Summary
Assembly of 30S ribosomal subunits from their protein and RNA components requires extensive refolding of the 16S rRNA and is assisted by 10–20 assembly factors in bacteria. We probed the structures of 30S assembly intermediates in E. coli cells, using a synchrotron X-ray beam to generate hydroxyl radical in the cytoplasm. Widespread differences between mature and pre-30S complexes in the absence of assembly factors RbfA and RimM revealed global reorganization of RNA-protein interactions prior to maturation of the 16S rRNA and showed how RimM reduces misfolding of the 16S 3′ domain during transcription in vivo. Quantitative 14N/15N mass spectrometry of affinity-purified pre-30S complexes confirmed the absence of tertiary assembly proteins and showed that N-terminal acetylation of proteins S18 and S5 correlates with correct folding of the platform and central pseudoknot. Our results indicate that cellular factors delay specific RNA folding steps to ensure the quality of assembly.
Keywords: hydroxyl radical footprinting, ribosome biogenesis, ribosomal protein modification, RNA folding, ESI mass spectrometry
Introduction
To safeguard the fidelity of protein synthesis, cells must ensure accurate ribosome biogenesis even under sub-optimal conditions. Though the RNA and protein components of the ribosome are sufficient for the self-assembly of its subunits in vitro, extra assembly factors must chaperone assembly in the cell. Here, we show that two such factors in E. coli change the folding pathway of the pre-rRNA by directly probing the structures of assembly intermediates in situ.
Nomura and co-workers showed by in vitro reconstitution that the 20 ribosomal proteins (r-proteins) that form the small (30S) subunit join the complex in a specific order. Primary r-proteins bind directly to the 16S rRNA, followed by secondary and tertiary assembly proteins (Held et al., 1973; Held et al., 1974). Structure probing showed that the hierarchy of protein addition arises from structural changes in the rRNA induced by binding of primary assembly proteins (Stern et al., 1989). In vitro, the modular 5′ (body), central (platform) and 3′ (head) domains of the 16S rRNA can assemble concurrently, leading to parallel assembly pathways populated with diverse intermediates that depend on the stabilities of the local rRNA and RNA-protein interactions (Adilakshmi et al., 2008; Mulder et al., 2010; Talkington et al., 2005).
In the cell, however, the rRNA folds and binds r-proteins as transcription proceeds (Chen and Williamson, 2013; Powers et al., 1993). Some rRNA nucleotides must wait several seconds for their base-pairing partners to be transcribed, increasing the potential for forming difficult-to-resolve non-native interactions. Moreover, the 5′ leader of the pre-rRNA, which is processed by a series of nucleases at a late stage of assembly (Deutscher, 2009), may alter the preferred folding pathway of the pre-rRNA (Balzer and Wagner, 1998; Besancon and Wagner, 1999). Finally, 30S ribosome biogenesis in E. coli is facilitated by 10–20 assembly factors that include modification enzymes, the Era and RsgA (yjeQ) GTPases, and chaperones such as RimM and RbfA that preferentially bind immature 30S subunits (Wilson and Nierhaus, 2007). These factors are thought to facilitate or proofread folding of the pre-rRNA and pre-30S assembly (Culver, 2003).
Here, we use X-ray-dependent hydroxyl radical footprinting to directly visualize the structures of pre-30S complexes in E. coli cells. Excitation of intracellular water by X-rays produces hydroxyl radical and aqueous electrons, which react with the solvent accessible surfaces of macromolecular assemblies (Ottinger and Tullius, 2000; Xu and Chance, 2007). The extent of RNA cleavage by hydroxyl radical reports RNA tertiary structure and RNA-protein interactions with nucleotide resolution (Tullius and Greenbaum, 2005), revealing structural details not visible by electron microscopy or crosslinking. Irradiation of whole cells produces cleavage patterns comparable to those obtained in vitro (Adilakshmi et al., 2006; Hayes et al., 1990), and avoids purification of fragile assembly intermediates. Quantitative mass spectrometry of pre-30S complexes showed that perturbations to the pre-rRNA structure correlated with differences in r-protein binding and modification.
We characterized pre-30S ribosomes that accumulate at low temperature in E. coli strains lacking the assembly factors RimM or RbfA, which act on different regions of the 30S subunit. RimM, a 21 kDa PRC-barrel domain protein widely conserved throughout bacteria, facilitates assembly of the 30S head or 16S 3′ domain (Bylund et al., 1998). Recent cryo-EM reconstructions of complexes from ΔrimM cells reveal particles lacking part or all of the head (Guo et al., 2013). Mutations in protein S19 or in 16S helices 31 and 33b suppress a rimM mutation (Lovgren et al., 2004), and RimM accelerates association of S19 with 30S complexes in vitro (Bunner et al., 2010).
RbfA, a 15 kDa type-II KH-domain protein (Huang et al., 2003), was identified as a high-copy suppressor of the cold-sensitive mutation C23U in 16S helix 1 (Dammel and Noller, 1995) and is thought to aid folding of 16S helix 1 and the central pseudoknot (helix 2). RbfA binds the neck of the 30S subunit, near the 5′ end of the mature 16S, where it blocks the tRNA binding sites and displaces the 3′ minor domain helix 44 (Datta et al., 2007).
Our footprinting and mass spectrometry results reveal pervasive structural differences between mature 30S ribosomes and the pre-30S complexes that accumulate in the absence of RimM or RbfA. Many differences map to the 3′ domain or “head” of the 30S ribosome, in agreement with recent cryo-EM studies of pre-30S particles (Jomaa et al., 2011, Guo et al., 2013). The details of RNA and protein interactions in these pre-30S complexes reported here suggest how RimM and RbfA facilitate co-transcriptional folding of the pre-rRNA in vivo by delaying certain interactions between RNA domains. The results additionally show that many RNA and protein interactions throughout the complex are remodeled at a late stage of assembly.
