Abstract
Urothelial bladder cancer (UBC) is heterogeneous at the clinical, pathological, and genetic levels. Tumor invasiveness (T) and grade (G) are the main factors associated with outcome and determine patient management (1). A discovery exome sequencing screen (n=17), followed by a prevalence screen (n=60), identified new genes mutated in this tumor coding for proteins involved in chromatin modification (MLL2, ASXL2, BPTF), cell division (STAG2, SMC1A, SMC1B), and DNA repair (ATM, ERCC2, FANCA). STAG2, a subunit of cohesin, was significantly and commonly mutated/lost in UBC, mainly in tumors of low stage/grade, and its loss was associated with improved outcome. Loss of expression was often observed in chromosomally-stable tumors and STAG2 knockdown in bladder cancer cells did not increase aneuploidy. STAG2 reintroduction in non-expressing cells led to reduced colony formation. Our findings indicate that STAG2 is a novel UBC tumor suppressor acting through mechanisms that are different from its role to prevent aneuploidy.
Keywords: exome sequencing, bladder cancer, STAG2, cohesin, aneuploidy, tumorsuppressor
The most commonly mutated oncogene in UBC is FGFR3 (50–60%): mutations are more frequent in non-muscle invasive bladder cancers (NMIBC) with a low risk of progression (stage Ta low grade tumors), here designated as "non-aggressive" (Online Methods) (2, 3). PIK3CA mutations occur in 15–20% of tumors and tend to associate with FGFR3 mutations (4). p53 and RB pathway inactivation have been associated with NMIBC with a high risk of progression (stage Ta or T1 high grade tumors) and with muscle-invasive bladder cancer (MIBC) (here designated as "aggressive") (5, 6). RAS mutations are less common and are mutually exclusive with FGFR3 mutations (7). There is now extensive evidence indicating that NMIBC of high grade are genomically similar to MIBC (8, 9): non-aggressive UBC are genomically stable whereas aggressive UBC are unstable (2, 8–10). Recently, exome sequencing has identified chromatin remodeling as an important pathway involved in UBC (11); this study focused mainly on MIBC.
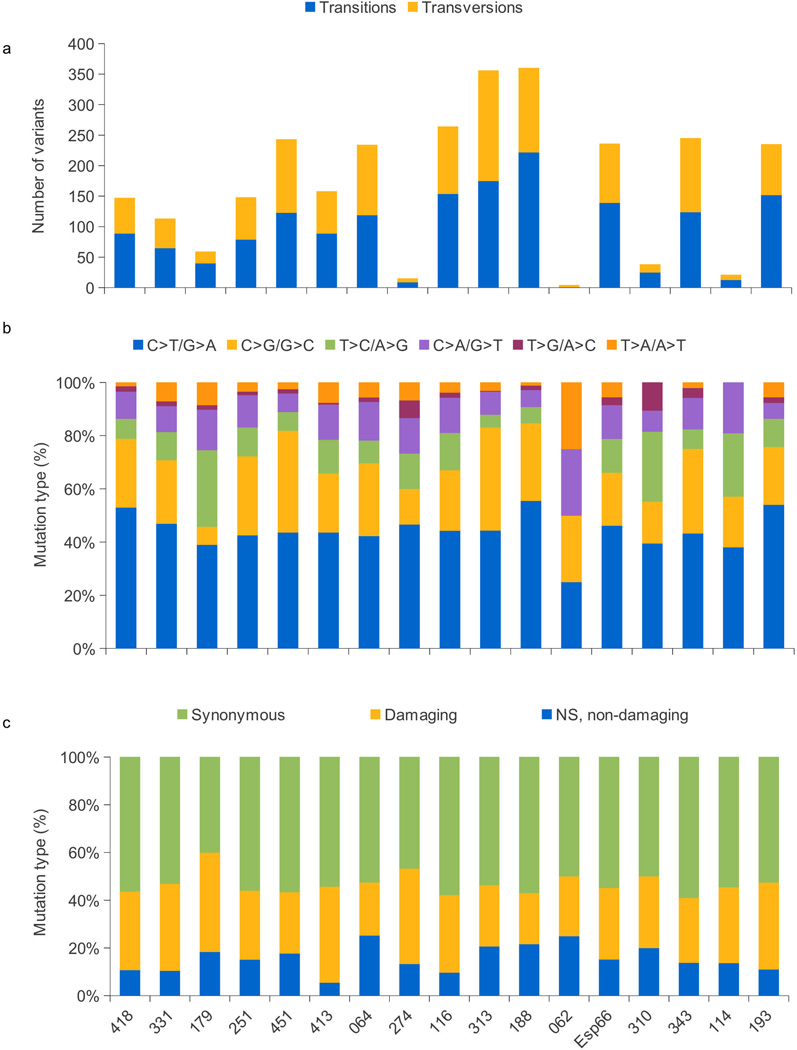
To discover new genes mutated in UBC, we sequenced the exome of 17 tumors of variable stage/grade and the corresponding normal leukocyte DNA; all neoplastic samples used had a tumor cellularity ≥70% (Supplementary Table 1). Because there are major initiatives on the sequencing of MIBC (i.e. the TCGA project), we have focused mainly on NMIBC. Metrics on enrichment and depth of coverage are shown in Supplementary Table 2: the mean coverage of tumor and leukocytes was 79±16 and 82±18, respectively. We identified 2927 somatic mutations, of which 1263 and 798 were predicted to be relevant (non-synonymous, NS) and damaging (predicted to have a functional effect) (Supplementary Table 3), respectively (Online Methods). The average number of somatic mutations per tumor was 169±114 with a wide interindividual variation (range 4–360) (Fig. 1a), a figure that falls in the mid-range of exome studies in solid tumors of the adult. C>T transitions were the most common nucleotide substitution (mean 44%), followed by C>G transversions (Fig. 1b). The ratio of non-synonymous:synonymous (NS:S) changes was <1 in 15/17 samples (Fig. 1c and Supplementary Fig. 1a). We compared the total number of single nucleotide variants (SNV), indels, transitions, transversions, S mutations, non-damaging NS mutations, and damaging NS mutations in "aggressive" vs. "non-aggressive" tumors; all variables were highly similar in both tumor groups. The same analysis was performed according to smoking status: the number of damaging mutations was higher in tumors from smokers vs. non-smokers but the differences did not reach statistical significance (P=0.09). The number of mutations in tumors from patients >60 years was also slightly, but non-significantly, higher than in younger patients (Supplementary Fig. 2). The ratio of NS:S mutations was similar regardless of aggressiveness and smoking status but it was slightly lower in patients diagnosed at age >60 (Supplementary Fig. 1b). It will be necessary to sequence more tumors to further investigate these relationships.
