Abstract
A major limitation to cardiac tissue engineering and regenerative medicine strategies is the lack of proliferation of postnatal cardiomyocytes. The extracellular matrix (ECM) is altered during heart development and studies suggest it plays an important role in regulating myocyte proliferation. Here, we studied the effects of fetal, neonatal, and adult cardiac ECM on the expansion of neonatal rat ventricular cells in vitro. At 24 hr, overall cell attachment was lowest on fetal ECM; however ∼80% of the cells were cardiomyocytes while many non-myocytes attached to older ECM and poly-L-lysine controls. After 5 days, the cardiomyocyte population remained highest on fetal ECM, with a 4-fold increase in number. Significantly more cardiomyocytes stained positively for the mitotic marker phospho-histone H3 on fetal ECM compared to other substrates at 5 days, suggesting that proliferation may be a major mechanism of cardiomyocyte expansion on young ECM. Further study of the beneficial properties of early developmental aged cardiac ECM could advance the design of novel biomaterials aimed at promoting cardiac regeneration.
Keywords: cardiac tissue engineering, cardiomyocyte, proliferation, extracellular matrix
1. Introduction
Congenital heart defects (CHDs) are the leading cause of mortality in live-born infants and young children [1]. Current surgical procedures to treat severe CHDs, such as hypoplastic left heart syndrome, are palliative and do not replicate native heart anatomy and function [2], and these patients often suffer from secondary complications throughout life [2, 3]. The ability to develop engineered cardiac tissue in vitro or stimulate cardiac regeneration in vivo has the potential to greatly improve treatments and outcomes in pediatric patients suffering from CHD. Although recent results with engineered vascular grafts in children are promising [4, 5], no engineered heart tissues have yet been used in the clinic [6]. A major challenge to moving cardiac tissue engineering towards clinical relevance is the limited proliferative capacity of postnatal cardiomyocytes [7].
While the timing of the process is species-dependent [8-10], mammalian cardiomyocytes undergo a switch from hyperplastic to hypertrophic growth after birth [8]. In contrast to their postnatal counterparts, embryonic and fetal cardiomyocytes are highly proliferative and have been shown to restore function to damaged or diseased hearts in animal models [11-16]. Although a number of factors can regulate myocyte proliferation in the developing heart such as cell-cell interactions [17, 18], growth factor signaling [18], and mechanical forces [19, 20], it is likely that the extracellular matrix (ECM) also plays an important role. Collagen synthesis [21] and Fibronectin expression [22] change with development and integrin isoforms change concurrently with the transition from proliferation to terminal differentiation [23]. Other studies have demonstrated a significant effect of ECM signaling on cardiomyocyte function. For example, Fibronectin and Collagen III, up-regulated by mouse embryonic fibroblasts, enhanced embryonic cardiomyocyte proliferation in response to growth factors [18, 24]. Periostin, an ECM protein expressed during fetal cardiac development [25, 26] was found to promote myocyte proliferation in vitro and improved heart function after myocardial infarction in adult rats [27]. Collagen resulted in better expansion of cardiac-like cells derived from mesenchymal stem cells compared to Collagen I [28], which is highly expressed in the adult heart [25]. While these findings point to a critical role for the developing ECM in promoting or mediating cardiomyocyte proliferation, none of the aforementioned studies investigated the cardiac ECM as a whole.
Decellularized organs can provide complex, tissue-specific cues and are thus attractive for tissue engineering and regenerative medicine approaches [29]. Indeed, adult cardiac tissues have been extensively studied and have shown promise for certain applications [30-35], such as providing mechanical support [35] or promoting neovascularization [30] in the adult heart. However, adult ECM may lack the necessary cues for myocyte proliferation, as the role of most signaling in the adult organ is to maintain homeostasis. The only known study to date that specifically investigated developmental age of the ECM showed that cells were better able to repopulate decellularized kidney sections from young rhesus monkey compared to adult, further supporting this concept [36, 37]. Since cardiomyocyte proliferation is highest during prenatal development, mimicking fetal ECM may be more appropriate for promoting cardiac regeneration but has not yet been explored.
The purpose of this study was to determine the effect of fetal cardiac ECM on the expansion of cardiomyocytes in vitro. Given the clinical need for novel tissue engineering and cell therapy-based treatments for the myocardium, we studied ventricular cell response to fetal, neonatal, and adult cardiac ECM. We found that cardiomyocytes had better adhesion and expansion on fetal ECM compared to older ECM, with myocytes remaining a large percentage of the cell population and increasing approximately 4-fold over 5 days in culture. Furthermore, we found that significantly more myocytes were positive for the mitosis marker phospho-histone H3 on fetal ECM compared to other substrates, suggesting that myocyte expansion is due to proliferation, at least partially. Our long term goal for this work is to develop biomaterials that mimic developmental cues to stimulate or maintain cardiomyocyte proliferation for tissue engineering or regenerative medicine approaches for treating CHD.
2. Materials and Methods
2.1. Heart harvest from fetal, neonatal, and adult rats
All animal procedures were performed in accordance with the Institutional Animal Care and Use Committee at Tufts University and the NIH Guide for the Care and Use of Laboratory Animals. Pregnant Sprague Dawley rats (∼3 months old) were deeply anesthetized with a mixture of ketamine (100mg/kg) and xylazine (10mg/kg) and then euthanized by harvest of the heart. Fetal pups (embryonic day 18-19) were isolated from the uterus, euthanized by decapitation, and then the hearts were isolated. Neonatal pups (postnatal day 2-4) were euthanized by conscious decapitation prior to heart harvest. Freshly isolated hearts from all three ages were fixed for assessment of native cardiac cell proliferation, processed for analysis of ECM composition, or decellularized for cell culture experiments, as described below.
2.2. Proliferation of cardiac cells in native hearts
To determine proliferation in native fetal, neonatal, and adult hearts, samples were immediately immersed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) after harvest and fixed at 4°C overnight. The samples were cryoprotected in 30% sucrose solution and then embedded and frozen in Tissue-Tek OCT compound (VWR). The hearts were sectioned into 7 μm-thick slices in the circumferential direction on a Cryostat (Leica CM 1950). The tissue slices were stored at -20°C until immunohistochemical staining. Samples were equilibrated to room temperature (RT), washed in phosphate buffered saline (PBS) to remove OCT and then blocked with 5% donkey serum and 0.1% bovine serum albumin (BSA) (Sigma) in PBS for 1hr at RT. Cells undergoing mitosis were labeled with phospho-histone H3 (PHH3) antibody (Ser 10-R, Santa Cruz Biotechnology, Santa Cruz, CA) for 1 hr at RT followed by incubation with Alexa Fluor 488 donkey anti-rabbit secondary antibody (Jackson ImmunoResearch, West Grove, PA) for 1 hr at RT. Cells of the cardiomyocyte lineage were then labeled with cardiac α-actin antibody (5C5; Santa Cruz) and Alexa Fluor 555 donkey anti-mouse secondary antibody (Invitrogen). Samples were mounted with Vectashield medium containing DAPI nuclear stain and imaged on an Olympus IX8I microscope using Metamorph Basic software (version 7.7.4.0, Molecular Devices).
2.3. Composition of ECM in fetal, neonatal, and adult hearts
To determine the composition of the ECM, hearts were first decellularized in sodium dodecyl sulfate (SDS) with concentrations of 0.05%, 0.5%, and 1% (wt/vol) in distilled water (diH2O) for fetal, neonatal, and adult hearts, respectively. Fetal and neonatal hearts were immersed in SDS with gentle agitation; adult hearts underwent perfusion decellularization [33] or were minced and soaked in SDS. Once the tissue turned white, samples were transferred to a wash in TritonX-100 (Amresco, Solon, OH) at the same concentration as the respective SDS solution for several hours. Next, the samples were washed with large volumes of diH2O at least three times to remove residual detergent. Samples were frozen at -20°C and then lyophilized overnight (Labconco, Kansas City, MO). Dry weights were determined and the samples were digested at a concentration of 5 mg/ml in a solution containing 5M urea, 2M thiourea, 50mM DTT, and 0.1% SDS in PBS [38] with constant stirring at 4°C for ∼48 hr. Afterward, samples were sonicated on ice (Branson Digital Sonifier, 20 sec pulses, 30% amplitude), protein was precipitated with acetone, and analyzed via liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Beth Israel Deaconess Medical Center Mass Spectrometry Core Facility. The 15 most abundant ECM components for each developmental age were identified from spectrum count data (N = 2 for each developmental age).