Results
ΔrimM and ΔrbfA strains accumulate pre-30S
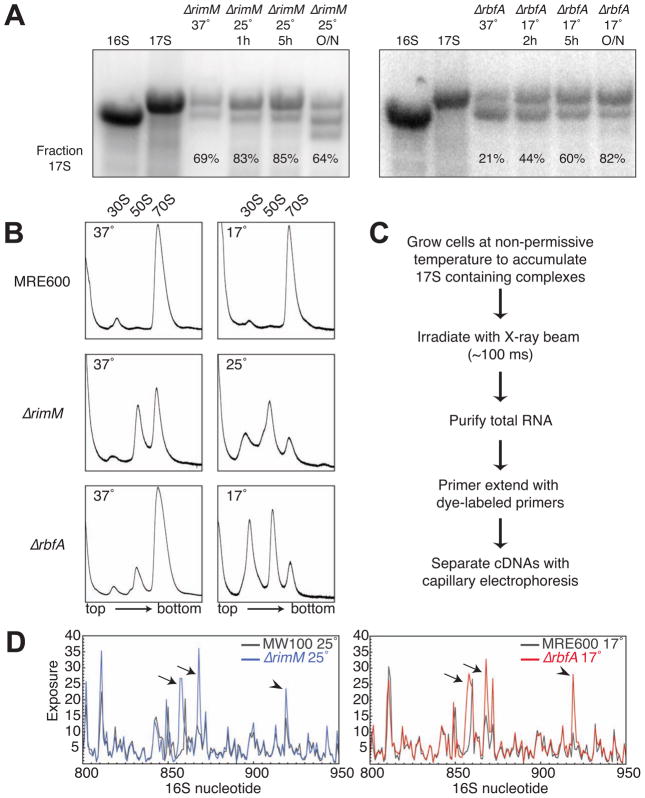
To probe the structure of pre-30S complexes in E. coli cells, we looked for growth conditions in which at least 80% of small ribosomal subunits contained 17S pre-rRNA, so that pre-rRNA signals dominate the footprinting signal. E. coli ΔrimM and ΔrbfA strains were reported to accumulate high levels of 17S pre-rRNA when grown at cold temperatures (Bylund et al., 1998; Inoue et al., 2003). The ΔrimM mutant MW37 (Persson et al., 1995) had a severe 16S processing defect, yielding 69% 17S rRNA at 37°C and 83% 17S after an hour at 25°C (Figure 1A, left panel). For footprinting, we incubated ΔrimM cells at 25°C for three hours to maximize the amount of 17S pre-rRNA, while minimizing a 16S fragment. The ΔrbfA mutant BX41 (Xia et al., 2003) contained 21% 17S rRNA at 37 °C (Figure 1A, right panel), and 80% 17S rRNA when grown overnight at 17°C.
Figure 1. ΔrimM and ΔrbfA strains are cold sensitive and accumulate 17S rRNA.
(A) Northern blots showing accumulation of 17S rRNA in ΔrimM and ΔrbfA strains grown at non-permissive temperatures, compared to in vitro 16S and 17S transcripts. Probe anneals to the 16S 5′ domain. The short fragment in ΔrimM (MW37) is consistent in size with cleavage just before the 3′ minor domain.
(B) Lysates from cells grown at permissive or non-permissive temperatures were centrifuged on 10–40% analyticalsucrose gradients and ribosome profiles (A 254) were recorded. ΔrimM and ΔrbfA strains accumulate excess free subunits at low temperatures. The 50S shoulder may be due to deficiency in L19 (rplS) that is downstream of ΔrimM.
(C) Experimental workflow for X-ray-dependent in vivo footprinting of pre-30S complexes.
(D) Sample footprinting data for the 16S 800–950 nt region, comparing control and deletion strains. Integrated peak areas for each nucleotide (y-axis) represent the average of three replicates and correlate qualitatively with relative exposure of the rRNA backbone. Exposures of helix 26 base (arrows) and A919 adjacent to the central pseudoknot (arrowhead) are indicated. See Figure S1 for further data.
Sedimentation profiles of ribosomal subunits confirmed that accumulation of pre-rRNA at low temperature correlated with defects in 30S subunit assembly. While our wild-type comparison strain (MRE600) showed no accumulation of free subunits at 17°C (Figure 1B, top), the ΔrimM strain contained a preponderance of free 30S and 50S subunits when grown at 25 °C, indicating a 30S subunit deficiency (Figure 1B, middle). A similar but less severe accumulation of free subunits was observed in ΔrbfA cells at 17 °C (Figure 1B, bottom).
X-ray-dependent in vivo hydroxyl radical footprinting
We next probed the structures of the pre-30S complexes that accumulate in the absence of RbfA or RimM by X-ray-dependent hydroxyl radical footprinting as outlined in Figure 1C (Adilakshmi et al., 2006). Frozen cells (5 μL) grown under non-permissive conditions were exposed to a synchrotron X-ray beam for 100 ms to generate hydroxyl radical in the cell (Figure S1A). Total RNA was purified from irradiated cells and the cleavage pattern read out by reverse transcription using a set of dye-labeled primers that provided complete coverage of the 16S rRNA (Table S1).
The frequency of backbone cleavage at each position of the 16S rRNA was inferred from the cDNA analysis and integration of cleavage products (Figure 1D; see Figure S1B for further data). We previously found that in vivo footprinting of ribosomes in MRE600 cells produced a cleavage pattern similar to that from in vitro footprinting, apart from residues that interact with the 50S subunit and tRNA (Adilakshmi et al., 2006; Merryman et al., 1999). A few residues could not be analyzed because of pauses in reverse transcription caused by base modifications or stable secondary structure.
Footprinting results from the two mutant strains were compared to wild-type control strains grown under the same conditions. We used MRE600 as a reference for the ΔrbfA results because MRE600 is used for many biochemical studies, while MW37 was compared to its parent MW100. Many 16S residues were cleaved to a similar extent in parental and deletion strains, indicating the quality of the peak assignments. Where differences existed, residues were often exposed in the mutant strain relative to the control strain, consistent with a more open pre-30S structure as illustrated by the 800–950 nt region (Figure 1D, arrows). Many differences were observed in both mutant strains (Figure S1B) and were greater than differences between the parental strains apart from a few locations (Figure S1C). Though the footprinting results reflected the average structures of the complexes present in each strain, these discrete structural perturbations suggested that without RimM or RbfA assembly stalls at specific steps, and that our strategy was able to reveal the features of the most prevalent pre-30S complexes.
Structures of pre-30S complexes
To identify structural differences in pre-30S and mature 30S ribosomes, we calculated the ratios of nucleotide accessibilities in each mutant strain versus its wild-type control and clustered residues according to their change in solvent accessibility (Figure S2). We only considered nucleotides with ratios less than 0.4 or greater than 1.8, which are appreciably greater than error (Supplemental Procedures). We also eliminated 9 residues from the ΔrbfA data and 15 residues from the ΔrimM data that changed structure when the parental strain (MRE600 or MW100) was grown at the non-permissive temperature (Figure 1D and Figure S1D). A few residues in MW37 (ΔrimM) data, such as nt 919 and the tip of helix 12, were not annotated because they were already exposed in the MW100 parental strain (Figure S1C).
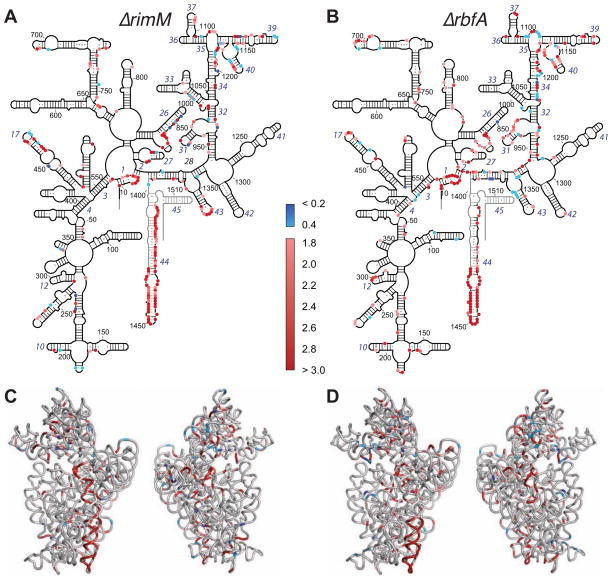
Pervasive differences between mature 30S ribosomes and pre-30S complexes indicated the extent to which the rRNA structure changes late in assembly. Nucleotides with altered solvent accessibility in the ΔrimM or ΔrbfA deletion strain were projected on the 16S secondary structure and the tertiary structure of the 16S rRNA in the 30S ribosome (Figure 2). The most perturbed regions were the 3′ major domain, that forms the 30S head, and helix 44, which lies along the subunit interface. This is consistent with a 5′ to 3′ order of assembly (Powers et al., 1993; Talkington et al., 2005), and the order of protein binding in the Nomura map and in vivo (Chen and Williamson, 2013). A subset of residues in the 16S 5′ domain (body) and central domain (platform) were also strongly exposed or protected in the absence of RimM or RbfA, indicating these regions also change structure late in assembly.