Figure 1.
Distribution of single nucleotide variants identified in the discovery screen through exome sequencing: total (a), according to type of nucleotide substitution (b), and according to the predicted effect (c).
We assessed the reliability of the exome analysis and somatic variant calling strategies using Sanger sequencing: we assayed 226 variants and verified 219, of which 214 were confirmed to be somatic (94.7%) (Supplementary Table 3). Table 1 shows the list of genes showing NS mutations in ≥3 tumors that are expressed in >30% of UBC based on Affymetrix expression analyses of an independent tumor sample series (n=43) covering the full spectrum of the disease. GO and KEGG pathway analyses identified chromatin modification, DNA repair and DNA damage response, apoptosis, and cell cycle among the most significant processes to which mutated genes were ascribed (Supplementary Table 4).
Table 1.
Genes frequently mutated in UBC assessed through exome sequencing or targeted Haloplex resequencing (n=77).
| DISCOVERY SCREEN | PREVALENCE SCREEN | DISCOVERY + PREVALENCE SCREEN**** | |||||
|---|---|---|---|---|---|---|---|
| GENE | Number of mutations (n=17) |
P-value* | Number of mutations (n=60) |
Number of mutations in all tumors (n=77) |
Number of "non- aggressive" mutant cases (n=29)** |
Number of "aggresssive" mutant cases (n=47)** |
P-value *** |
| ARID1A | 7 | 0.0001 | 3 | 10 | 3 | 7 | 0.732 |
| STAG2 | 3 | 0.019 | 9 | 12 | 6 | 5 | 0.315 |
| KDM6A | 4 | 0.019 | 6 | 10 | 3 | 7 | 0.732 |
| PDZD2 | 3 | 0.019 | 0 | 3 | 0 | 2 | 0.521 |
| MYCBP2 | 3 | 0.061 | 2 | 5 | 2 | 2 | 0.999 |
| LPHN3 | 3 | 0.096 | 0 | 3 | 0 | 2 | 0.521 |
| CREBBP | 2 | 0.098 | 9 | 11 | 4 | 7 | 1 |
| EP300 | 2 | 0.098 | 5 | 7 | 3 | 4 | 1 |
| ATM | 3 | 0.138 | 6 | 9 | 4 | 4 | 0.702 |
| TP53 | 3 | 0.2117 | 8 | 11 | 2 | 9 | 0.188 |
| RREB1 | 3 | 0.237 | 0 | 3 | 1 | 1 | 1 |
| PIK3CA | 6 | 0.239 | 4 | 10 | 5 | 4 | 0.289 |
| WHSC1L1 | 2 | 0.241 | 1 | 3 | 1 | 2 | 1 |
| MYO5B | 3 | 0.430 | 0 | 3 | 0 | 2 | 0.521 |
| MLL2 | 2 | 0.636 | 13 | 15 | 6 | 5 | 0.315 |
| FGFR3 | 2 | 0.659 | 12 | 14 | 10 | 4 | 0.011 |
| TEX15 | 3 | 0.778 | 0 | 3 | 0 | 2 | 0.521 |
| BRAF | 1 | NA | 6 | 7 | 2 | 5 | 0.701 |
| ERCC2 | 0 | NA | 8 | 8 | 5 | 1 | 0.040 |
| MAPK8IP3 | 1 | NA | 4 | 5 | 3 | 2 | 0.363 |
| MLL | 1 | NA | 5 | 6 | 3 | 2 | 0.363 |
| NUP93 | 1 | NA | 4 | 5 | 1 | 3 | 1 |
| STAG1 | 0 | NA | 5 | 5 | 0 | 3 | 0.282 |
| RB1 | 1 | NA | 3 | 4 | 1 | 3 | 1 |
| FANCA | 0 | NA | 4 | 4 | 0 | 3 | 0.282 |
| MLL3 | 1 | NA | 4 | 5 | 2 | 3 | 1 |
| NOTCH1 | 0 | NA | 4 | 4 | 0 | 3 | 0.282 |
| ASXL2 | 3 | NA | 1 | 4 | 1 | 1 | 1 |
P-value calculations based on the mutations identified in the discovery screen (see Online Methods).
One sample with a mutation in STAG2 is excluded from the "non-aggressive" vs. "aggressive" tumor comparison due to insufficient information for classification.
P-value refers to the frequency of mutant tumors with "non-aggressive" vs. "aggressive" features.
The discrepancies between numbers of mutations and numbers of mutant tumors result from the occurrence of ≥2 mutations in the same gene in a given tumor sample.
To extend our findings we performed a mutation prevalence screen (n=60) (Supplementary Table 5) using HaloPlexTM Target Enrichment System followed by sequencing. We included selected genes that were recurrently mutated in the discovery screen as well as additional genes from the pathways in which they participate (Supplementary Table 6). We identified 260 SNV: 200 were predicted to be relevant and 143 of them were predicted to be damaging. We analyzed 95 mutations identified by HaloPlex by Sanger sequencing; 73 were verified (76.13%) and 72 of these (98.6%) were confirmed to be somatic.