2.4. Second harmonic generation microscopy
Images of decellularized heart tissues were acquired on a Leica TCS SP2 confocal microscope equipped with a Ti:sapphire laser (Spectra Physics, Mountain View, CA). With the laser tuned to 800 nm, second harmonic generation (SHG) images were collected in the forward direction using a photomultiplier tube (PMT) with a 400(±10) nm bandpass filter. Two-photon excited fluorescence emission was also simultaneously collected between 500-550nm from a non-descanned PMT in the epi-direction. Using a 63× water immersion objective (1.2 NA), a series of image slices (512 × 512 pixels, 238 × 238 μm2 field of view) were acquired within the first 180-200 μm from the outer surface of the tissue at 2.5-5μm increments. Image intensities were normalized for PMT gain and laser power as previously described [39], and the mean SHG signal was computed from each image volume. Average intensity projections in the z-direction are displayed in false color with the same normalized intensity scale.
2.5. Optimization of fetal heart decellularization
The use of 1% SDS for adult hearts is well established [30, 33, 34], but few studies have reported decellularization of younger organs [36]. Therefore, we explored SDS and TritonX-100 at various concentrations ranging from 0.1-1% for fetal heart decellularization to determine the optimal conditions. DNA was measured using a DNA Quantitation Kit (Sigma) per the manufacturer's instructions to assess cellular content in the fetal hearts after decellularization. Equal dry weights of native tissue controls and decellularized samples were tested. In addition, a sub-set of ECM proteins was analyzed using a modified ELISA [40] to determine ECM preservation after decellularization. Approximately 20 μg of ECM per well, with each sample run in triplicate, was adsorbed in a 96 well plate. After washing with PBS, samples were blocked with 5% milk in PBST for 1 hr at RT. Samples were then incubated with Collagen I (M-19), Collagen (E-8) or Fibronectin (A-11) primary antibodies (all from Santa Cruz) for 1 hr at RT. After washing with PBS, samples were incubated with appropriate secondary antibodies conjugated to HRP (Invitrogen) for 45 min. Samples were then reacted with TMB SureBlue (KPL, Gaithersburg, MD) for 20 min at RT. The reaction was stopped with the addition of an equal volume of 1M HCl. Immediately afterward, 100 μl of each sample was transferred to a new 96-well plate and read at 450 nm on a plate reader (SpectraMax M2, Molecular Devices).
2.6. Solubilization of cardiac ECM for cell culture studies
To generate ECM for cell culture studies, hearts were decellularized as described above. After lyophilization, samples were solubilized with pepsin (Sigma) at 10:1 tissue:pepsin ratio in 0.1M HCl (Sigma), similar to previously described methods [41]. Samples were mixed with a magnetic stir bar at ∼700 rpm at RT. The tissue was allowed to digest until no solid tissue pieces were visible and the solution was homogeneous (∼1 hr for fetal, ∼2-4 hr for neonatal, and 24-48 hr for adult hearts). Assuming complete solubilization, the final concentration of the ECM solutions was 5 mg/ml. The solution was then neutralized with 10% (v/v) 1N NaOH, aliquoted, and stored at -20°C (< 24 hr) or used immediately. For the cell culture studies described in this paper, fetal hearts were E18-E19, neonatal hearts were P3-P4, and adult hearts were from the pregnant dams. Our preliminary studies indicated no difference in cell response when cultured on pregnant vs. non-pregnant female cardiac ECM (data not shown); therefore, we used the pregnant dam hearts for adult cardiac ECM to minimize the number of animals sacrificed for the experiments.
2.7. Preparation of ECM-coated substrates
Solubilized ECM was adsorbed onto 48-well tissue culture polystyrene (TCPS) dishes at 50 μg/cm2 and allowed to air dry uncovered overnight in a sterile tissue culture cabinet. ECM was diluted in sterile DMEM such that 37.5 μg ECM/100 μl DMEM per well was used. Poly-L-lysine (PLL) (0.01% solution, Sigma), which served as a control substrate for cell adhesion, was coated at 100 μl per well and incubated for 5 min at RT. Excess solution was then aspirated and the wells were allowed to air dry overnight per the manufacturer's instructions. Prior to cell seeding, the substrates were washed twice with sterile PBS.
2.8. Primary cardiac cell isolation and culture
Cardiac cells were isolated from P2-P3 neonatal pups according to previously described methods [42]. After euthanasia, hearts were harvested and the atria and ventricles were separated. Ventricular tissue was minced into 3-4 pieces per heart and underwent 7 × 7 min digestions in Collagenase type 2 (Worthington Biochemical Corp, Lakewood, NJ) in sterile PBS supplemented with 20mM glucose. Cells were counted with a hemocytometer and seeded at a density of 100,000 cells/cm2 on the pre-coated 48-well plates in serum free media containing a 50/50 mixture of DMEM and Ham's F12 Nutrient Mix (Invitrogen), 0.2% BSA (wt/vol) (Sigma), 0.5% Insulin-Transferrin-Selenium-X (Invitrogen), and 1% penicillin-streptomycin (Invitrogen), with 0.1 mM L-ascorbic acid (Sigma) added fresh at every feeding [43]. Cells were fed on days 1 and 3 of culture, and fixed in 100% methanol on days 1 and 5 for the analyses described below. For a positive control of cell proliferation, a set of cells on PLL were seeded and cultured with DMEM containing 15% FBS. Refer to Suppl. Fig. 1A for a schematic of the experimental set-up.
2.9. Live/dead assay
A live/dead assay (Invitrogen) was performed after 5 days in culture according to the manufacturer's instructions. Samples were incubated with 2 μM calcein AM and 4 μM ethidium homodimer-1 in culture medium for 30 min. Samples were then immediately imaged and the number of live and dead cells was determined for each condition using a custom-written pipeline in CellProfiler automated image analysis software (r11710) (The Broad Institute, Cambridge, MA).
2.10. Cell number and proliferation assays
Immunofluorescent staining methods were similar to those described above for native tissue. Cardiomyocytes were identified by cardiac α-actin staining for image analysis; in addition, some samples were stained for sarcomeric α-actinin (clone EA-53, Sigma) to visualize sarcomeric structure on ECM at 24 hr. Nuclei were identified by staining with Hoechst 33258 (Invitrogen). To assess proliferation, cells were stained for the mitotic marker PHH3. Image acquisition was automated using Metamorph's multidimensional acquisition settings and a motorized stage. Ten random fields per sample were imaged, with three samples per condition. The total number of cell nuclei, cardiac α-actin+ cells, and PHH3+ cells were determined using customized pipelines in CellProfiler (see Suppl. Fig. 1B for workflow and example outputs). More than 900 and 3000 cells per sample were analyzed for 24 hr and 5 day time points, respectively.
2.11. Statistical analysis
Statistical analysis was performed using a one-way analysis of variance (ANOVA) and the Tukey's post-hoc test with SigmaPlot 12.0 software. Differences were considered statistically significant for p < 0.05.
3. Results
3.1. Cardiac cell proliferation and ECM composition in native rat hearts
As the primary goal of this work was to investigate the effects of cardiac ECM on myocyte/precursor cell expansion, we were first interested in determining if there was a correlation between native cardiac cell proliferation and ECM composition in fetal, neonatal, and adult rat hearts. We studied mitosis of cardiac cells via PHH3 staining in late fetal (E19), early postnatal (P3) and adult (∼3 months) heart tissue sections. About 1% of cells were undergoing mitosis in fetal and neonatal hearts (0.9 ± 0.1% and 1.2 ± 0.2%, respectively; not significantly different), but mitosis declined significantly in the adult heart (0.02 ± 0.03%, p < 0.001) (Fig. 1A, B). Of the mitotic cells identified, 82 ± 3.6% and 71 ± 3% were cardiac α- actin+ in fetal and neonatal hearts, respectively; in the adult heart, no mitotic myocytes were detected (Fig. 1C). These results show a decline in the proliferation of cells of the cardiomyocyte lineage with age, as others have found [11, 44].