Figure 2. In vivo footprinting reveals widespread differences in the structures of mature and immature 16S rRNA.
(A) Map of altered rRNA solvent accessibility in ΔrimM on the 16S secondary structure. Residues with ≥ twofold change in cleavage in MW37 (ΔrimM) relative to MW100 were clustered (histogram, Figure S2A). Red, nucleotides exposed in ΔrimM pre-30S complexes (1.8 – 10X); blue, protected (0.4-2X); black, changed less than 0.4 – 1.8X; light gray, data not available. Helix numbers are in blue italics.
(B) Changes in pre-30S complexes from the ΔrbfA strain as in (A). See Figure S2B for a histogram of cleavage intensity ratios.
(C) Three-dimensional mature 16S structure (PDB 2I2P) showing conformational changes in the ΔrimM strain colored as in (A). Left, 50S interface side; right, solvent side.
(D) Changes in the ΔrbfA strain, as in (C).
Many over-exposed residues (pink and red; Figure 2) such as the tetraloop in nt 1166–1170 participate in RNA tertiary interactions in the mature 30S subunit, indicating these interactions are not present in the pre-30S complexes. Interactions in the 16S 5′ domain appeared to toggle between two alternative conformations (Ramaswamy and Woodson, 2009) that alternately expose the tips of helices 9 (nt 190) and 17 (nt 468) in the absence of RbfA or over-protect it in the absence of RimM. Finally, many residues on the solvent side of the 30S head were exposed in the deletion strains (Figure 2C and 2D), indicating that pre-30S complexes lack the r-proteins that bind these helices.
Pre-30S complexes are partially active
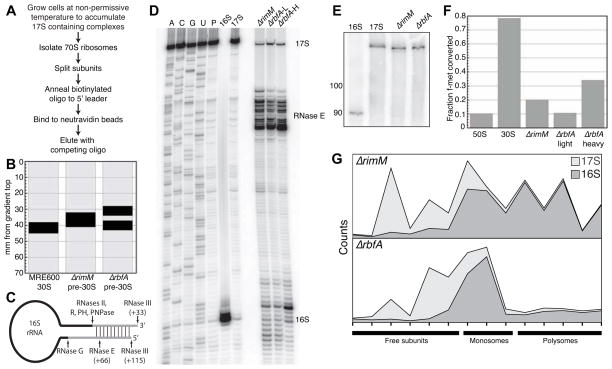
To correlate the rRNA structure with protein binding and activity, pre-30S complexes were purified from ΔrimM and ΔrbfA deletion strains using a biotinylated oligonucleotide annealing to the 16S 5′ leader as outlined in Figure 3A. The ΔrbfA pre-30S complexes sedimented more slowly than mature 30S ribosomes and formed discrete “light” and “heavy” bands on sucrose gradients, while ΔrimM subunits sedimented at an intermediate rate (Figure 3B).
Figure 3. Isolated pre-30S complexes are partially processed and competent for low levels of translation.
(A) Affinity purification of pre-30S complexes from total ribosomes.
(B) Relative mobility of pre-30S complexes in preparative sucrose gradients, measured by the distance (mm) from the top of the gradient.
(C) The 17S rRNA is processed by endonucleases RNases E and G at the 5′ end (Deutscher, 2009), and four exonucleases at the 3′ end (Sulthana and Deutscher, 2013).
(D) Primer extension of pre-16S 5′ end. Lanes ACGU, sequencing ladders; P, reverse transcriptase pausing control; 16S and 17S, in vitro transcripts. Product lengths for mature 16S, 16.3S (RNase E), and 17S rRNA (RNase III) are indicated on the right.
(E) Immature 3′ end of rRNA from pre-30S complexes were mapped by northern hybridization (probe anneals 3′ of nt 1486) after oligonucleotide-directed RNase H cleavage. The mature 16S rRNA yields a 90 nt fragment, while the 17S pre-rRNA product is 123 nt. The in vitro 17S marker in lane 2 includes an extra 3 nt.
(F) Some pre-30S complexes catalyze di-peptide formation. Fraction of f-met converted to f-met-phe di-peptide after 300 s in the presence of MRE600 50S subunits alone (50S), with MRE600 30S subunits (30S) or with purified pre-30S complexes in (B). Precision of the assay was ± 10%.
(G) Presence of 17S pre-rRNA in monosomes and polysomes. Stacked plot of the quantity of 17S (light) and 16S (dark) rRNA in deletion strains at non-permissive temperatures. Polysomes were analyzed on 10–40% sucrose gradients after treatment with chloramphenicol (Figure S3A). The 17S and 16S rRNA in each gradient fraction was determined by primer extension (Figure S3B). In cell lysates, less than 2% rRNA in all fractions was cleaved at the RNase E site (not shown).
We mapped the 5′ and 3′ ends of the pre-rRNA, which are processed by different ribonucleases (Figure 3C). The cellular pre-rRNA was cleaved by RNase III but not by RNase E (17S), though rRNA from the isolated complexes was slightly shorter than 17S (Figure S3A) due to imprecise cleavage in the vicinity of the RNase E site during purification (Figure 3D). Northern analysis showed that all the rRNA 3′ ends were immature (Figure 3E).
Although the isolated pre-30S complexes were expected to be inactive, they catalyzed the formation of dipeptides in vitro in proportion to their sedimentation velocity: light complexes were inactive, but pre-30S complexes from the ΔrimM strain and the heavy pre-30S from the ΔrbfA strain produced 25% and 40% as much dipeptide as mature 30S subunits, respectively (Figure 3F). This peptidyl transferase activity correlated with the presence of pre-30S complexes in polysomes. Polysomes from MRE600 grown at 37°C contained less than 1% 17S rRNA, as judged by primer extension (data not shown). When the ΔrimM strain was grown at the non-permissive temperature, a broad peak around 70S contained 8% 17S pre-rRNA (light gray, Figure 3G and Figure S3A). More strikingly, polysome fractions contained up to 23% 17S pre-rRNA in the absence of RbfA (Figure 3G). Thus, contrary to early reports (Lindahl, 1975), a fraction of immature complexes participate in protein synthesis in the absence of these assembly factors.
