449325. [DOI] [PubMed] [Google Scholar]
- 75. Ahmad-Nejad P, Denz C, Zimmer W, Wacker J, Bugert P et al. (2011) The presence of functionally relevant toll-like receptor polymorphisms does not significantly correlate with development or outcome of sepsis. Genet Test Mol Biomarkers 15: 645-651. doi: 10.1089/gtmb.2010.0258. PubMed: 21721932. [DOI] [PubMed] [Google Scholar]
- 76. Carregaro F, Carta A, Cordeiro JA, Lobo SM, Silva EH et al. (2010) Polymorphisms IL10-819 and TLR-2 are potentially associated with sepsis in Brazilian patients. Mem Inst Oswaldo Cruz 105: 649-656. doi: 10.1590/S0074-02762010000500008. PubMed: 20835611. [DOI] [PubMed] [Google Scholar]
- 77. Feterowski C, Emmanuilidis K, Miethke T, Gerauer K, Rump M et al. (2003) Effects of functional Toll-like receptor-4 mutations on the immune response to human and experimental sepsis. Immunology 109: 426-431. doi: 10.1046/j.1365-2567.2003.01674.x. PubMed: 12807489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Najmi N, Kaur G, Sharma SK, Mehra NK (2010) Human Toll-like receptor 4 polymorphisms TLR4 Asp299Gly and Thr399Ile influence susceptibility and severity of pulmonary tuberculosis in the Asian Indian population. Tissue Antigens 76: 102-109. PubMed: 20403143. [DOI] [PubMed] [Google Scholar]
- 79. Newport MJ, Allen A, Awomoyi AA, Dunstan SJ, McKinney E et al. (2004) The toll-like receptor 4 Asp299Gly variant: no influence on LPS responsiveness or susceptibility to pulmonary tuberculosis in The Gambia. Tuberculosis (Edinb) 84: 347-352. doi: 10.1016/j.tube.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 80. Sánchez D, Lefebvre C, Rioux J, García LF, Barrera LF (2012) Evaluation of Toll-like receptor and adaptor molecule polymorphisms for susceptibility to tuberculosis in a Colombian population. Int J Immunogenet 39: 216-223. doi: 10.1111/j.1744-313X.2011.01077.x. PubMed: 22221660. [DOI] [PubMed] [Google Scholar]
- 81. Selvaraj P, Harishankar M, Singh B, Jawahar MS, Banurekha VV (2010) Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis (Edinb) 90: 306-310. doi: 10.1016/j.tube.2010.08.001. PubMed: 20797905. [DOI] [PubMed] [Google Scholar]
- 82. Rosas-Taraco AG, Revol A, Salinas-Carmona MC, Rendon A, Caballero-Olin G et al. (2007) CD14 C(-159)T polymorphism is a risk factor for development of pulmonary tuberculosis. J Infect Dis 196: 1698-1706. doi: 10.1086/522147. PubMed: 18008256. [DOI] [PubMed] [Google Scholar]
- 83. Akil I, Ozkinay F, Onay H, Canda E, Gumuser G et al. (2012) Assessment of Toll-like receptor-4 gene polymorphism on pyelonephritis and renal scar. Int J Immunogenet 39: 303-307. doi: 10.1111/j.1744-313X.2012.01090.x. PubMed: 22308961. [DOI] [PubMed] [Google Scholar]
- 84. Ertan P, Berdeli A, Yilmaz O, Gonulal DA, Yuksel H (2011) LY96, UPKIB mutations and TLR4, CD14, MBL polymorphisms in children with urinary tract infection. Indian J Pediatr 78: 1229-1233. doi: 10.1007/s12098-011-0399-8. PubMed: 21390520. [DOI] [PubMed] [Google Scholar]
- 85. Karoly E, Fekete A, Banki NF, Szebeni B, Vannay A et al. (2007) Heat shock protein 72 (HSPA1B) gene polymorphism and Toll-like receptor (TLR) 4 mutation are associated with increased risk of urinary tract infection in children. Pediatr Res 61: 371-374. doi: 10.1203/pdr.0b013e318030d1f4. PubMed: 17314700. [DOI] [PubMed] [Google Scholar]
- 86. Goepfert AR, Varner M, Ward K, Macpherson C, Klebanoff M et al. (2005) Differences in inflammatory cytokine and Toll-like receptor genes and bacterial vaginosis in pregnancy. Am J Obstet Gynecol 193: 1478-1485. doi: 10.1016/j.ajog.2005.03.053. PubMed: 16202743. [DOI] [PubMed] [Google Scholar]
- 87. Reddy BH, Jayakumar ND, Akula SR, Sharma R, Kaarthikeyan G et al. (2011) Analysis of association between TLR-4 Asp299Gly and Thr399Ile gene polymorphisms and chronic periodontitis in a sample of south Indian population. J Indian Soc Periodontol 15: 366-370. doi: 10.4103/0972-124X.92571. PubMed: 22368361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zintzaras E (2012) Gamma-aminobutyric acid A receptor, alpha-2 (GABRA2) variants as individual markers for alcoholism: a meta-analysis. Psychiatr Genet 22: 189-196. doi: 10.1097/YPG.0b013e328353ae53. PubMed: 22555154. [DOI] [PubMed] [Google Scholar]