Figure 1. Changes in cardiac cell proliferation and ECM composition with developmental age.
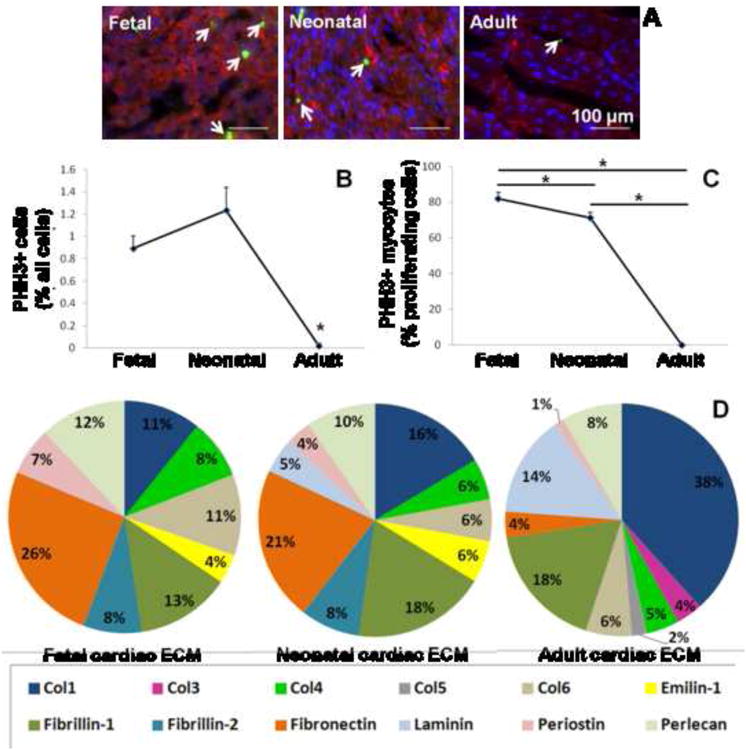
(A) Example images of fetal, neonatal, and adult heart tissue sections at 20× magnification. Tissue sections are stained for nuclei with Hoechst (blue), the mitosis marker phospho-histone H3 (green), and cardiac α-actin (red). (B) Total cell proliferation drops significantly in the adult heart. (C) Cardiomyocyte-specific proliferation is highest in the fetal heart. *indicates significant difference for p < 0.05. Scale bars = 100 μm. (D) ECM composition is represented as the percentage of the 15 most abundant proteins detected by LC-MS/MS at each developmental age.
Changes in ECM composition were also determined at the corresponding developmental ages by LC-MS/MS (Fig. 1D). The most abundant ECM protein in fetal and neonatal hearts was Fibronectin (26% and 21% of the top 15 ECM components, respectively) while in the adult heart, Collagen I dominated (38%). Other relatively abundant proteins in the fetal cardiac ECM included Fibrillin-1 (13%), Perlecan (12%), and Collagen I (11%). Neonatal hearts were similar but tended to have higher relative abundance of Fibrillin-1 (18%) and Collagen I (16%). Of note, fetal cardiac ECM was more abundant in Periostin (7%) compared to neonatal (4%) and adult hearts (∼1%). After Collagen I, adult cardiac ECM was mostly composed of Fibrillin-1 (18%) and Laminin (14%), but had relatively low levels of Fibronectin (4%). Overall, Fibronectin, Collagen IV, Emilin-1, and Periostin decreased with age while Collagen I, Collagen III, and Laminin increased with age. Taken together, these findings show that changes in cardiomyocyte proliferation occur with changes in the composition of the cardiac ECM and provide rationale for our further studies in vitro.
3.2. Optimization of fetal heart decellularization
Decellularization of adult organs is well described in the literature but ECM from younger developmental time points has been described by only one other group [36, 37]. Therefore, prior to cell culture experiments we optimized the processing procedures for fetal hearts. Decellularization was considered complete when the tissue turned white (Fig. 2A, B). We then studied the decellularized hearts using SHG microscopy which clearly revealed fibers in decellularized fetal, neonatal, and adult hearts (Fig. 2C). Two photon excited fluorescence images acquired simultaneously did not indicate cellular remnants (Suppl. Fig. 2). Fetal ECM was characterized by small diameter fibers that collectively exhibited a relatively weak average SHG signal that was 1.81 % of the mean intensity of adult tissue. Neonatal ECM demonstrated larger diameter fibers than fetal tissue and produced a SHG signal with 39.61% of the mean intensity of adult tissue. Adult tissue demonstrated increased collagen fiber organization into larger collagen fiber bundles. The increased SHG signal was partially due to increases in collagen content. However, SHG signal intensity on a per pixel basis was also increased over time possibly as a result of increased fibril size and organization within fiber bundles [45].
Figure 2. Decellularization of cardiac ECM.
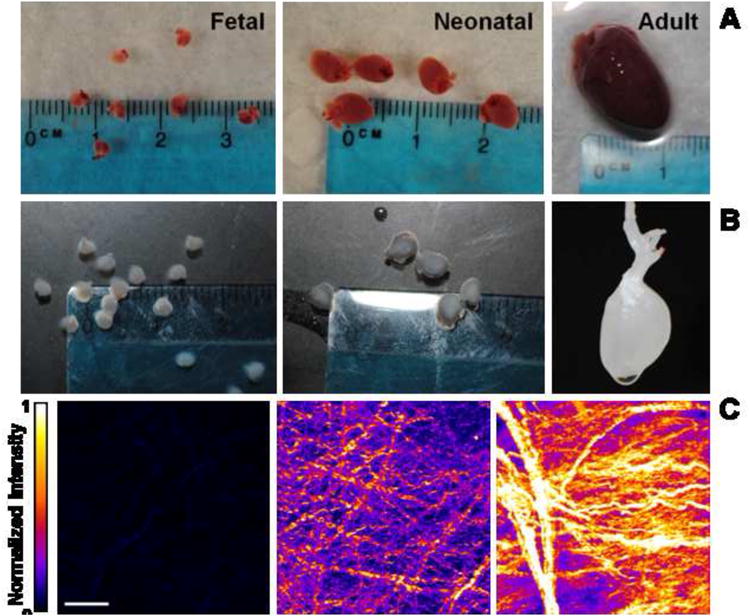
(A) Native hearts are shown from fetal pups at E18, neonatal pups at P2, and an adult rat. Fetal and neonatal hearts are shown with a dime while the adult heart is shown with fetal hearts for size comparison. (B) Hearts after decellularization with SDS and TritonX-100 wash. Tissue has turned white, indicating removal of cells. (C) Second harmonic generation image volume projections of decellularized hearts. Fetal ECM has smaller fibers and lower average signal compared to neonatal and adult hearts. Scale bar = 50 μm.
In our initial experiments we found that 1% SDS, which is standard for adult hearts [30, 33, 34], was too harsh on fetal hearts and nearly fully solubilized the tissue. Given our findings from SHG microscopy indicating that the ECM fibers were smaller and the collagen matrix was less dense, this was not surprising. Therefore, we investigated SDS at lower concentrations, as well as TritonX-100, another common decellularization agent [46], to determine the method that would result in adequate cell removal while preserving the ECM. We found that although both detergents at low concentrations preserved gross morphology of the heart (Fig. 3A), decellularization with SDS was completed in less than 24 hours while TritonX-100 took up to several days. Furthermore, the hearts did not fully turn white in TritonX-100, suggesting incomplete removal of cells.
Figure 3. Optimization of fetal heart decellularization.