Protein complement of pre-30S complexes
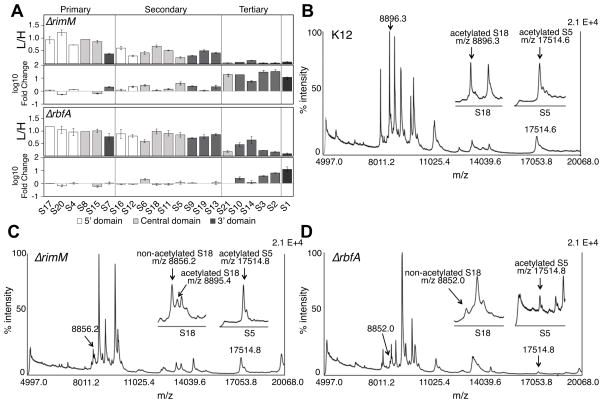
Based on the in vivo footprinting results, we expected that pre-30S complexes from ΔrimM and ΔrbfA strains lack certain r-proteins. To determine which r-proteins were present in our affinity-purified pre-30S complexes, we analyzed total 30S proteins (TP30) by quantitative mass spectrometry (Figure 4A). The ΔrbfA TP30 sample represented a mixture of r-proteins from the light and heavy pre-30S complexes. Equal amounts of TP30 from purified pre-30S complexes were mixed with 15N-labeled TP30 prepared using mature 30S ribosomes from MRE600 cells grown in heavy medium, and the mixture was analyzed by data-dependent LC-MS/MS (Sykes et al., 2010; Gouw et al., 2011). To facilitate comparison of the various strains, the 14N/15N (L/H) ratios were normalized to that of primary binding protein S8 and to K12 TP30 controls (Figure 4A).
Figure 4. Mass spectrometry of isolated pre-30S proteins.
(A) Relative quantities of r-proteins in ΔRimM and ΔRbfA pre-30S complexes. For each protein, the ratio of unlabeled (L) peptide in the test sample to 15N-labeled (H) peptide from MRE600 TP30 was determined by data-dependent LC-MS/MS (Experimental Procedures) and normalized to that of protein S8. Error bars represent the standard deviation of three technical replicates. Lower panels compare the relative pre-30S proteins (L/H) to L/H values from wild type (K12) 30S ribosomes. L/H values in the K12 samples ranged from 60–130% and reflected the intrinsic variation in recovery of tryptic peptides (data not shown). R-proteins are organized by their position in the Nomura map and colored by location: white, 5′ domain (body); light gray, central domain (platform); dark gray, 3′ domain (head); black, S1. Protein S1 was readily detected in K12 ribosomes but depleted in pre-30S complexes, consistent with their reduced activity.
(B–D) MALDI-TOFMS of intact proteins from (B) wild type K12 TP30, (C) ΔrimM TP30 and (D) ΔrbfA TP30. Details of the quantification of S5 acetylation are in Figure S4.
Pre-30S complexes formed in the absence of RbfA lacked the final proteins in the Nomura map (S2, S3 and S21) and had reduced quantities of other tertiary assembly proteins (Figure 4A, bottom). A likely explanation for partial stoichiometries is that the heaviest, most active ΔrbfA subunits contain more r-proteins than lighter, inactive complexes. By contrast, pre-30S complexes that formed in the absence of RimM lacked all the tertiary r-proteins (≤10%) and many secondary assembly proteins (Figure 4A, middle). Proteins S7 and S9 that bind the 3′ domain were less also less abundant than 5′ and central domain proteins (Figure 4A, dark gray bars). These results agree with previous studies showing that RimM facilitates assembly of the 30S head (Bunner et al., 2010; Bylund et al., 1998; Guo et al., 2013; Lovgren et al., 2004).
Ribosomal protein modification
Finally, we determined whether r-proteins are modified in pre-30S complexes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS) of intact 30S proteins. Proteins S5 and S18 are modified by the N-acetyltransferases RimJ and RimI, respectively (Yoshikawa et al., 1987). Though S5 and S18 were fully acetylated in 30S proteins isolated from E. coli K12 (Figure 4B, Table S2), S18 was mostly unmodified in the ΔrimM complexes (Figure 4C, left inset), and mostly acetylated in ΔrbfA complexes (Figure 4D, left inset).
Because MALDI-TOFMS could not resolve the S5 modification, we used LC-MSE (Silva et al., 2006; Geromanos et al., 2009) to determine the relative abundance of the modified and unmodified forms of S5 (Figure S4 A–H). S5 was 88% acetylated in TP30 from ΔrimM (Figure S4 A–B, Table S2) and 66% acetylated in ΔrbfA TP30 (Figure S4 E–F, Table S2). Over-expressed S5 was only 4% acetylated, confirming that RimJ acts on the pre-30S complex rather than free protein (Cumberlidge and Isono, 1979). As discussed below, the extent of S18 and S5 modification correlates with the formation of specific rRNA interactions during assembly.
Discussion
Widespread differences in the structures of immature and mature 30S subunits in vivo revealed by our footprinting data indicate that specific RNA and protein interactions throughout the complex are remodeled in the final stages of 30S biogenesis. Although the in vivo footprinting results reflect a mixture of intracellular complexes, features shared by the most abundant assembly intermediates are readily apparent owing to the high fraction of pre-30S complexes at non-permissive temperatures. Assembly is more likely to stall at large conformational rearrangements at low temperature. Below we discuss how RimM and RbfA help avoid or overcome stalling by delaying the formation of specific rRNA interactions.
Several observations suggest the pre-30S complexes that accumulate in the absence of RimM and RbfA represent common rather than atypical assembly intermediates. First, our footprinting and mass spectrometry results show that the hierarchy of protein addition follows the Nomura assembly map, in agreement with pulse labeling of r-proteins (Chen and Williamson, 2013). Thus, the order in which proteins join the complex does not change although the RNA folding path may change. Second, the rapid accumulation of pre-30S complexes once cells are shifted to a lower temperature indicates they are not immediately targeted for destruction. Further work is needed to determine what fraction of pre-30S complexes reach maturity under various conditions.
Common structural features of pre-30S subunits
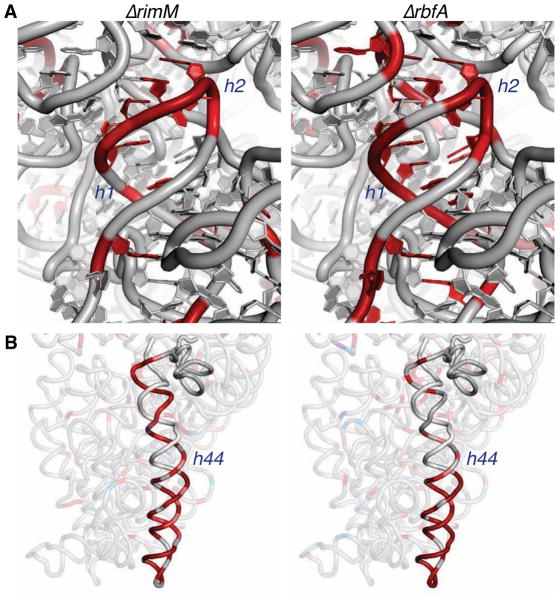
A comparison of the solvent accessibility of the rRNA backbone in ΔrimM and ΔrbfA cells (Figure 2) revealed structural perturbations we propose are shared by all pre-30S complexes containing 17S pre-rRNA. First, nucleotides in helices 1 and 2 that comprise the central pseudoknot were strongly exposed in ΔrimM and ΔrbfA cells (Figure 5A). This finding agrees with in vivo and in vitro chemical modification studies showing that the central pseudoknot forms late in assembly (Besancon and Wagner, 1999; Holmes and Culver, 2004; Powers et al., 1993) and with cryoEM reconstructions showing perturbations to these helices (Guo et al., 2013; Jomaa et al., 2011).