- 89. Zintzaras E, Doxani C, Rodopoulou P, Bakalos G, Ziogas DC et al. (2012) Variants of the MTHFR gene and susceptibility to acute lymphoblastic leukemia in children: a synthesis of genetic association studies. Cancer Epidemiol 36: 169-176. doi: 10.1016/j.canep.2011.10.002. PubMed: 22094326. [DOI] [PubMed] [Google Scholar]
- 90. Tian T, Jin S, Dong J, Li G (2013) Lack of association between Toll-like receptor 4 gene Asp299Gly and Thr399Ile polymorphisms and tuberculosis susceptibility: a meta-analysis. Infect Genet Evol 14: 156-160. doi: 10.1016/j.meegid.2012.11.009. PubMed: 23200920. [DOI] [PubMed] [Google Scholar]
- 91. Murray HW, Zhang Y, Zhang Y, Raman VS, Reed SG et al. (2013) Regulatory Actions of TLR2 and TLR4 in Leishmania donovani Infection in the Liver. Infect Immun 81: 2318-2326. doi: 10.1128/IAI.01468-12. PubMed: 23589575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Moreno-Eutimio MA, Tenorio-Calvo A, Pastelin-Palacios R, Perez-Shibayama C, Gil-Cruz C et al. (2013) Salmonella Typhi OmpS1 and OmpS2 porins are potent protective immunogens with adjuvant properties. Immunology. 139: 459-471. doi: 10.1111/imm.12093. PubMed: 23432484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Verma A, Prasad KN, Cheekatla SS, Nyati KK, Paliwal VK et al. (2011) Immune response in symptomatic and asymptomatic neurocysticercosis. Med Microbiol Immunol 200: 255-261. doi: 10.1007/s00430-011-0198-x. PubMed: 21533784. [DOI] [PubMed] [Google Scholar]
- 94. Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M et al. (2007) TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci U S A 104: 16645-16650. doi: 10.1073/pnas.0704828104. PubMed: 17925445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mori Y, Yoshimura A, Ukai T, Lien E, Espevik T et al. (2003) Immunohistochemical localization of Toll-like receptors 2 and 4 in gingival tissue from patients with periodontitis. Oral Microbiol Immunol 18: 54-58. doi: 10.1034/j.1399-302X.2003.180109.x. PubMed: 12588460. [DOI] [PubMed] [Google Scholar]
- 96. Ren L, Leung WK, Darveau RP, Jin L (2005) The expression profile of lipopolysaccharide-binding protein, membrane-bound CD14, and toll-like receptors 2 and 4 in chronic periodontitis. J Periodontol 76: 1950-1959. doi: 10.1902/jop.2005.76.11.1950. PubMed: 16274315. [DOI] [PubMed] [Google Scholar]
- 97. Wang PL, Ohura K, Fujii T, Oido-Mori M, Kowashi Y et al. (2003) DNA microarray analysis of human gingival fibroblasts from healthy and inflammatory gingival tissues. Biochem Biophys Res Commun 305: 970-973. doi: 10.1016/S0006-291X(03)00821-0. PubMed: 12767925. [DOI] [PubMed] [Google Scholar]
- 98. Kinane DF, Peterson M, Stathopoulou PG (2006) Environmental and other modifying factors of the periodontal diseases. Periodontol 2000 40: 107-119. doi: 10.1111/j.1600-0757.2005.00136.x. PubMed: 16398688. [DOI] [PubMed] [Google Scholar]
- 99. Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW et al. (2004) Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun 72: 5041-5051. doi: 10.1128/IAI.72.9.5041-5051.2004. PubMed: 15321997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mahanonda R, Pichyangkul S (2007) Toll-like receptors and their role in periodontal health and disease. Periodontol 2000 43: 41-55. doi: 10.1111/j.1600-0757.2006.00179.x. PubMed: 17214834. [DOI] [PubMed] [Google Scholar]
- 101. Kinane DF, Shiba H, Stathopoulou PG, Zhao H, Lappin DF et al. (2006) Gingival epithelial cells heterozygous for Toll-like receptor 4 polymorphisms Asp299Gly and Thr399ile are hypo-responsive to Porphyromonas gingivalis. Genes Immun 7: 190-200. doi: 10.1038/sj.gene.6364282. PubMed: 16437123. [DOI] [PubMed] [Google Scholar]
- 102. van Well GT, Sanders MS, Ouburg S, van Furth AM, Morré SA (2012) Polymorphisms in Toll-like receptors 2, 4, and 9 are highly associated with hearing loss in survivors of bacterial meningitis. PLOS ONE 7: e35837. doi: 10.1371/journal.pone.0035837. PubMed: 22662111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nuolivirta K, Hurme M, Halkosalo A, Koponen P, Korppi M et al. (2009) Gene polymorphism of IFNG +874 T/A and TLR4 +896 A/G and recurrent infections and wheezing in toddlers with history of bronchiolitis. Pediatr Infect Dis J 28: 1121-1123. doi: 10.1097/INF.0b013e3181af37ee. PubMed: 19773677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
(DOC)
Flow diagram of meta-analysis.
(DOCX)