(A) Representative images of fetal hearts after decellularization with SDS or TritonX-100 at different concentrations. (B) DNA assay shows significant decrease in cellular content after decellularization. ELISA results for (C) Collagen I, (D) Collagen V, and (E) Fibronectin show differences in ECM preservation with different decellularization methods. *indicates significant difference for p < 0.05.
Cellular content was determined using a DNA assay with native tissue serving as a control. Assuming that native DNA content was 100%, all four methods resulted in a significant decrease in cellular material, with 0.05% SDS demonstrating the best cell removal (reduced to 3.1 ± 2.6% of native levels) and 1% TritonX-100 performing the worst (19.6 ± 1.5% of native levels) (Fig. 3B). Next we examined a sub-set of ECM proteins to determine preservation of the ECM. Collagens I and were significantly lower in samples treated with 0.1% SDS compared to other decellularization methods, while Fibronectin was lower with TritonX-100 (Fig. 3C-E). As 0.05% SDS resulted in greatest cell removal with highest ECM content compared to the other decellularization methods tested on fetal hearts, it was used for subsequent experiments involving fetal ECM.
3.3. Ventricular cell populations on cardiac ECM at 24 hr
The mixed ventricular cell population (containing various cell types, predominantly myocytes and cardiac fibroblasts) [47] was seeded onto PLL and cardiac ECM-coated surfaces at equal cell densities. PLL was used as a control substrate for non-integrin-mediated cell adhesion [18], and 15% FBS was used as a positive control for cell proliferation and survival [48]. After 24 hr, cells were well spread with defined sarcomeres in the FBS condition; cells tended to be rounder in serum-free conditions on PLL and fetal ECM but became more spread on older ECM, and sarcomeres were evident even in the round cells (Fig. 4A). Total cell adhesion was highest on PLL controls (with similar cell densities for FBS and serum-free conditions: 714 ± 85 cells/mm2 and 743 ± 49 cells/mm2, respectively) compared to cardiac ECM. Cell density was lowest on fetal ECM (181 ± 26 cells/mm2) (Fig. 4B blue). However, of these cells, 142 ± 18 were cardiac α-actin+ (Fig. 4B brown), resulting in 79 ± 2% of the total cell population being myocytes (Fig. 4C). Although significantly more myocytes adhered with FBS compared to serum free conditions in terms of total cell numbers, they represented only 47 ± 1.7% of the adherent population. The cardiomyocyte % population was lowest on PLL under serum-free conditions (27 ± 1.6%, p < 0.001), and there was a trend in decreasing cardiomyocyte percentages for increasing developmental age (Fig. 4C). Taken together, these data show that fetal cardiac ECM selectively promoted the cardiomyocyte population over non-myocytes compared to PLL and older ECM.
Figure 4. Cell adhesion on cardiac ECM at 24 hr.
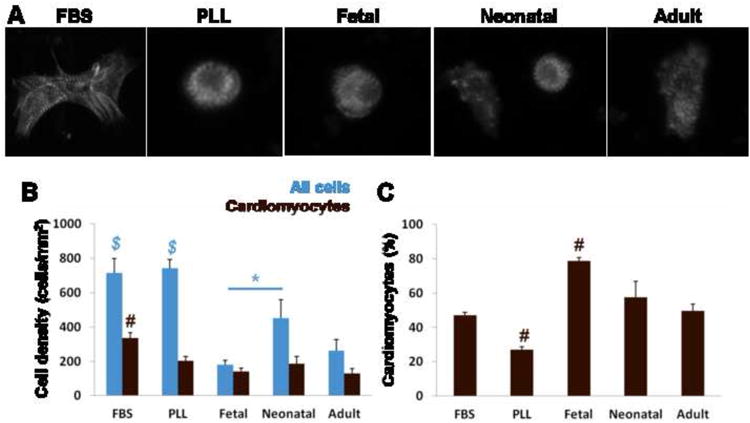
(A) Representative images of cardiomyocytes stained for sarcomeric α-actinin on PLL and ECM-coated substrates. (B) Cell density (blue) was highest on PLL substrates and lowest on fetal ECM. Cardiomyocyte density (brown) was highest for FBS conditions, but the majority of cells were myocytes on fetal ECM. (C) The cardiomyocyte population as a percentage of all cells shows that specific adhesion was greatest on fetal ECM and lowest on PLL serum free. $ = significant difference vs. all ECM. # = significant difference vs. all other conditions.
3.4. Ventricular cell populations on cardiac ECM at 5 days
After 5 days in culture cells were generally well spread on all substrates, although many rounded cells (single and clustered) were observed via bright field microscopy (data not shown). We performed a viability assay and found that the single rounded cells were dead while spread cells were live; the cell clusters contained a mixture of live and dead cells (Suppl. Fig. 3A). Based on this finding, we excluded small rounded cells from our image analysis by setting a threshold for size at 5 days in order to focus on the live population. Cell viability was significantly higher with FBS treatment; there was no difference among the serum free groups on PLL and ECM (Suppl. Fig. 3B), indicating that ECM at each developmental age had a similar ability to support cell survival.
The overall cell density was highest with FBS treatment compared to serum free conditions on PLL and ECM (1904 ± 341 cells/mm2) (Fig. 5A, B blue). Cell densities were similar on PLL serum-free and ECM, although there were significantly more cells on neonatal ECM compared to adult. Cardiomyocyte density was significantly higher on fetal and neonatal ECM (546 ± 62 and 708 ± 99 cells/mm2, respectively) compared to both PLL with FBS stimulation and serum free conditions as well as adult ECM (Fig. 5B brown). Similar to the 24 hr time point, the cell population was mostly cardiac α-actin+ on fetal ECM (71 ± 14%) (Fig. 5C). The cardiomyocyte populations on FBS-treated and serum-free PLL controls dropped to 20.5 ± 1.7% and 30.7 ± 5.5% of the total cell population, respectively. Cardiomyocytes remained at 58 ± 12% of the total population on neonatal ECM and dropped to 35 ±2.7% on adult ECM. In summary, the data show that fetal and neonatal ECM better promoted cardiomyocyte numbers compared to PLL and adult ECM, with a more pure myocyte population being maintained on fetal ECM.
Figure 5. Cell response on cardiac ECM at 5 days.
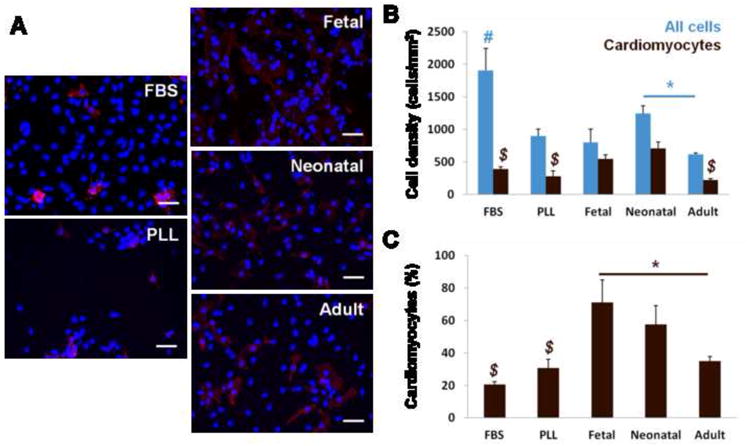
(A) Representative images of ventricular on PLL and ECM-coated substrates. Nuclei stained with Hoechst (blue) and cells of the myocyte lineage stained with cardiac α-actin (red). (B) Total cell density (blue) is highest on PLL with FBS stimulation. Cardiomyocyte density (brown) was significantly higher on fetal and neonatal ECM compared to FBS, PLL, and adult ECM. (C) The cardiomyocyte population as a percentage of all cells shows that fetal ECM maintained high purity of myocytes over time in culture. # = significant difference vs. all other conditions. $ = significant difference vs. fetal and neonatal ECM.