Figure 5. Perturbations to helix 44 and the central pseudoknot in pre-30S complexes in vivo.
(A) The central pseudoknot (helices 1 and 2) is unfolded in both strains. Red indicates exposed segments of the rRNA backbone, helix numbers are given in navy italics (see Figure 2).
(B) Helix 44 is undocked in both ΔrimM and ΔrbfA strains. Due to 16S modifications, this region was not fully covered by our primer extensions (light gray; Figure 2A).
The stability of helix 1 directly correlates with 30S function, because the central pseudoknot is essential for protein synthesis by the ribosome (Brink et al., 1993; Poot et al., 1998). In the pre-rRNA, helix 1 competes with an alternative secondary structure formed by leader sequences (Dammel and Noller, 1993), which reduces the stability of the central pseudoknot. When RbfA is present, delayed folding of the pseudoknot may help keep immature subunits out of the translation cycle (Roy-Chaudhuri et al., 2010). When RbfA is missing, the inactive structure may equilibrate more easily, allowing some pre-30S complexes to enter translation (Figure 3G).
Second, we observe that both strands of helix 44 are strongly cleaved, indicating the entire helix is exposed in pre-30S complexes (Figure 5B). These cleavages extend beyond residues protected by the 50S subunit (Merryman et al., 1999) and cannot be explained by free subunits in the mutants (Figure S5). Analysis of 17S rRNA from unirradiated mutant cells showed no evidence of degradation products corresponding to cleavage of helix 44 (Figure 3E).
Several observations indicate assembly factors prevent helix 44 from docking in vivo. Helix 44 docks with the 30S body during in vitro reconstitution (Holmes and Culver, 2004; Adilakshmi et al., 2008), demonstrating these contacts are intrinsically stable when r-proteins are present. By contrast, helix 44 was exposed in the absence of RimM (Figure 5B, left) and disordered in certain cryo-EM reconstructions of ΔrimM pre-30S complexes (Guo et al., 2013), showing that it remains undocked when assembly stalls in vivo. Cryo-EM reconstructions show that RbfA and the RsgA GTPase displace helix 45 and the top of helix 44 (Datta et al., 2007; Jomaa et al., 2011) and may act together in vivo (Goto et al., 2011). The KsgA methylase also displaces helix 44, likely at a later stage of maturation (Boehringer et al., 2012).
Third, proteins S2, S3 and S21 were missing in our pre-30S complexes, indicating that assembly stalls in vivo before these tertiary proteins can bind. Residues in helices 26, 35, 37 and 40 that contact protein S2 in the 30S neck were highly exposed in both strains, showing that S2 binding site is disordered in both pre-30S complexes (Figure 6A). S2 is one of the last proteins to join the 30S complex in vitro (Talkington et al., 2005), and early association of S2 has been implicated in a kinetically trapped intermediate (Mulder et al., 2010).
Figure 6. Incomplete assembly of the head and platform in pre-30S ribosomes.
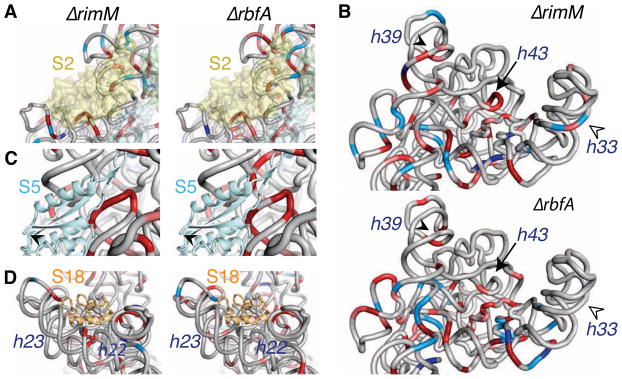
(A) Perturbations to the head (3′ major domain) in the absence of RimM (top) or RbfA (bottom). Exposure and protection of the rRNA backbone relative to the parental strain is colored as in Figure 2. The head is less folded without RimM, with more extensive changes in helix 39 (closed arrowhead), helix 33 “beak” (open arrowhead), and core helix 43 (arrow).
(B) Exposure of the binding site for protein S2 (semi-transparent yellow surface).
(C) Exposure of the central pseudoknot and residues contacted by protein S5 (cyan). The N-terminus of S5 is only partially acetylated in pre-30S complexes (arrowhead).
(D) Exposure of helices 22 and 23 in the S18 binding site in ΔrimM cells compared to ΔrbfA cells.
Assembly of the 3′ domain
The structural differences between ΔrimM and ΔrbfA complexes suggest that RimM facilitates assembly of the 30S head by changing the folding pathway of the 16S 3′ major domain. A substantial energy barrier impedes assembly of the 3′ domain. In vitro, tertiary assembly r-proteins (S2, S3, S10, S14, S21) only bind the 16S rRNA after a heat-dependent conformational change (Traub and Nomura, 1969). These proteins were also depleted in our pre-30S complexes, and bind 16S 3′ domain helices that are most structurally altered in our footprinting data (Figure 6B). These deficiencies were more severe in the absence of RimM. For example, the 30S “beak” (helices 33b and 33c) was folded in the absence of RbfA but not in the absence of RimM (Figure 6B, open arrowhead).
RimM interacts with protein S19 and is thought to contact helix 32 in the 50S interface (Lovgren et al., 2004). Surprisingly, helices facing the 50S subunit folded correctly in the absence of RimM, in agreement with the presence of proteins S7, S13 and S19 that bind this region (Figure 4A). By contrast, the helix 34-35-38 junction on the solvent side of the head, and helices 31 and 43 that form the core of the 3′ domain, folded improperly in the absence of RimM. These rRNA interactions create the S10 and S14 binding sites (Figure 6B, filled arrowhead), explaining why these proteins do not bind in the absence of RimM (Figure 4A).
Together, these results suggest that RimM does not simply recruit S19 to the complex, as previously thought (Bunner et al., 2010; Lovgren et al., 2004). Instead, we propose that RimM (via its interactions with S19 and helix 32) holds the 16S 3′ domain in an open conformation that permits core helices 31 and 43 to fold correctly. This is supported by the observation that RimM changes the order in which the core and solvent-side helices fold. When RimM is present in vivo (ΔrbfA strain), helix 43 folds properly yet the solvent-side is more open. In vitro or in the absence of RimM, helix 43 folds slowly but the solvent-side helices fold rapidly (Adilakshmi et al., 2008; Holmes and Culver, 2004). This reversal in the folding order may be needed in vivo because core helix 43 is the last helix of the 3′ major domain to be transcribed.