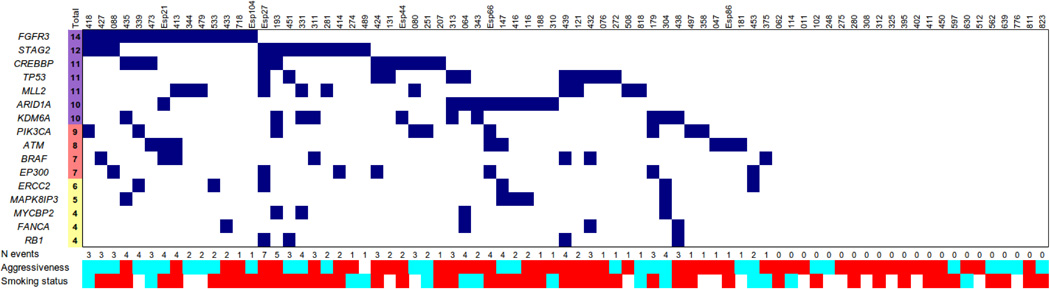
Table 1 and Fig. 2 show the joint distribution of mutations in the discovery and prevalence screens. Among the genes recurrently mutated, we identify novel genes involved in chromatin remodeling (MLL2, ASXL2, BPTF). BPTF binds H3K4me3 and has been found mutated in hepatocarcinoma (12) but little is known about its function in cancer. We show that BPTF knockdown led to a marked reduction in colony formation in 3 UBC lines tested (Supplementary Fig. 3), suggesting that it plays a role in cancer cell proliferation. We confirm recurrent mutations in ARID1A, KDM6A/UTX, CREBBP, EP300, MLL, and MLL3 (11), in agreement with recent reports implicating mutations in a wide range of chromatin remodelers in human cancer (12–15). Importantly, we identify recurrent, previously unreported, somatic mutations in genes involved in DNA repair (ATM, ERCC2, and FANCA, among others) and in the cohesin subunits STAG2, STAG1, SMC1A, and SMC1B, indicating that these pathways play an important role in UBC. FGFR3, TP53, PIK3CA, and RB1 are among the recurrently mutated genes, providing evidence of the representativeness of the tumors analyzed.
Figure 2.
Distribution of mutations in genes recurrently mutated in UBC that are expressed in >30% of tumors; joint analysis of the discovery and prevalence screens. In 22/77 tumors (28.6%), none of the genes listed in this Figure was found to be mutated.
We have focused on STAG2 because it is significantly mutated in our exomes (Table 1), one additional mutation was found in 9 published UBC exomes (11), and we identified 2 mutations among 21 MIBC exomes from the TCGA Consortium [overall damaging mutation rate, 13 % (6/47)]. Our prevalence screen identified 9 additional somatic mutations predicted to be damaging. Altogether, we have identified damaging somatic STAG2 mutations in 12/77 (15.6%) tumors (5 nonsense, 4 exon junction, 2 missense, and 1 indel) (Supplementary Fig. 4 and Supplementary Table 7) and 9/11 were verified by Sanger sequencing (Supplementary Fig. 5). Damaging mutations were found in both "non-aggressive" (6/29, 20.7%) and "aggressive" tumors (5/47, 10.6%). STAG2 inactivating mutations leading to loss of protein expression have recently been reported in non-epithelial tumors (16). STAG2 expression was low/undetectable in 6/7 (85%) UBC with damaging mutations and in 3/34 (9%) wild type tumors (P=0.0001) (Fig. 3, Supplementary Fig. 5 and Supplementary Table 7). Together with the exome significance analysis, these data indicate that STAG2 is a new gene commonly mutated in UBC.
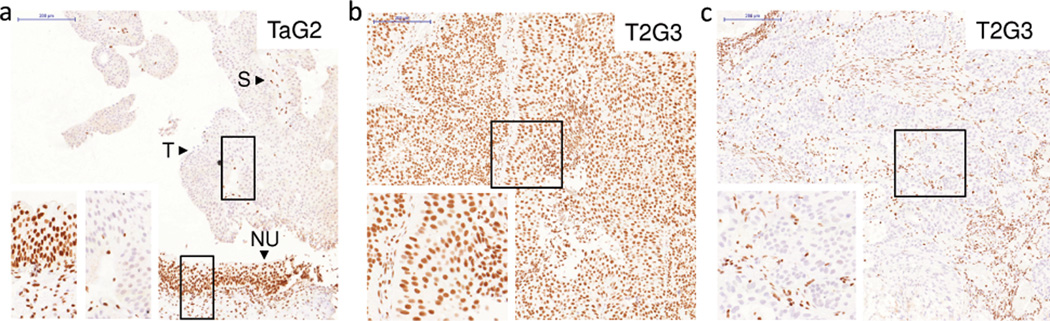
Figure 3.
Immunohistochemical analysis of STAG2 expression in bladder tumors of different stage and grade: 2 of the tumors show lack of STAG2 expression (a, c) and one tumor shows strong expression (b). Of note the strong STAG2 protein expression in normal urothelium of a patient with a STAG2-negative tumor (a) and in the stroma of all tumor samples. (T, tumor; S, stroma; NU, normal urothelium). Scale bar: 200 µm.