3.5. Proliferation on cardiac ECM
To determine whether cell numbers increased, decreased, or were maintained on cardiac ECM, we looked at fold-changes in the cell populations from 24 hr to 5 days. Interestingly, the greatest increases in cell numbers were on fetal ECM (4.4 fold). Cell numbers did not change significantly on PLL (1.2 fold) and increased roughly 2-3 fold for FBS and older ECM (Fig. 6A blue). Changes in the cardiomyocyte population tended to follow the overall cell population, with the greatest increase on fetal ECM (3.8 fold), followed by neonatal ECM (2.7 fold), and then adult ECM (1.7 fold). Cardiomyocyte numbers did not change appreciably in the FBS-treated control (1.1 fold) and only 1.4 fold for the serum-free PLL control. In general, cardiac ECM better promoted increases in the cardiomyocyte population over time, with younger ECM resulting in the greatest increases in cell numbers.
Figure 6. Proliferation on ECM.
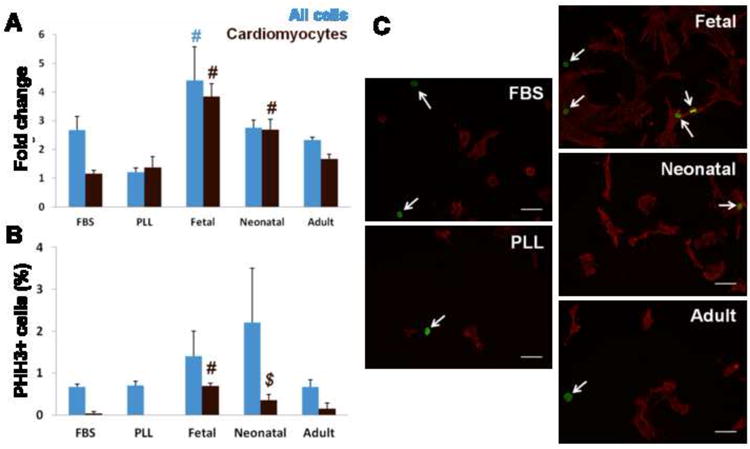
(A) Fold change in total cell numbers (blue) and cardiomyocytes (brown) from 24 hr to 5 days. Cardiomyocyte numbers had greatest increase on fetal ECM compared to all other conditions, followed by neonatal ECM. (B) Staining for PHH3 shows no significant difference in the overall proliferating cell population (blue); however, significantly more myocytes (brown) were PHH3+ on fetal ECM compared to other conditions. (C) Representative images of cells stained for PHH3 (green) and cardiac α-actin (red) on PLL and ECM (arrows indicate PHH3+ nuclei). # = significant difference vs all other conditions. $ = significant difference vs FBS stimulated and serum-free PLL controls.
To determine whether the increase in cardiomyocytes could be due to proliferation, we investigated mitosis by staining for PHH3. At 24 hr, proliferation was similar on all substrates, with 5-10% of the cells undergoing mitosis, though very few cardiomyocytes were PHH3+ (not shown). At 5 days, there was no statistically significant difference in proliferation of the overall cell population (Fig. 6B blue), although there was a trend in increased mitosis on young ECM. However, significantly more cardiomyocytes were PHH3+ on fetal ECM compared to all other conditions (0.7 ± 0.07% of the total cell population), followed by neonatal ECM (0.35 ± 0.14%, significantly higher compared to FBS-treated and serum-free PLL controls) (Fig. 6B brown, C). No PHH3+ cells were observed on PLL under serum-free conditions, and only a small fraction of myocytes proliferated in response to FBS (0.04 ± 0.4%). Given the increase in cardiomyocyte numbers and significant labeling of PHH3 on fetal ECM, our data suggest that early developmental age cardiac ECM may promote cardiomyocyte expansion via proliferation.
3.6. Reproducibility of ECM effects on cardiomyocytes
Particularly when working with primary cells and fresh ECM isolations, it is expected that there will be batch-to-batch variability. To determine whether we could obtain consistent results in expanding the cardiomyocyte population on fetal ECM, we ran repeat experiments with different batches of ECM and different primary cell isolations. Although the initial cell population varied (not shown) and absolute cell numbers were different (as some batches of cells were more proliferative than others), we generally found that ECM had relatively consistent effects on the cardiomyocyte population. An example of the results at 5 days of a repeat experiment is shown in Figure 7. The cardiomyocyte population both in terms of absolute cell numbers (Fig. 7A) and the percentage of the population (Fig. 7B) was highest on fetal ECM, followed by neonatal ECM. The fold change in the cardiomyocyte population was significantly higher on fetal ECM (2.1 fold) compared to PLL (0.4 fold, indicating loss of myocytes) and adult ECM (0.8 fold), and not significantly different from FBS (2.4) and neonatal ECM (1.4 fold). On fetal ECM, myocytes made up a significant percentage of the proliferating cell population, with 22% of the PHH3+ cells being myocytes. Taken together, the data shows that although there is some variability among experiments, young cardiac ECM can have consistent effects on promoting the cardiomyocyte population compared to PLL and older ECM.
Figure 7. Consistent effects of ECM on cardiomyocytes.
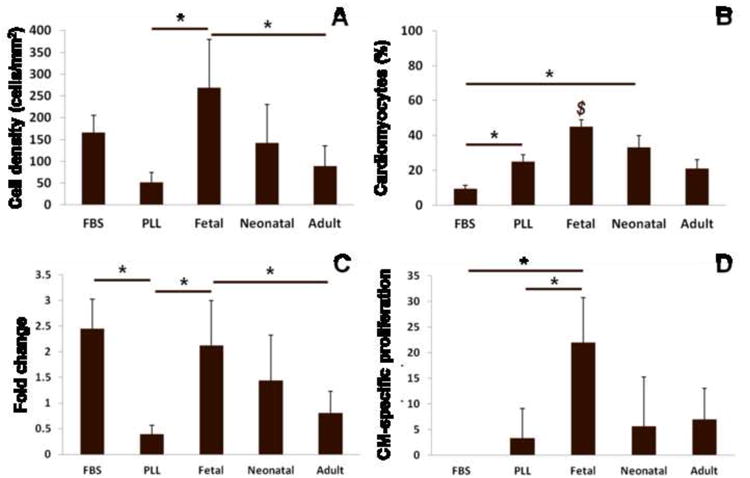
Data from a repeat experiment with different isolations of ECM and cells. (A) At 5 days, cardiomyocyte density is significantly higher on fetal ECM compared to PLL serum free and adult ECM. (B) Cardiomyocytes made up a greater percentage of the cell population on fetal ECM compared to FBS, PLL, and adult ECM. (C) Fold changes in cardiomyocyte number from 24 hr to 5 days were higher on fetal ECM compared to PLL serum-free and adult ECM. (D) Cardiomyocytes were a significantly greater portion of the proliferating cell population on fetal ECM compared to FBS and PLL. $ = significant difference vs FBS, PLL, and adult ECM.
4. Discussion
Pediatric patients with severe CHDs would greatly benefit from novel tissue engineering and regenerative medicine strategies that aim to recapitulate healthy heart anatomy and function. However, postnatal cardiomyocytes have limited capacity for significant regeneration [11, 44, 49] and stem cell differentiation toward the cardiomyocyte lineage is still inefficient, even from cardiac progenitor cells [50]. Given that fetal cardiac cells are highly proliferative [15, 16] and the cardiac ECM changes during development [21, 22, 25, 51], fetal ECM likely plays a major role in mediating cardiomyocyte proliferation and thus may be useful in approaches to grow engineered cardiac tissue or promote regeneration in vivo. In this study, we investigated the effects of fetal, neonatal, and adult cardiac ECM on the expansion of neonatal rat cardiac cells in vitro. We found that fetal cardiac ECM promoted cardiomyocytes in terms of the percentage of the total cell population and increases in cell numbers, which was significantly greater than older ECM and PLL controls. Furthermore, we found that cardiomyocyte mitosis was significantly higher on fetal ECM, suggesting that proliferation is a mechanism of cardiomyocyte expansion in response to the ECM.