Assembly factor binding sites
In addition to RimM and RbfA, at least five other assembly factors plus 15 modifying enyzmes contribute to 30S biogenesis in E. coli (Culver, 2003; Wilson and Nierhaus, 2007). Strong protection of specific nucleotides in the deletion strains may reflect factors bound to pre-30S complexes in the cell, although these factors did not remain bound during our stringent pre-30S purification. For example, protection of nt 927 (helix 28) in both strains may be due to the Era GTPase, which contacts helix 28 in addition to protein S7 and the 16S 3′ end (Sharma et al., 2005; Tu et al., 2011). In the ΔrbfA strain, a cluster of protected residues in helices 32 and 34 (nt 960, 988, 1244) around S19 suggests a candidate binding site for RimM. Protection of the rRNA backbone in helix 34 may be due to RimM or to RsmC, which methylates G1207 in helix 34.
Protein modification correlates with remodeled RNA-protein interactions
Although the rRNA must fold for proteins to bind, certain r-protein binding sites change conformation after the r-protein binds. The most astonishing example is protein S5, which uses an extended β-hairpin (loop 2) to contact helix 1 and the central pseudoknot in the mature ribosome. This region of the rRNA is completely unfolded in pre-30S complexes, exposing nt 920, 922, 923, 1081 that contact S5 directly (Figure 6C), even though S5 is fully present (Figure 4A). We propose that S5 itself changes structure at a later step of 30S maturation when the central pseudoknot is formed. A conformational change in S5 is plausible because loop 2 folds back against the N-terminal domain in free S5 protein (Ramakrishnan and White, 1992). Interestingly, a G25D mutation in loop 2 causes defects in 30S biogenesis due to a failure to refold an alternative structure in the 17S pre-rRNA (Roy-Chaudhuri et al., 2010).
Reorganization of S5-rRNA interactions may be triggered by the RimJ acetylase, which itself functions as an assembly factor (Roy-Chaudhuri et al., 2008). S5 was more acetylated in ΔrimM pre-30S complexes, than in ΔrbfA pre-30S complexes that lack the stabilizing influence of RbfA on the “neck” region (Figure 4C and 4D). Thus S5 modification occurs on the ribosome (Poot et al., 1997) and correlates with the stage of assembly.
The connection between modification and assembly is reinforced by under-acetylation of S18 in the ΔrimM strain (Figure 4C). Unmodified S18 correlated with perturbations in helices 22 and 23 near the S18 binding site in the central domain (Figure 6C) and reduced recovery of S18 in pre-30S complexes (Figure 4A). Because the N-terminus of S18 is disordered in 70S crystals, it seems unlikely that acetylation directly increases S18 affinity. Instead, we infer that RimI either acetylates S18 after the correct protein-rRNA interactions are in place, or facilitates rRNA folding, resulting in more stable association of S18. In either case, the remarkable correlation between folding of the pre-rRNA and acetylation of S5 and S18 suggests these modifications report the formation of specific RNA-protein contacts during assembly.
A model for 30S assembly in vivo
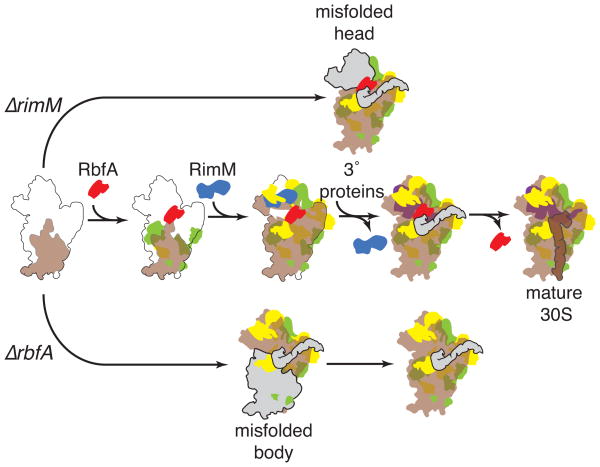
Our results and previous biochemical and genetic data show how RbfA and RimM chaperone distinct steps of 30S assembly by altering the conformation of the pre-rRNA (Figure 7). First, RbfA supports refolding of the 16S leader (Dammel and Noller, 1995) and facilitates a conformational change in the 5′ domain necessary for forming the central pseudoknot. The presence of light and heavy pre-30S complexes in ΔrbfA cells is consistent with RbfA acting on assembly of the 5′ domain (body) at an early stage of 30S assembly, and on the central pseudoknot at a later stage of assembly. RbfA may act together with the RsgA GTPase and the RimJ S5 acetylase to prevent helix 44 from docking against the body before the central pseudoknot has formed.
Figure 7. A model for the roles of RimM and RbfA in 30S assembly in vivo.
Folding of the pre-rRNA during transcription in vivo (Nierhaus, 1991) is accompanied by binding of primary (green) and secondary (yellow) assembly proteins (middle row). Tertiary assembly proteins (purple) join the complex after conformational changes in the head and neck. Brown represents properly folded rRNA and solid gray represents non-native structures. RimM (blue) facilitates proper folding of the 16S 3′ domain during assembly of the head (top row), while RbfA (red) acts early to promote 5′ domain assembly, and later to promote formation of the central pseudoknot (bottom row). Our results suggest that maturation of the 16S 5′ and 3′ ends is preceded by reorganization of interdomain interactions and helix 44 docking, which may be triggered by release of RbfA and other assembly factors.
Next, we propose that RimM binds the 3′ major domain during transcription (Figure 7), together with protein S19 (Bunner et al., 2010; Lovgren et al., 2004), holding the 3′ domain in a conformation that allows the helix 33 (beak) and helix 43 to fold correctly. In the absence of RimM, misfolding of these helices traps pre-30S complexes in an intermediate that cannot recruit the tertiary assembly proteins S10, S14 and S3. Based on chemical probing experiments (Holmes and Culver, 2004), we suggest a similar folding trap stalls in vitro reconstitution at low temperature. Our data show that RimM influences assembly of the platform and acetylation of S18 in vivo. The Era GTPase, which interacts with both domains (Sharma et al., 2005), may communicate structural changes in the head to the platform, based on its position on the 30S subunit.
Finally, structural features of pre-30S complexes from ΔrimM and ΔrbfA strains point to a common bottleneck that precedes reorganization of domain interfaces in the final stages of 30S maturation. In both complexes, the 5′ leader and 3′ trailer are unprocessed except by RNase III, the central pseudoknot is unfolded, helix 44 is undocked, and proteins S2 and S3 are missing. As some of these interactions form readily in vitro (such as docking helix 44), we hypothesize that assembly factors bound to the pre-30S complex block this reorganization in vivo, delaying final maturation of the subunit as proposed for yeast 40S biogenesis (Strunk et al., 2011). Our results reveal steps of 30S self-assembly prone to failure at low temperature, and suggest that assembly factors act not by recruiting r-proteins to the complex, but by delaying specific folding steps and by sensing the remodeling of RNA and protein interactions.
Experimental Procedures
Bacterial Strains
MRE600 (F−, rna) was used as the wild-type unless stated otherwise. The ΔrimM strain MW37 (Hfr P4X ΔrimM-2 16K::nptI) (Persson et al., 1995) was compared to its parental strain MW100 (Hfr P4X) (Wikstrom et al., 1988). The ΔrbfA strain was BX41 (F− araD139 Δ(argF-lac)U169 ptsF25 deoC1 relA1 flbB5301 rspL150−rbfA::kan) (Xia et al., 2003). For mass spectrometry, pre-30S proteins were compared to TP30 from E. coli K12 (ATCC).