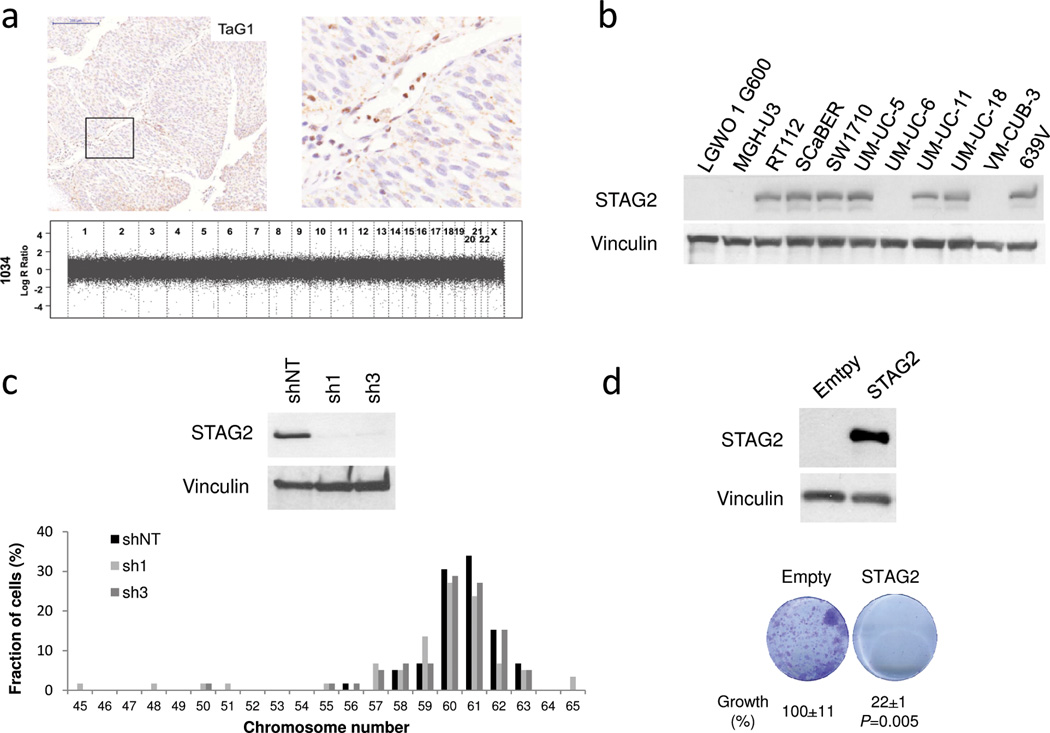
We then analyzed STAG2 expression in tissue microarrays of incident tumors representative of the disease spectrum (Supplementary Tables 8 and 9) (3, 17). STAG2 tumor loss, defined as a histoscore ≤50 with detectable stromal expression, was observed in 197/671 (29.3%) tumors (Fig. 3, Supplementary Fig. 5). STAG2 loss was significantly associated with multicentricity (P= 0.011), tumor size (P= 0.002), low stage (P= 5.7×10−15), and low grade (P= 1.96×10−15) (Supplementary Table 10). Abnormal STAG2 expression patterns included focal losses within otherwise positive tumors and a predominant cytoplasmic distribution of the protein (Supplementary Fig. 6). Because "non-aggressive" tumors are more differentiated, lack of STAG2 might reflect urothelial cell maturation. Arguing against this possibility, STAG2 expression was detected in all cell layers of normal urothelium (Supplementary Fig. 7). We also analyzed whether mechanisms other than mutation might account for loss of STAG2 expression. Using SNP arrays, we found STAG2 losses in 1 of 18 (5%) TaG1/G2, STAG2-negative, tumors from male patients. Similar findings have been reported in leukemia (16, 18–20) (Supplementary Fig. 8).
STAG2 encodes a subunit of cohesin, a complex that mediates sister chromatid cohesion to ensure accurate chromosome segregation and DNA repair. Cohesin also regulates gene expression through mechanisms involving DNA looping and interactions with transcriptional regulators such as Mediator and CTCF (21, 22). Somatic human cells contain two versions of this complex consisting of SMC1A, SMC3, RAD21, and either STAG2 or STAG1 (21). While there is still an incomplete understanding of the functional redundancy of both complexes, differential roles in centromeric vs. telomeric cohesion have been proposed, as well a preferential implication of STAG1 in transcriptional control (23, 24). Using immunohistochemistry, we show that STAG1 is expressed in normal urothelium and in the majority of UBC, including most tumors lacking STAG2 (Supplementary Fig. 7 and Supplementary Fig. 9). Interestingly, all 6 tumors that lost both STAG1 and STAG2 were of high grade and FGFR3 wild type, suggesting partial functional compensation in the maintenance of an integral chromosome segregation machinery. The more frequent loss of STAG2 in tumors (Supplementary Fig. 9a) may reflect that only one hit is required for its inactivation, given its location on the X chromosome.
Recently, STAG2 mutations in glioblastoma, melanoma, and Ewing sarcoma have been proposed to participate in tumor development by promoting aneuploidy (16). This hypothesis is at odds with our finding that loss of STAG2 expression occurs mainly in "non-aggressive" UBC that are genomically stable. To address this issue, we analyzed chromosome number changes in a panel of 23 TaG1/TaG2 tumors using high resolution SNP or BAC arrays. Of 11 tumors without STAG2 expression, 9 lacked aneuploidy and 2 showed loss of one copy of chromosome 9; similarly, 9/12 tumors expressing STAG2 showed normal chromosomal content (Fig. 4a, Supplementary Fig. 10, and Supplementary Table 11). Consistent with these findings, mutations in STAG2 and other cohesin genes have recently been reported not to be associated with aneuploidy in acute myeloid leukemia (18–20). We next knocked down STAG2 in 3 UBC lines displaying a broad range of phenotypes. As shown in Fig. 4b-c, Supplementary Fig. 11, and Supplementary Table 12, efficient knockdown was achieved in the 3 lines but there were no consistent effects on chromosome number/metaphase, unlike previously reported in HCT116 cells (16). This discrepancy may reflect that different cell types show variable tolerance to aneuploidy. We also introduced STAG2 cDNA in 3 cell lines lacking protein expression: UM-UC-6 cells harbor a R305X stop gain mutation (exon 11) and a F1228L mutation (exon 33), VM-CUB-3 harbour a 10 bp deletion in exon 6, and LGW0 1 G600 have a wild type sequence in exons 3–35. In the 3 cell lines, we observed a significant decrease in colony formation upon STAG2 lentiviral expression (Fig. 4d and Supplementary Fig. 12). Intringuingly, STAG2 knockdown was also associated with reduced colony formation in 5 different cell lines (Supplementary Fig. 13). Similar effects have been reported upon of knockdown of the tumor suppressor ARID1A in pancreatic and bladder cancer cells (25, 26).