Similar to what others have reported [9, 11, 44], we found a significant decrease in total cell and cardiomyocyte-specific proliferation in the native adult heart compared to fetal and neonatal hearts. Concurrent with these decreases in cell proliferation, the composition of the ECM changed considerably. The most abundant ECM protein in fetal and neonatal hearts, which contained proliferating myocytes, was Fibronectin. Fetal hearts also had a relatively high abundance of Periostin, similar to other studies [26, 52]. Adult cardiac ECM was mostly composed of Collagen I, also in agreement with the literature [53]. In line with these results, we found that although adult hearts could be decellularized with 1% SDS, fetal hearts required a less harsh treatment of 0.05% SDS and significantly shorter digestion times in pepsin-HCl (1 hr vs. up to 48 hr for the adult heart). This was likely due to their overall composition as determined by LC-MS/MS, as well as smaller fibers and less dense collagen matrix as shown by SHG microscopy. Furthermore, it is known that collagen cross-linking increases with developmental age [54], and this may also play a role in the rapid of solubilization of the fetal ECM, although our characterization methods did not investigate this directly. Determining the optimal parameters for fetal ECM processing has been critical to this study, especially since there is little data available in the literature: only one other group has reported decellularization of fetal animal organs (rhesus monkey kidney) [36, 37].
At 24 hr, cardiac ECM in general promoted adhesion of the cardiomyocyte population compared to PLL. Cell adhesion to PLL is not mediated through integrins [18], which is the case for adhesion to ECM [55]. Thus, our findings suggest that integrin-mediated adhesion is especially critical for cardiomyocytes. Furthermore, although cell adhesion was lowest on fetal ECM, cardiomyocyte purity was highest compared to other conditions, at ∼80% of the adherent cell population at 24 hr. These cells increased ∼4 fold by 5 days in culture on fetal ECM while maintaining their high purity (∼70% of the population). For our studies, the full cardiac population was seeded without any pre-plating to enrich for myocytes. Studies have shown that even with pre-plating techniques that initially yield 90% myocyte purity, the more proliferative fibroblast population tends to overtake the culture within several days [18]. We also observed increased proliferation of non-myocyte lineage cells and limited expansion of cardiomyocytes in FBS- stimulated controls. However, the fibroblast population did not out-compete the myocyte population on cardiac ECM, and was especially low for the fetal condition. Considering the data all together, it is possible that a sub-set of cardiomyocytes with proliferative potential had selective adhesion to the fetal ECM. Characterization of cell phenotype and integrin expression on fetal cardiac ECM will be of interest in our future work.
Given the increases in the cell numbers, we studied PHH3 staining in the cell population at our time points of interest. At 24 hr, there was no significant difference in cell proliferation among the conditions, although it was highly variable, and very few to no myocytes stained for PHH3. At 5 days, however, significantly more myocytes were PHH3+ on fetal cardiac ECM compared to other substrates, with ∼50% of the PHH3+ cell population comprised of myocytes. The data suggests that proliferation of the cardiomyocytes is at least one mechanism of their expansion on fetal cardiac ECM. Another potential mechanism could be expansion of myocyte precursors, such as c-kit+ cells which are important in cardiomyogenesis in the developing heart [56]. Indeed, either or both mechanisms would greatly benefit cardiac tissue engineering or cell therapy strategies, and there is much hope for CHD patients as evidence points to greater cardiomyogenic potential in the young [50, 57-59]. Recent work by French et al showed that adult c-kit+ progenitor cells could proliferate and express early cardiomyocyte markers on adult cardiac ECM [60]; it will be interesting to explore how older cardiac cell populations and stem cells respond to fetal cardiac ECM. We further note that the studies presented here used late fetal ECM (E18); younger cardiac ECM may have an even greater effect on promoting cardiomyocyte proliferation.
Given the variability of biological systems, and thus expecting that each batch of ECM and/or cells will have some inherent differences, we presented data from another experiment in which the cells were less proliferative (2 fold increase on fetal ECM vs 4 fold – see Figure 7). The initial cell population at 24 hr was also different (data not shown), with a less pure population of myocytes of 40-50% on ECM. However, the general outcome on the cardiomyocyte population after 5 days showed similar trends, with more myocytes on young ECM, greater increases in number, and greater proliferation. It is possible that the initial cell population may have a significant effect on cardiomyocyte response over time. For example, interactions with other cell types (either via direct contact or paracrine signaling) could alter myocyte proliferation [17, 18], and fibroblasts could be synthesizing and/or remodeling the ECM [61]. These potential effects were not studied here but will be of interest in future studies. In addition, to get a better sense of the direct effects of the ECM on specific cell types in the cardiac population, it will be necessary to study purified or enriched populations of myocytes or precursor cells. Development of a synthetic mimic of the fetal ECM should also lead to improved consistency; this will require identification of the key players in the fetal ECM that promote cardiomyocyte proliferation.
Our current study focused on ventricular cell response to cardiac ECM. This cell population is of interest because tissue engineering and cell therapy efforts are directed toward regenerating the ventricular myocardium. In regards to pediatric patients, potential applications would include the development of a functional tissue patch for reconstruction of the right ventricular outflow tract in Tetralogy of Fallot or an injectable material for promoting regeneration in vivo and improving function in cardiomyopathy or heart failure. It should be noted that in order to develop cardiac tissue in vitro using human cells, it will be necessary to use stem cells. The effect of cardiac ECM on human cardiac progenitors has yet to be determined and is currently under investigation in our lab.
Our studies of the ECM were performed under serum-free conditions to isolate its effects on cell response and were carried out only to 5 days in culture. Interestingly, fetal ECM had a greater effect on cardiomyocyte expansion compared to FBS stimulation of cells on PLL, further implying the critical role of integrin-mediated signaling in cardiomyocyte proliferation. Indeed, studies have shown that ECM proteins can significantly enhance fetal cardiomyocyte proliferation in response to growth factors [18]. Further exploration and optimization of culture conditions on fetal cardiac ECM should enhance its potential use for tissue engineering and cell therapy strategies in the future.
Imaging techniques have been well-established for the analysis of native cardiac tissues, particularly for scarce and valuable samples such as those obtained from humans [10, 62]. Our image analysis approach offered some unique advantages that allowed us to assay various cell populations and features, such as quantifying cell adhesion/cell density and measuring PHH3+ myocytes. We found these methods useful as our sample sizes were limited by the yield of fetal cardiac ECM. However, our study also had limitations. Our automated image analysis approach provides an estimate of cell numbers, as there will be some small error due to image artifacts and/or difficulty distinguishing cells that are very close together. However, all of our cell numbers were determined by nuclear counts and co-staining (or lack thereof) with cardiac α-actin and/or PHH3; therefore the trends of our data should be preserved with this approach. Another limitation of our study is that the standard method for preparing ECM for LC-MS/MS analysis is different from the standard solubilization techniques used in our cell studies and by many others. Therefore the protein targets that could be suggested by our proteomics data may not necessarily be those responsible for the effects on proliferation of cardiomyocytes, and will require further investigation.
5. Conclusions
In summary, fetal cardiac ECM significantly promoted the adhesion and expansion of neonatal cardiomyocytes compared to PLL, neonatal ECM, and adult ECM. Cardiomyocytes were a majority of the cell population on fetal ECM both at 24 hr and 5 days, and increased 4 fold over this time period. Additionally, we observed significantly more PHH3+ cardiomyocytes on fetal ECM compared to PLL and older ECM, suggesting that proliferation is a mechanism of cardiomyocyte expansion on fetal ECM. Mimicking the properties of fetal cardiac ECM in future biomaterials design may lead to improved expansion of immature cardiomyocytes for tissue engineering or cell therapy strategies for the young.
Supplementary Material
(A) Overview of the experimental set-up. (B) Example workflow and outputs from CellProfiler for image analysis. First, all nuclei were identified. The corresponding cardiac α-actin stained image was converted to a binary “mask”. The mask was then used to identify cardiomyocyte nuclei.