In vivo hydroxyl radical footprinting
MRE600 cells were prepared for footprinting as previously described (Adilakshmi et al., 2006). 50 mL cultures were grown at 37°C to OD600 = 0.5–0.6, rapidly chilled, harvested by centrifugation, washed, and resuspended in 500 μL TM (10 mM Tris–HCl, pH 7.5, 1 mM MgCl2). Aliquots (5 μL) were frozen in 0.2 mL PCR tubes (Brand catalog #781305) and stored at −80°C. Deletion strains and parental controls were grown to OD600= 0.2 before shifting to the non-permissive temperature. ΔrimM cultures were shifted to 25°C for 3 h, and ΔrbfA cells were shifted to 17°C for 12–16 h, based on northern analysis of 17S pre-rRNA (see Supplemental Procedures).
Frozen samples were irradiated for 100 ms at beamline X28C, NSLS, as described in (Adilakshmi et al., 2006). For RNA analysis, pellets were thawed in 500 μL RNAprotect Bacteria Reagent (Qiagen), and total RNA extracted with RNeasy Mini using the lysozyme lysis protocol (Qiagen). Total 16S rRNA from irradiated cells and unexposed controls was analyzed by extension with dye-labeled primers and ShapeFinder (Vasa et al., 2008) as described in Supplemental Procedures, to yield integrated intensities for most positions in the 16S rRNA. The standard deviation in reported values was ±15–20%. Data are available from the authors upon request.
Pre-30S purification and activity assay
Affinity purification of pre-30S complexes using an oligonucleotide complementary to the rRNA 5′ leader was adapted from (Schnapp et al., 1998) and carried out as described in Supplemental Procedures. Di-peptide synthesis was measured as previously described (Zaher and Green, 2010), using MRE600 30S ribosomes or ΔrimM and ΔrbfA pre-30S complexes, and 50S subunits from MRE600 cells. Reaction products were separated by cellulose-TLC electrophoresis (Youngman et al., 2004) and the fraction of radioactivity in each band was quantified. 30S complexes and polysomes were analyzed on 10–40% sucrose gradients as previously described (Maki et al., 2002) using a piston fractionator (Biocomp).
5′ and 3′ end mapping
The 16S 5′ end was mapped by primer extension with 4 pmol unmodified rRNA and 6.6 pmol 32P-end-labeled primer 46 (Table S1), using SuperScript III reverse transcriptase. The cDNA products were separated on an 8% polyacrylamide gel. Band volumes were quantified with a Phosphorimager and normalized to the sum of the major products.
The 16S 3′ end was mapped by site-directed RNase H cleavage as described in (Yu et al., 1997), using 1 pmol rRNA and 25 pmol C16S3 (Table S1) 2′-O-methyl-RNA oligonucleotide. Cleavage products were separated on a 8% polyacrylamide gel and transferred to Nytran SPC membrane using a semi-dry electroblotter. The blot was hybridized with 32P-end-labeled primer 1486 (Table S1) in ULTRAhyb-oligo (Ambion) overnight at 42°C, washed and imaged.
Quantitative mass spectrometry
15N-labeling and data-dependent LC-MS/MS on a Synapt G2 mass spectrometer coupled to a nanoAcquity UPLC (Waters, Milford, MA) was used to determine the relative amounts of proteins associated with mature 30S and pre-30S complexes (Sykes et al., 2010; Gouw et al., 2011). For peptide identification and quantitation, raw mass spectral data from data-dependent acquisition mode were searched against E. coli tryptic peptides in the UniProtKB/Swiss-Pro protein database (Supplemental Procedures). A minimum of two light-heavy peptide pairs were used for quantification of the proteins, while manual inspection of the data was done for proteins with only one light-heavy peptide pair to assess data quality.
Endoproteinase Arg-C (Sigma, St. Louis, MO) digestion and alternate scanning mode, LC-MSE, of protein S5 was performed to determine its modification status as described in Supplemental Procedures. MALDI-TOFMS of total 30S proteins (TP30) from wild type K12, ΔrimM, or ΔrbfA was performed on a MDS SCIEX 4800 MALDI TOF/TOF analyzer equipped with Nd:YAG 200 Hz laser (Applied Biosystems, Framingham, MA) as described previously (Suh and Limbach, 2004).
Supplementary Material
Highlights.
Pre-30S ribosomes were visualized in situ by X-ray footprinting on whole cells.
Pre-30S structures reveal widespread refolding of pre-rRNA late in assembly.
RimM and RbfA change folding paths of the 16S 5′ and 3′ domains.
Quantitative mass spectrometry correlates protein acetylation with rRNA structure.
Acknowledgments
The authors thank Masayori Inoue and Mikael Wikström for bacterial strains, Sayan Gupta and Rhijuta D’Mello for help with X-ray footprinting, Julie Brunelle and Rachel Green for assistance with the di-peptide assay, and Ken Greis and Therese Rider (UC Proteomics Core Facility) for access to the MALDI-TOF mass spectrometer. This work was supported by grants from the NIH (GM60819 to SAW; GM58843 and RR027671 to PAL, and P30-EB-00998 to the Center for Synchrotron Biosciences X28C). Use of the National Synchrotron Light Source was supported by the U.S. DOE under Contract DE-AC02-98CH10886.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adilakshmi T, Lease RA, Woodson SA. Hydroxyl radical footprinting in vivo: mapping macromolecular structures with synchrotron radiation. Nucleic Acids Res. 2006;468:e64. doi: 10.1093/nar/gkl291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer M, Wagner R. Mutations in the leader region of ribosomal RNA operons cause structurally defective 30 S ribosomes as revealed by in vivo structural probing. J Mol Biol. 1998;276:547–557. doi: 10.1006/jmbi.1997.1556. [DOI] [PubMed] [Google Scholar]
- Besancon W, Wagner R. Characterization of transient RNA-RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA. Nucleic Acids Res. 1999;27:4353–4362. doi: 10.1093/nar/27.22.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, O’Farrell HC, Rife JP, Ban N. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J Biol Chem. 2012;287:10453–10459. doi: 10.1074/jbc.M111.318121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink MF, Verbeet MP, de Boer HA. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993;12:3987–3996. doi: 10.1002/j.1460-2075.1993.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunner AE, Nord S, Wikstrom PM, Williamson JR. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J Mol Biol. 2010;398:1–7. doi: 10.1016/j.jmb.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund GO, Wipemo LC, Lundberg LA, Wikstrom PM. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Williamson JR. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J Mol Biol. 2013;425:767–779. doi: 10.1016/j.jmb.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM. Assembly of the 30S ribosomal subunit. Biopolymers. 2003;68:234–249. doi: 10.1002/bip.10221. [DOI] [PubMed] [Google Scholar]
- Cumberlidge AG, Isono K. Ribosomal protein modification in Escherichia coli. I A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J Mol Biol. 1979;131:169–189. doi: 10.1016/0022-2836(79)90072-x. [DOI] [PubMed] [Google Scholar]
- Dammel CS, Noller HF. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 1995;9:626–637. doi: 10.1101/gad.9.5.626. [DOI] [PubMed] [Google Scholar]