Figure 4.
STAG2 loss is not associated with aneuploidy in primary tumors and in 639V bladder cancer cells. Effects of STAG2 reconstitution on cell growth. (a) SNP array genomic plots show lack of chromosomal changes (aneuploidy) in a tumor lacking STAG2 expression (note strong STAG2 labeling of normal stroma). Scale bar: 200 µm. (b) Western blotting analysis of STAG2 in UBC lines shows undetectable expression in 4 of 11 lines used for functional studies. (c) STAG2 overexpression in UM-UC-6 cells leads to reduced colony formation efficiency. (d) Efficient STAG2 knockdown in 639V cells demonstrated by western blotting does not lead to consistent changes in chromosome number (quantification shown in Supplementary Table 12).
To place these findings in the context of the known pathways of UBC progression, we assessed the association of STAG2 alterations with FGFR3 mutation/overexpression, p53 nuclear accumulation, and Ki67 expression (Supplementary Tables 13–16) (2). In NMIBC, loss of STAG2 was significantly more common among tumors with mutant FGFR3 (42.7% vs. 27.2%, P=0.001), tumors lacking p53 overexpression (P=0.002), and those with a low Ki67 index (P=0.002). These results indicate that loss of STAG2 is associated with less aggressive tumors. Within the low-risk NMIBC subgroup, STAG2 loss was associated with FGFR3 mutant status (P=0.059) and with low p53 expression (P=0.011). Among patients with high-risk NMIBC, STAG2 loss was associated with high FGFR3 expression (P=0.037), FGFR3 mutation (P=0.12), and low Ki67 index (P=0.049). Among both high-risk NMIBC and MIBC, there was no association with p53 immunohistochemical expression (Supplementary Tables 13–16).
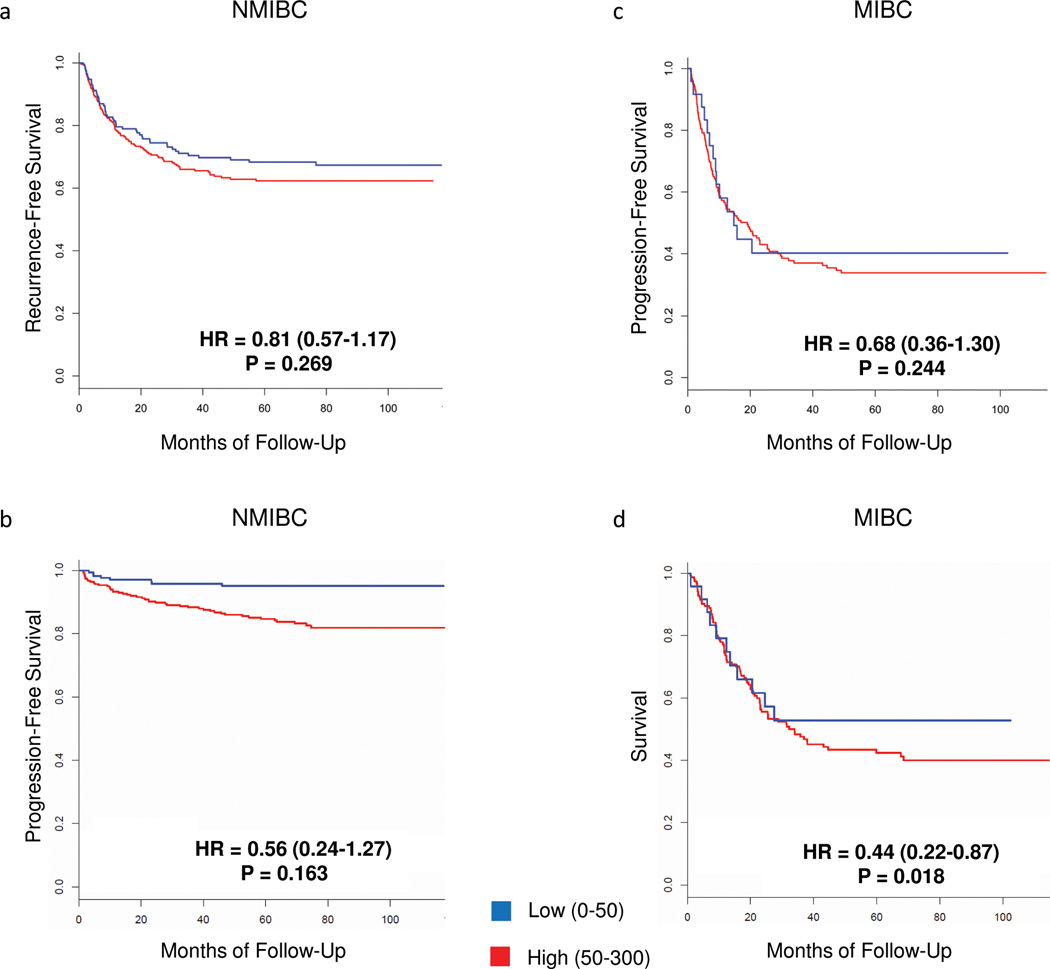
We then analyzed the association of STAG2 loss with recurrence and progression among patients with NMIBC, and progression and mortality in MIBC. We applied both Kaplan-Meier curves and multivariable Cox regression analyses. The large sample size of our study allowed performing a more informative stratified analysis. Loss of STAG2 expression was associated with a lower risk of tumor recurrence and progression among patients with NMIBC (Fig. 5a, b). However, in multivariable analyses, STAG2 expression was not an independent predictor of recurrence or progression after adjusting for T, G, and FGFR3 mutations (Supplementary Tables 17 and 18) since these parameters were highly correlated. Among patients with MIBC, STAG2 loss was associated with a lower risk of progression (HR=0.68, P=0.244) and it was an independent predictor of survival in the multivariable analysis (HR=0.44, P=0.018) (Fig. 5c, d) (Supplementary Tables 19 and 20). Therefore, we conclude that STAG2 loss is associated with better prognosis in patients with both NMIBC and in MIBC; additional studies are required to determine its clinical value.