(A) Second harmonic generation at 400nm (red) and two-photon excited fluorescence emission at 500-550nm (green) enabled simultaneous detection of collagen fibers and cells within native heart tissue. (B) Following decellularization, cell autofluorescence is not apparent, and small regions of residual autofluorescence may be associated with remaining ECM proteins. Images were taken 50μm from the tissue surface with 800nm excitation light. Scale bar represents 50μm.
(A) Representative images show live (green) and dead (red) staining on PLL controls and ECM. Individual small, rounded cells were dead; cell clusters contained mixtures of live and dead cells. (B) Image analysis reveals significantly greater viability with FBS; there were no differences among the serum-free conditions. # = significant difference vs. all other conditions.
Acknowledgments
This work was funded by NRSA individual postdoctoral fellowships F32 HL112538 to CW and F32 AR061933 to KPQ, R01EB007542 to IG, and the NIH Pathway to Independence Award R00 HL093358 to LDB. We are very grateful to Professor John Asara (Beth Israel Deaconess Medical Center) and the Mass Spectrometry Core Laboratory for the proteomics analysis of the ECM.
Footnotes
Disclosures: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, Daniels SR, et al. Noninherited risk factors and congenital cardiovascular defects: Current knowledge: A scientific statement from the American Heart Association council on cardiovascular disease in the young: Endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- 2.Stumper O. Hypoplastic left heart syndrome. Heart. 2010;96:231–6. doi: 10.1136/hrt.2008.159889. [DOI] [PubMed] [Google Scholar]
- 3.Fredenburg TB, Johnson TR, Cohen MD. The fontan procedure: anatomy, complications, and manifestations of failure. Radiographics. 2011;31:453–63. doi: 10.1148/rg.312105027. [DOI] [PubMed] [Google Scholar]
- 4.Shin'oka T, Matsumara G, Hibino N, Naito Y, Watanabe M, Konuma T, et al. Midterm clinical result of tissue-engineered vascular autografts seeded with autologous bone marrow cells. J Thorac Cardiovasc Surg. 2005;129:1330–2338. doi: 10.1016/j.jtcvs.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 5.Hibino N, McGillicuddy E, Matsumura G, Ichihara Y, Naito Y, Breuer C, et al. Late-term results of tissue-engineered vascular grafts in humans. J Thorac Cardiovasc Surg. 2010;139:431–6. doi: 10.1016/j.jtcvs.2009.09.057. [DOI] [PubMed] [Google Scholar]
- 6.Miyagawa S, Roth M, Saito A, Sawa Y, Kostin S. Tissue-engineered cardiac constructs for cardiac repair. Ann Thorac Surg. 2011;91:320–9. doi: 10.1016/j.athoracsur.2010.09.080. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann WH, Cesnjevar R. Cardiac tissue engineering: Implications for pediatric heart surgery. Pediatr Cardiol. 2009;30:716–23. doi: 10.1007/s00246-009-9405-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertropy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–46. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 9.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H9. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 10.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci. 2013;110:1446–51. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soonpaa MH, Field LJ. Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res. 1998;83:15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 12.Roell W, Lu ZJ, Bloch W, Siedner S, Tiemann K, Xia Y, et al. Cellular cardiomyoplasty improves survival after myocardial injury. Circulation. 2002;105:2435–41. doi: 10.1161/01.cir.0000016063.66513.bb. [DOI] [PubMed] [Google Scholar]
- 13.Roell W, Lewalter T, Sasse P, Tallini YN, Choi BR, Breitbach M, et al. Engraftment of connexin 43- expressing cells prevents post-infarct arrhythmia. Nature. 2007;450:819–24. doi: 10.1038/nature06321. [DOI] [PubMed] [Google Scholar]
- 14.Scorsin M, Hagege A, Vilquin JT, Fiszman M, Marotte F, Samuel JL, et al. Comparison of the effects of fetal cardiomyocyte and skeletal myoblast transplantation on postinfarction left ventricular function. J Thorac Cardiovasc Surg. 2000;119:1169–75. doi: 10.1067/mtc.2000.104865. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto KL, Clause KC, Liu LJ, Tinney JP, Verma S, Wagner WR, et al. Engineered fetal cardiac graft preserves its cardiomyocyte proliferation within postinfarcted myocardium and sustains cardiac function. Tissue Eng Part A. 2011;17:585–96. doi: 10.1089/ten.tea.2010.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drenckhahn JD, Schwarz QP, Gray S, Laskowski A, Kiriazis H, Ming Z, et al. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev Cell. 2008;15:521–33. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh PCH, Davis ME, Lisowski LK, Lee RT. Endothelial-cardiomyocyte interactions in cardiac development and repair. Annu Rev Physiol. 2006;68 doi: 10.1146/annurev.physiol.68.040104.124629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–44. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clause KC, Tinney JP, Liu LJ, Keller BB, Tobita K. Engineered early embryonic cardiac tissue increases cardiomyocyte proliferation by cyclic mechanical stretch via p38-MAP kinase phosphorylation. Tissue Eng Part A. 2009;15:1373–80. doi: 10.1089/ten.tea.2008.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tobita K, Garrison JB, Liu LJ, Tinney JP, Keller BB. Three-dimensional myofiber architecture of the embryonic left ventricle during normal development and altered mechanical loads. Anat Rec A: Discov Mol Cell Evol Biol. 2005;283A:193–201. doi: 10.1002/ar.a.20133. [DOI] [PubMed] [Google Scholar]
- 21.Mays PK, McAnulty RJ, Campa JS, Laurent GJ. Age-related changes in collagen synthesis and degradation in rat tissues. Biochem J. 1991;276:307–13. doi: 10.1042/bj2760307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farhadian F, Contard F, Corbier A, Barrieux A, Rappaport L, Samuel JL. Fibronectin expression during physiological and pathological cardiac growth. J Mol Cell Cardiol. 1995;27:981–90. doi: 10.1016/0022-2828(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 23.Maitra N, Flink IL, Bahl JJ, Morkin E. Expression of alpha and beta integrins during terminal differentiation of cardiomyocytes. Cardiovasc Res. 2000;47:715–25. doi: 10.1016/s0008-6363(00)00140-1. [DOI] [PubMed] [Google Scholar]
- 24.deAlmeida A, Sedmera D. Fibroblast growth factor-2 regulates proliferation of cardiac myocytes in normal and hypoplastic left ventricles in the developing chick. Cardiol Young. 2009;19:159–69. doi: 10.1017/S1047951109003552. [DOI] [PubMed] [Google Scholar]
- 25.Bowers SLK, Banerjee I, Baudino TA. The extracellular matrix: At the center of it all. J Mol Cell Cardiol. 2010;48:474–82. doi: 10.1016/j.yjmcc.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruzynska-Frejtag A, Machnicki M, Rogers R, Markwald RR, Conway SJ. Periostin (an osteoblast- specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev. 2001;103:183–8. doi: 10.1016/s0925-4773(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn B, Monte Fd, Hajjar RJ, Chang YS, Lebeche D, Arab S, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nature Medicine. 2007;13:962–9. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 28.Tan G, Shim W, Gu Y, Qian L, Chung YY, Lim SY, et al. Differential effect of myocardial matrix and integrins on cardiac differentiation of human mesenchymal stem cells. Differentiation. 2010;79:260–71. doi: 10.1016/j.diff.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Badylak SF. The extracellular matrix as a biologic scaffold material. Biomaterials. 2007;28:3587–93. doi: 10.1016/j.biomaterials.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 30.Singelyn JM, DeQuach JA, Seif-Naraghi SB, Littlefield RB, Schup-Magoffin PJ, Christman KL. Naturally derived myocardial matrix as an injectable scaffold for cardiac tissue engineering. Biomaterials. 2009;30:5409–16. doi: 10.1016/j.biomaterials.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seif-Naraghi SB, Salvatore MA, Schup-Magoffin PJ, Hu DP, Christman KL. Design and characterization of an injectable pericardial matrix gel: A potentially autologous scaffold for cardiac tissue engineering. Tissue Eng Part A. 2010;16:2017–27. doi: 10.1089/ten.tea.2009.0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B, Borazjani A, Tahai M, Curry ALDJ, Simionescu DT, Guan J, et al. Fabrication of cardiac patch with decellularized porcine myocardial scaffold and bone marrow mononuclear cells. J Biomed Mater Res. 2010;94A:1100–10. doi: 10.1002/jbm.a.32781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nature Medicine. 2008;14:213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 34.Akhyari P, Aubin H, Gwanmesia P, Barth M, Hoffmann S, Huelsmann J, et al. The quest for an optimized protocol for whole heart decellularization: A comparison of three popular and a novel decellularization technique and their diverse effects on crucial extracellular matrix qualities. Tissue Eng Part C Methods. 2011;17:915–26. doi: 10.1089/ten.TEC.2011.0210. [DOI] [PubMed] [Google Scholar]