- Dammel CS, Noller HF. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- Datta PP, Wilson DN, Kawazoe M, Swami NK, Kaminishi T, Sharma MR, Booth TM, Takemoto C, Fucini P, Yokoyama S, Agrawal RK. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell. 2007;28:434–445. doi: 10.1016/j.molcel.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–391. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC- MS with data dependent LC- MS/MS. Proteomics. 2009;9:1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- Goto S, Kato S, Kimura T, Muto A, Himeno H. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. EMBO J. 2011;30:104–114. doi: 10.1038/emboj.2010.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw JW, Tops BB, Krijgsveld J. Metabolic labeling of model organisms using heavy nitrogen (15N) Methods Mol Biol. 2011;753:29–42. doi: 10.1007/978-1-61779-148-2_2. [DOI] [PubMed] [Google Scholar]
- Guo Q, Goto S, Chen Y, Feng B, Xu Y, Muto A, Himeno H, Deng H, Lei J, Gao N. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 2013;41:2609–2620. doi: 10.1093/nar/gks1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JJ, Kam L, Tullius TD. Footprinting protein-DNA complexes with gamma-rays. Methods Enzymol. 1990;186:545–549. doi: 10.1016/0076-6879(90)86148-o. [DOI] [PubMed] [Google Scholar]
- Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- Held WA, Mizushima S, Nomura M. Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components. J Biol Chem. 1973;248:5720–5730. [PubMed] [Google Scholar]
- Holmes KL, Culver GM. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol. 2004;11:179–186. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Swapna GV, Rajan PK, Ke H, Xia B, Shukla K, Inouye M, Montelione GT. Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli. J Mol Biol. 2003;325:521–536. doi: 10.1016/s0022-2836(03)00061-5. [DOI] [PubMed] [Google Scholar]
- Inoue K, Alsina J, Chen J, Inouye M. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol Microbiol. 2003;48:1005–1016. doi: 10.1046/j.1365-2958.2003.03475.x. [DOI] [PubMed] [Google Scholar]
- Jomaa A, Stewart G, Martin-Benito J, Zielke R, Campbell TL, Maddock JR, Brown ED, Ortega J. Understanding ribosome assembly: the structure of in vivo assembled immature 30S subunits revealed by cryo-electron microscopy. RNA. 2011;17:697–709. doi: 10.1261/rna.2509811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J Mol Biol. 1975;92:15–37. doi: 10.1016/0022-2836(75)90089-3. [DOI] [PubMed] [Google Scholar]
- Lovgren JM, Bylund GO, Srivastava MK, Lundberg LA, Persson OP, Wingsle G, Wikstrom PM. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA. 2004;10:1798–1812. doi: 10.1261/rna.7720204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki JA, Schnobrich DJ, Culver GM. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol Cell. 2002;10:129–138. doi: 10.1016/s1097-2765(02)00562-2. [DOI] [PubMed] [Google Scholar]
- Merryman C, Moazed D, McWhirter J, Noller HF. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J Mol Biol. 1999;285:97–105. doi: 10.1006/jmbi.1998.2242. [DOI] [PubMed] [Google Scholar]
- Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- Ottinger LM, Tullius TD. High-Resolution in Vivo Footprinting of a Protein DNA Complex Using γ-Radiation. J Am Chem Soc. 2000;122:5901–5902. [Google Scholar]
- Persson BC, Bylund GO, Berg DE, Wikstrom PM. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J Bacteriol. 1995;177:5554–5560. doi: 10.1128/jb.177.19.5554-5560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot RA, Jeeninga RE, Pleij CW, van Duin J. Acetylation of ribosomal protein S5 affected by defects in the central pseudoknot in 16S ribosomal RNA? FEBS Lett. 1997;401:175–179. doi: 10.1016/s0014-5793(96)01467-6. [DOI] [PubMed] [Google Scholar]
- Poot RA, van den Worm SH, Pleij CW, van Duin J. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 1998;26:549–553. doi: 10.1093/nar/26.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J Mol Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, White SW. The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature. 1992;358:768–771. doi: 10.1038/358768a0. [DOI] [PubMed] [Google Scholar]
- Ramaswamy P, Woodson SA. S16 throws a conformational switch during assembly of 30S 5′ domain. Nat Struct Mol Biol. 2009;6:438–445. doi: 10.1038/nsmb.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Chaudhuri B, Kirthi N, Culver GM. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci U S A. 2010;107:4567–4572. doi: 10.1073/pnas.0912305107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Chaudhuri B, Kirthi N, Kelley T, Culver GM. Suppression of a cold-sensitive mutation in ribosomal protein S5 reveals a role for RimJ in ribosome biogenesis. Mol Microbiol. 2008;68:1547–1559. doi: 10.1111/j.1365-2958.2008.06252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp G, Rodi HP, Rettig WJ, Schnapp A, Damm K. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 1998;26:3311–3313. doi: 10.1093/nar/26.13.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Barat C, Wilson DN, Booth TM, Kawazoe M, Hori-Takemoto C, Shirouzu M, Yokoyama S, Fucini P, Agrawal RK. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol Cell. 2005;18:319–329. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MJ, Limbach PA. Investigation of methods suitable for the matrix-assisted laser desorption/ionization mass spectrometric analysis of proteins from ribonucleoprotein complexes. Eur J Mass Spectrom (Chichester, Eng) 2004;10:89–99. doi: 10.1255/ejms.626. [DOI] [PubMed] [Google Scholar]
- Sulthana S, Deutscher MP. Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA) J Biol Chem. 2013;288:12574–12579. doi: 10.1074/jbc.C113.459172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes MT, Shajani Z, Sperling E, Beck AH, Williamson JR. Quantitative proteomic analysis of ribosome assembly and turnover in vivo. J Mol Biol. 2010;403:331–345. doi: 10.1016/j.jmb.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. VI Mechanism of assembly of 30 S ribosomes studied in vitro. J Mol Biol. 1969;40:391–413. doi: 10.1016/0022-2836(69)90161-2. [DOI] [PubMed] [Google Scholar]
- Tu C, Zhou X, Tarasov SG, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X. The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3′ end of 16S rRNA. Proc Natl Acad Sci U S A. 2011;108:10156–10161. doi: 10.1073/pnas.1017679108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr Opin Chem Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom PM, Bystrom AS, Bjork GR. Non-autogenous control of ribosomal protein synthesis from the trmD operon in Escherichia coli. J Mol Biol. 1988;224:141–152. doi: 10.1016/0022-2836(88)90098-8. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- Xia B, Ke H, Shinde U, Inouye M. The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli. J Mol Biol. 2003;332:575–584. doi: 10.1016/s0022-2836(03)00953-7. [DOI] [PubMed] [Google Scholar]
- Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A, Isono S, Sheback A, Isono K. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol Gen Genet. 1987;209:481–488. doi: 10.1007/BF00331153. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol Cell. 2010;39:110–120. doi: 10.1016/j.molcel.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.