Figure 5.
Kaplan-Meier plots of the association of STAG2 expression with outcome in patients with UBC. (a) Recurrence in patients with STAG2-high (n=309) vs. STAG2-low (n=171) NMIBC. (b) Progression in patients with STAG2-high (n=309) vs. STAG2-low (n=171) NMIBC. (c) Progression in patients with STAG2-high (n=158) vs. STAG2-low (n=24) MIBC. (d) Cancer-specific survival of patients with STAG2-high (n=158) vs. STAG2-low (n=24) MIBC. P-values correspond to the results of the multivariable analysis. Details on results and variables used for adjustment are shown in Supplementary Tables 17–20.
In summary, we find that both previously reported and newly identified genes coding for proteins involved in chromatin modification are recurrently mutated in UBC. In addition, we identify mutations in genes involved in cell cycle, DNA repair, and regulation of apoptosis. The frequent alteration of genes in these pathways may provide opportunities for novel therapies, including those based on synthetic lethality. STAG2 is significantly mutated in UBC; mutations and loss of expression are common, particularly among tumors of low stage and grade, and are associated with patient outcome. In "non-aggressive" tumors, STAG2 alterations occur in the absence of chromosomal instability. Our findings strongly suggest that STAG2 is a novel tumor suppressor in UBC through mechanisms that are different from its role in cohesion to prevent aneuploidy.
Online Methods
Patients and samples
Patients and samples came from the Epicuro/Spanish Bladder Cancer Study (SBCS) (3, 27) and from the Integrated Study of Bladder Cancer (ISBLAC) (Supplementary Tables 1 and 5). STAG2 expression was analyzed using tissue microarrays containing tumors from the Epicuro/SBCS, including patients with newly diagnosed UBC. Staging, grading, and follow-up were performed as described (3, 27). Expert pathologists reviewed diagnostic slides from all tumor blocks. We categorized TaG1 and TaG2 tumors as "low-risk NMIBC" or "non-aggressive"; TaG3, T1G2, and T1G3 tumors were categorized as "high-risk NMIBC"; ≥T2 tumors were MIBC. The latter two groups were pooled as "aggressive" tumors. Supplementary Tables 8 and 9 summarize patient characteristics. Among patients with NMIBC, recurrence was defined as the reappearance of a NMIBC following a negative follow-up medical evaluation. Progression was defined as transition from NMIBC to MIBC or the development of new local or metastatic tumors after primary treatment for patients with MIBC. Median follow-up was 62.6 months (range 1–98). All deaths were recorded but only UBC-related deaths were considered for survival analysis. Cases dying from other causes were censored at the time of death. Survival was computed as the period comprised between diagnosis and death or last control. All patients provided written informed consent (3). The Ethics Committees of all participating institutions approved both studies.
Exome sequencing, targeted resequencing, bioinformatic analyses, and mutation verification
The Agilent SureSelect Human All Exon plus v3 50MB (samples 114, 116, 193, 251, 310, 331, 413, 418, and Esp66) or v4 51MB (samples 062, 064, 179, 188, 274, 313, 343, and 451) were used for library preparation and enrichment. Libraries were applied to an Illumina flowcell; sequencing was performed on HiSeq2000 instruments using paired-end 75 bp reads.
Base calling and quality control were performed on the Illumina RTA analysis pipeline. Sequence reads were trimmed until the first base with a quality >10 and mapped to Human genome build hg19 (GRCh37) using GEM, allowing ≤4 mismatches. Reads not mapped by GEM (~4%) were submitted to a last round of mapping with BFAST. Results were merged; only uniquely mapping non-duplicate read pairs were used. SAM tools suite version 0.1.18 with default settings was used to call SNVs and short indels. Variants on regions with low mappability, read depth <10, tail distance bias P<0.05, or strand bias P<0.001 were filtered out. Somatic mutations were called by comparing tumor and blood exomes; Fisher's exact test was performed using variant supporting read counts. Only variants with Fisher test P-value <0.0001 were considered.
All SNV in exon junctions or leading to a non-synonymous change were considered "relevant". SNV leading to an amino acid substitution were evaluated using MutationAssessor (28) and SIFT (29) to predict their damaging effect; both scores were normalized into the 0–1 range. The P-values from SIFT were subtracted from 1. MutationAssessor predictions were scored as follows: high damage risk was assigned 1, medium was assigned 0.7, and low was assigned 0.5. When both predictions were available, scores were averaged; if one prediction was missing, the other score was used. Variants with a final score >0.8 were considered as "damaging". Stop gains and frameshifts were considered "damaging" if they ablated >30% of the sequence or some protein domain annotated in InterPro. Variants close to an exon boundary were considered "relevant" and "damaging" if the distance from the exon junction was 8 bases into the intron or 2 bases into the exon of donor junctions, or 8 bases into the intron, or 3 bases into the exon of acceptor junctions. The scores from both methods were used as input to calculate P-value of the associated genes. We used the Oncodrive-fm approach (30) combining recurrence and functional impact (Table 1).
Statistical analyses within R Software (2.15.1) were performed on stage, smoking status, and age groups using Mann-Whitney U test (Fig. 1). Fisher exact test (non-aggressive vs. aggressive) was assessed in recurrent genes (Table 1); P-values <0.05 were considered statistically significant.
Pathway analysis was performed as reported (31). First, a gene list was selected. We processed different combinations of three lists: (A) all relevant genes (those with mutations leading to non-synonymous substitutions or affecting exon junctions); (B) damaged genes (those with mutations predicted to be damaging), and (C) recurrent genes (those with relevant mutations in ≥2 samples). The resulting lists were examined for enrichment in terms from Gene Ontology (biological process) and Kegg pathways. For the latter, pathways associated to diseases were filtered out, as reported (32). Enrichment analysis was based on a hypergeometric test. P-values were adjusted using Benjamini-Hochberg’s false discovery rate (FDR); only FDR<0.1 were considered. A correction for genes in overlapping clusters was applied.