- 35.Kochupura PV, Azeloglu EU, Kelly DJ, Doronin SV, Badylak SF, Krukenkamp IB, et al. Tissue-engineered myocyardial patch derived from extracellular matrix provides regional mechanical function. Circulation. 2005;112:I-444–I-149. doi: 10.1161/CIRCULATIONAHA.104.524355. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama KH, Batchelder CA, Lee CCI, Tarantal AF. Renal tissue engineering with decellularized rhesus monkey kidneys: age-related differences. Tissue Eng Part A. 2011;17:2891–901. doi: 10.1089/ten.tea.2010.0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama KH, Batchelder CA, Lee CI, Tarantal AF. Decellularized rhesus monkey kidney as a three-dimensional scaffold for renal tissue engineering. Tissue Eng Part A. 2010;16:2207–16. doi: 10.1089/ten.tea.2009.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ngoka LCM. Sample prep for proteomics of breast cancer: proteomics and gene ontology reveal dramatic differences in protein solubilization preferences of radioimmunoprecipitation assay and urea lysis buffers. Proteome Science. 2008;6:30. doi: 10.1186/1477-5956-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinn KP, Bellas E, Fourligas N, Lee K, Kaplan DL, Georgakoudi I. Characterization of metabolic changes associated with the functional development of 3D engineered tissues by non-invasive, dynamic measurement of individual cell redox ratios. Biomaterials. 2012;33:5341–8. doi: 10.1016/j.biomaterials.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sazonova OV, Lee KL, Isenberg BC, Rich CB, Nugent MA, Wong JY. Cell-cell interactions mediate the response of vascular smooth muscle cells to substrate stiffness. Biophys J. 2011;101:622–30. doi: 10.1016/j.bpj.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeQuach JA, Mezzano V, Miglani A, Lange S, Keller GM, Sheikh F, et al. Simple and high yielding method for preparing tissue specific extracellular matrix coatings for cell culture. PLoS One. 2010;5:e13039. doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye KY, Sullivan KE, Black LD. Encapsulation of cardiomyocytes in a fibrin hydrogel for cardiac tissue engineering. J Vis Exp. 2011;19:pii–3251. doi: 10.3791/3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–87. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Nadiarynkh O, Plotnikov S, Campagnola PJ. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nature Protocols. 2012;7:654–69. doi: 10.1038/nprot.2012.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials. 2006;27:3675–83. doi: 10.1016/j.biomaterials.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 47.Naito H, Melnychenko I, Didie M, Schneiderbanger K, Schubert P, Rosenkranz S, et al. Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle. Circulation. 2006;1114:I-72–I-8. doi: 10.1161/CIRCULATIONAHA.105.001560. [DOI] [PubMed] [Google Scholar]
- 48.Lorts A, Schwanekamp JA, Elrod JW, Sargent MA, Molkentin JD. Genetic manipulation of periostin expression in the heart does not affect myocyte content, cell cycle activity, or cardiac repair. Circ Res. 2009;104:e1–e7. doi: 10.1161/CIRCRESAHA.108.188649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mishra R, Vijayan K, Colletti EJ, Harrington DA, Mattiesen TS, Simpson D, et al. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 2011;123:364–73. doi: 10.1161/CIRCULATIONAHA.110.971622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klewer SE, Krob SL, Kolker SJ, Kitten GT. Expression of type VI collagen in the developing mouse heart. Dev Dynamics. 1998;211:248–55. doi: 10.1002/(SICI)1097-0177(199803)211:3<248::AID-AJA6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 52.Norris RA, Kern CB, Wessels A, Moralez EI, Markwald RR, Mjaatvedt CH. Identification and detection of the periostin gene in cardiac development. Anat Rec A: Discov Mol Cell Evol Biol. 2004;281A:1227–33. doi: 10.1002/ar.a.20135. [DOI] [PubMed] [Google Scholar]
- 53.Bishop JE, Greenbaum R, Gibson DG, Yacoub M, Laurent GJ. Enhanced deposition of predominantly type I collagen in myocardial disease. J Mol Cell Cardiol. 1990;22:1157–65. doi: 10.1016/0022-2828(90)90079-h. [DOI] [PubMed] [Google Scholar]
- 54.Lovvorn HN, Cheung DT, Nimni ME, Perelman N, Estes JM, Adzick NS. Relative distribution and crosslinking of collagen distinguish fetal from adult sheep wound repair. J Pediatr Surg. 1999;34:218–23. doi: 10.1016/s0022-3468(99)90261-0. [DOI] [PubMed] [Google Scholar]
- 55.Bokel C, Brown NH. Integrins in development: Moving on, responding to, and sticking to the extracellular matrix. Dev Cell. 2002;3:311–21. doi: 10.1016/s1534-5807(02)00265-4. [DOI] [PubMed] [Google Scholar]
- 56.Ferreira-Martins J, Ogorek B, Cappetta D, Matsuda A, Signore S, D'Amario D, et al. Cardiomyogenesis in the developing heart is regulated by c-kit-positive cardiac stem cells. Circ Res. 2012;110:701–15. doi: 10.1161/CIRCRESAHA.111.259507. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Tallini Y, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, et al. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc Natl Acad Sci. 2009;106:1808–13. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jesty SA, Steffey MA, Lee FK, Breitbach M, Hesse M, Reining S, et al. c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci. 2012;109:13380–5. doi: 10.1073/pnas.1208114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simpson DL, Mishra R, Sharma S, Goh SK, Deshmukh S, Kaushal S. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation. 2012;126(suppl 1):S46–S53. doi: 10.1161/CIRCULATIONAHA.111.084699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.French KM, Boopathy AV, DeQuach JA, Chingozha L, Lu H, Christman KL, et al. A naturally derived cardiac extracellular matrix enhances cardiac progenitor cell behavior in vitro. Acta Biomaterialia. 2012;8:4357–64. doi: 10.1016/j.actbio.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Porter KE, Turner NA. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther. 2009;123:255–78. doi: 10.1016/j.pharmthera.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Urbanek K, Quaini F, Tasca G, Torella D, Castaldo C, Nadal-Ginard B, et al. Intense myocyte formation from cardiac stem cells in human cardiac hypertrophy. Proc Natl Acad Sci. 2003;100:10440–5. doi: 10.1073/pnas.1832855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Overview of the experimental set-up. (B) Example workflow and outputs from CellProfiler for image analysis. First, all nuclei were identified. The corresponding cardiac α-actin stained image was converted to a binary “mask”. The mask was then used to identify cardiomyocyte nuclei.
(A) Second harmonic generation at 400nm (red) and two-photon excited fluorescence emission at 500-550nm (green) enabled simultaneous detection of collagen fibers and cells within native heart tissue. (B) Following decellularization, cell autofluorescence is not apparent, and small regions of residual autofluorescence may be associated with remaining ECM proteins. Images were taken 50μm from the tissue surface with 800nm excitation light. Scale bar represents 50μm.
(A) Representative images show live (green) and dead (red) staining on PLL controls and ECM. Individual small, rounded cells were dead; cell clusters contained mixtures of live and dead cells. (B) Image analysis reveals significantly greater viability with FBS; there were no differences among the serum-free conditions. # = significant difference vs. all other conditions.