For targeted resequencing, the HaloPlexTM Target Enrichment System (Agilent) was used in an independent tumor series (n=60) following manufacturer's instructions. Five cases from the discovery screen were included for targeted resequencing. A library of genomic DNA fragments was created by digestion with eight restriction reactions and hybridized with probes against target regions incorporating Illumina paired-end sequencing motifs and index sequences. DNA was captured, PCR amplified with KAPA HiFi HotStart polymerase or Herculase II Fusion Enzyme, and products were purified using AMPure XP beads. Amplicons were sequenced in multiplex using Illumina protocols. Adapters and primers were removed from both ends of the reads with FAR (http://sourceforge.net/projects/theflexibleadap/). Trimmed reads were mapped; SNPs and indels were called as described above, without excluding duplicates and filtering by tail distance bias P<0.05 or with strand bias P<0.001. Variants out of the regions selected for enrichment, with low mappability, read depth <10, occurring in >1% of the reads in blood, or annotated in 1000 Genomes project as SNP (release 20110521) were filtered out. Variants with Fisher test P-value for somatic comparison <0.0001 were considered. Somatic mutations were called by comparing tumor and blood. Damaging and effect annotations were performed as described for exomes.
Mutations were verified by Sanger sequencing. Using this bioinformatics pipeline we verified 94.7% of the SNV called as somatic mutations by exome sequencing.
STAG2 sequencing of UBC lines
Cell lines were authenticated by gene mutation analyses; all cultures were Mycoplasma-free. Exons 3–7, 11–31 and 33–35 were sequenced from overlapping amplicons generated from cDNA; exons 8–10 were sequenced from genomic DNA. The predominant transcript lacks exon 32 (Ensemble variant STAG2-0001) and codes for a 1231 residue protein.
Immunohistochemistry
STAG2 was detected using clone J-12 (Santa Cruz Biotechnology, sc-81852) and affinity-purified rabbit polyclonal antibodies raised against a synthetic peptide (DPASIMDESVLGVSMF) (23). Both antibodies yielded concordant results in 92% of tumors. To detect STAG1, we used affinity-purified rabbit polyclonal antibodies raised against a synthetic peptide (EDDSGFGMPMF) (23). Antibodies D5/16B4 and Ks20.8 detecting KRT5/6 and KRT20, respectively, were from Dako. Antigen retrieval and reactions were performed as described (27, 33). A histoscore was calculated as the product of staining intensity (0–3) and percentage of positive cells (0–100%). Unsupervised clustering analysis was performed using scores and the heatmap.2 function of the gplots package within the R 2.15.1 statistical environment.
Gene copy number analyses
Copy number changes were analyzed using manually microdissected fresh tissue samples containing ≥60% tumor cells (n=55). DNA was hybridized to Illumina HumanHap 1M BeadChip SNP arrays; 20 tumors were TaG1/G2. Copy number changes were called as described (34). An additional 76 samples were analyzed using Human 2.0 BAC arrays (UCSF Cancer Center) (35, 36).
STAG2 functional assays
To knockdown STAG2, control or STAG2-targeting lentiviral particles were produced in HEK-293T using Sigma Mission plasmids. Viral supernatants were used to infect RT112, UM-UC-5, 639V, SW1710, and UM-UC-11 cells; after 3 rounds of infection cells were selected for 48h in medium containing puromycin (2 µg/ml). To overexpress STAG2, the human cDNA (b isoform, 1231 residues) was amplified by PCR (Addgene pEGFP-STAG2 plasmid, ref. 31972) and subcloned into pLVX-puro lentiviral vector. After 3 rounds of infection, cells were selected for 48h with puromycin (2 µg/ml). Western blotting was performed as described (27).
For colony formation assays, 8×103 puromycin-selected cells were seeded; 7 days later, cells were methanol-fixed and crystal violet-stained; after elution (10% acetic acid), 680 nm absorbance was measured.
For chromosome analyses, puromycin-selected knockdown cells were arrested with colcemid (0.1 mg/ml) for 6h, harvested, swollen in 75mM KCl for 15 (RT112), 25 (639V) or 30 (UM-UC-11) min at 37 °C, and fixed. Metaphases were captured and chromosomes/metaphase were counted (Axioplan II Imaging MetaSystem Microsoft and Ikaros software, Metasystems, Altlussheim, Germany). Chromosome number was compared using the Wilcoxon rank sum test.
Other statistical analyses
Categorical data were reported as numbers and percentages. Associations between STAG2 and patient characteristics were assessed using the chi-square test. Associations between markers were evaluated using the chi-square test and the odds ratio (OR) and 95% confidence interval (95%CI) as a measure of association between categorical variables.
Outcomes considered were recurrence-free and progression-free survival (NMIBC) and progression-free and cancer-specific mortality (MIBC). Survival was represented using Kaplan-Meier curves; the differences between curves were assessed with the log-rank test. Cox proportional hazards models were applied for multivariable analysis. The adjusting factors used are indicated in the Supplementary Tables. Statistical significance was considered at 0.05. R Software (version 2.14, available at http://www.r-project.org/) was used for statistical analysis.
Supplementary Material
Acknowledgements
This work was supported, in part, by grants from Ministerio de Economia y Competitividad, Madrid (grants Consolíder ONCOBIO, Consolider INESGEN, SAF-2010-21517, and SAF2011-15934-E), Instituto de Salud Carlos III [grants G03/174, 00/0745, PI051436, PI061614, G03/174, PI080440, PI120425, and Red Temática de Investigación Cooperativa en Cáncer (RTICC)]; Asociación Española Contra el Cáncer, EU-FP7-201663, and NIH RO-1 (CA089715). C.B.M. is recipient of a La Caixa International PhD Fellowship. E. L. is supported by a grant from Fundación Banco Santander Postdoctoral Programme. We thank F. Algaba, Y. Allory, A Cuadrado, C. González, E. López, P. Lapunzina, T. Lobato, M. Malumbres, S. Remeseiro, V. J. Sánchez-Arévalo, F. Waldman, and the CNIO core facilities for valuable contributions. We also thank The Cancer Genome Atlas investigators for providing unpublished information for analysis.
Footnotes
The authors declare no competing financial interests.
Data Access
Sequencing data have been deposited at the European Genome-phenome archive. SNP array data have been deposited at the Gene Expression Omnibus.
Author contribution
CBM, EL, and LR designed and performed in vitro functional studies; CBM performed immunohistochemical analysis of tumor samples; AS and CBM designed and performed mutation validation analyses; AS designed and prepared Haloplex libraries for sequencing; ECdSP, MV, FCG, SB, and DGP processed and analyzed exome sequencing and targeted resequencing data; JE, ELK, DR, and SC performed gene copy number analyses of tumors; MM coordinated patient and sample data management; AC, MK, JAL, and AT contributed to patient recruitment and data collection; JH performed statistical analyses; XL provided technical support with patient samples; MB and MG coordinated library preparation and sequencing; OD contributed to library preparation and sequencing; JCC and RNS contributed to the in vitro analysis of the effects of STAG2 knockdown on aneuploidy; NJ and JL performed pathological review of samples; IG coordinated exome sequencing and targeted resequencing; IG, SH, and AV supervised bioinformatics analyses; AL provided scientific insight and contributed with reagents; NM coordinated patient recruitment, and collection of clinical and pathological data, and supervised clinical-pathological-molecular association and outcome analyses; FXR and NM conceived the study; FXR supervised the overall conduct of the study. FXR wrote the paper with NM; CBM, AS, ECdSP, AV, AL, and IG contributed to manuscript writing.
Editorial Summary: Francisco X. Real and colleagues report exome sequencing in urothelial bladder tumors. They show that STAG2, a subunit of the cohesin complex, is recurrently mutated and they provide evidence that STAG2 loss does not lead to increases in aneuploidy.
References
- 1.Sylvester RJ. Natural history, recurrence, and progression in superficial bladder cancer. ScientificWorldJournal. 2006;6:2617–2625. doi: 10.1100/tsw.2006.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luis NM, Lopez-Knowles E, Real FX. Molecular biology of bladder cancer. Clin. Transl. Oncol. 2007;9:5–12. doi: 10.1007/s12094-007-0003-x. [DOI] [PubMed] [Google Scholar]
- 3.Hernández S, et al. FGFR3 mutations as a prognostic factor in non-muscle invasive urothelial bladder carcinomas: results of a prospective study. J. Clin. Oncol. 2006;24:3664–3671. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 4.López-Knowles E, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66:7401–7404. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- 5.Real FX. p53: it has it all, but will it make to the clinic as a marker in bladder cancer? J. Clin. Oncol. 2007;25:5341–5344. doi: 10.1200/JCO.2007.13.1904. [DOI] [PubMed] [Google Scholar]
- 6.López-Knowles E, et al. The p53 pathway and outcome among patients with T1G3 bladder tumors. Clin. Cancer Res. 2006;12:6029–6036. doi: 10.1158/1078-0432.CCR-06-0206. [DOI] [PubMed] [Google Scholar]
- 7.Jebar AH, et al. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–5225. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 8.Lindgren D, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 9.Hoglund M. The bladder cancer genome; chromosomal changes as prognostic makers, opportunities, and obstacles. Urol. Oncol. 2012;30:533–540. doi: 10.1016/j.urolonc.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Blaveri E, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin. Cancer Res. 2005;11:7012–7022. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
- 11.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 13.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson BG, Roberts CW. SWI/SNF nucleosome remodelers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 16.Solomon DA, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amaral AFS, et al. Plasma 25-hydroxyvitamin D3 levels and bladder cancer risk according to tumor stage and FGFR3 status: a mechanism-based epidemiological study. J. Natl. Cancer Inst. 2012;104:1897–1904. doi: 10.1093/jnci/djs444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter MJ, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc. Natl. Acad. Sci. USA. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter MJ, et al. Clonal architecture of secondary acute myeloid leukemia. N. Eng. J. Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 22.Remeseiro S, Losada A. Cohesin, a chromatin engagement ring. Curr. Opin. Cell Biol. 2013;25:63–71. doi: 10.1016/j.ceb.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Remeseiro S, et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remeseiro S, et al. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012;31:2090–2102. doi: 10.1038/emboj.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shain AH, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2012;109:E252–E259. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balbás-Martínez C, et al. ARID1A alterations are associated with FGFR3 wild type, poor-prognosis, urothelial bladder tumors. PLoS ONE. 2013;8:e62483. doi: 10.1371/journal.pone.0062483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaral AFS, et al. Plasma 25-hydroxyvitamin D3 levels and bladder cancer risk according to tumor stage and FGFR3 status: a mechanism-based epidemiological study. J. Natl. Cancer Inst. 2012;104:1897–1904. doi: 10.1093/jnci/djs444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reva B, et al. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P, et al. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez A, et al. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012;40:e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez M, et al. Chapter 14: cancer genome analysis. PLoS Comput. Biol. 2012;8:e1002824. doi: 10.1371/journal.pcbi.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 33.López-Knowles E, et al. The p53 pathway and outcome among patients with T1G3 bladder tumors. Clin. Cancer Res. 2006;12:6029–6036. doi: 10.1158/1078-0432.CCR-06-0206. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez-Santiago B, et al. Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am. J. Hum. Genet. 2010;87:129–138. doi: 10.1016/j.ajhg.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snijders AM, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat. Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 36.Blaveri E, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin. Cancer Res. 2005;11:7012–7022. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.