Abstract
Microglia, the principal neuroimmune sentinels of the brain, continuously sense changes in their environment and respond to invading pathogens, toxins and cellular debris. Microglia exhibit plasticity and can assume neurotoxic or neuroprotective priming states that determine their responses to danger. We used direct RNA sequencing, without amplification or cDNA synthesis, to determine the quantitative transcriptomes of microglia of healthy adult and aged mice. We validated our findings by fluorescent dual in-situ hybridization, unbiased proteomic analysis and quantitative PCR. We report here that microglia have a distinct transcriptomic signature and express a unique cluster of transcripts encoding proteins for sensing endogenous ligands and microbes that we term the “sensome”. With aging, sensome transcripts for endogenous ligand recognition are downregulated, whereas those involved in microbe recognition and host defense are upregulated. In addition, aging is associated with an overall increase in expression of microglial genes involved in neuroprotection.
Keywords: Microglia, Sensome, Direct RNA Sequencing, Aging, Macrophages, classical activation, Microbes, Alternative activation, quantitative transcriptome
Microglia, the principal resident immune cells of the brain, constitute 5–12% of brain cells depending on the region studied 1. They are important in homeostatic functions in the brain, in host defense against infectious pathogens and in neurodegenerative diseases and traumatic brain injury 2. The role of microglia as mononuclear phagocytes has been recognized for decades and their involvement in inflammatory and necrotizing processes are beginning to be elucidated 3.
In the past decade we witnessed an explosion of work to understand the role of microglia in the developing, healthy, aging, and diseased brain. Seminal work showed that a major function of these cells is to constantly survey their environment sensing any changes in that environment and responding to them 4. The genes that encode the microglial sensing apparatus are not defined.
Microglia, like other mononuclear phagocytes such as macrophages, exhibit plasticity and can assume neuroprotective or neurotoxic phenotypes 5–7. In advanced stages of neurodegenerative disorders like Alzheimer’s disease, multiple sclerosis and amyotrophic lateral sclerosis, microglia become neurotoxic 7–9. Microglia also exhibit various priming states that determine their responses to subsequent injury or infection of the brain. Two of these priming states include the classical priming state which occurs in response to stimulation with Interferon-γ, and the alternative priming state associated with exposure of these cells to Interleukin (IL) 4 or IL138, 10, 11. Classically-primed mononuclear phagocytes are neurotoxic whereas alternatively-primed cells promote axonal elongation and sprouting and are considered neuroprotective 8, 12.
Aging dramatically affects the gene expression profile of the brain 13. Transcriptome profiling of the whole brain using microarrays found downregulation of genes involved in energy production, protein synthesis and protein transport with aging and upregulation of many genes that regulate proliferation 14. Studies looking at aging-associated changes in individual cells have been limited 15 and quantitative changes that occur in the microglial transcriptome with aging have not been defined.
Several approaches to analyze the transcriptomes of tissues and cells have been developed. These include quantitative PCR (qPCR) 16, microarrays 17, 18, Nanostring™ 19 technology and deep sequencing (RNA-Seq) 20. Valuable information can be obtained from each of these approaches. While qPCR and Nanostring are quantitative, they are limited by the number of transcripts that can be measured. Microarrays are very useful, but only provide a semi-quantitative assessment of the transcriptome. aAvailable RNA-Seq platforms are quantitative and provide a snapshot of the transcriptome of cells at a specific time point, but vary in their requirements of the amount of RNA as a starting material and require generation of cDNA and amplification if the sample size is small 21. Because the number of microglia that can be harvested is limited, we were interested in an approach that utilizes small amounts of RNA, that does not require cDNA synthesis or amplification, and that gives robust quantitative results.
To analyze the transcriptome of microglia, we selected a recently developed technology, Direct RNA Sequencing or DRS, because it allows the quantification of mRNA without the need for amplification or cDNA synthesis 22 thereby providing several advantages over existing techniques. Essentially, poly(A) mRNA is hybridized to a surface coated with poly(dT), unbound RNA (non mRNA) is washed away and the remainder of the poly(A) tail is blocked, followed by sequencing as described in supplementary methods. Using this approach, we determined the unbiased quantitative transcriptome of microglia isolated from adult 5-month old mice and compared it to that of peritoneal macrophages isolated from the same mice and to whole brain. The datasets generated allowed us to identify a cluster of genes that constitute the microglia sensing apparatus, which we term “sensome”. We also identified a unique microglial signature that distinguishes them from macrophages. Furthermore, we determined changes that occur in the microglial transcriptome and sensome during normal aging using microglia isolated from 24-month old animals. With aging, the microglial overall gene expression profile shows an upregulation of genes involved in host defense and neuroprotection. Our dataset is the first quantitative transcriptome of normal microglia in adulthood and aging.
RESULTS
In order to have a standardized source of cells we used C57BL6 mice obtained from the national Institute of Aging mouse colony (NIA, Bethesda MD). Microglia were isolated from 5-months old adult mice by enzymatic digestion, as previously described 7, and peritoneal macrophages were isolated by peritoneal lavage 23. Cells were stained with fluorescent antibodies to CD11b and CD45, two well established microglia and macrophage markers 2, 24 and supplementary Figure 1a). Microglia and macrophages were subsequently isolated by Fluorescence Activated Cell Sorting (FACS) and RNA was extracted. Supplementary Figure 1b-c displays the CD11b/CD45 expression patterns used to collect a distinct population of microglia (high CD11b, low-to-intermediate CD45 2, 24) and peritoneal macrophages (high CD11b and high CD45).
Identification of the microglial Sensome by DRS
We performed DRS on RNA isolated from whole brain (3 samples from 2 mice each) and microglia isolated from 3 pools comprising 22, 10 and 20 mice respectively. We developed programs in MolBioLib to annotate the various sequences obtained 25 and then analyzed the data via GSEA 26 and edgeR27 A total of 21025 different coding transcripts were detected (Supplementary Figure 1d). To standardize data presentation and allow comparison between different datasets we computed the data for each transcript as copies of mRNA per million mapped reads (CMMR).
Microglia processes constantly move in the area surrounding the cell body sensing any changes in the environment 4. Such changes could be caused by microbial invasion, cell injury or death associated with neurodegeneration, deposition of neurotoxic substances such as amyloid β 23, 28 or inflammatory molecules such as chemokines and cytokines. Additional milieu alterations could include changes in pH, in the composition or integrity of the extracellular matrix or in extracellular metabolites such as nucleotides or amino acids. A large armamentarium of proteins and receptors may be used by a given cell for sensing such changes in the environment. The full spectrum of the cellular receptors and proteins used by microglia for this purpose are not known. In this paper we define these proteins as the “Sensome” of microglia.
We used the DAVID 29 Gene Ontology (GO) analysis software as well as manual annotation and data mining through PubMed.gov to identify candidate sensome genes, regardless of their level of expression. These potential sensome genes included all transmembrane proteins and receptors, such as integrins, purinoceptors, lectins, transporters and CD antigens, but not genes encoding for secreted proteins or proteins expressed only in nuclear, mitochondrial or endoplasmic reticulum membranes. This analysis resulted in identifying 1299 candidate sensome genes.
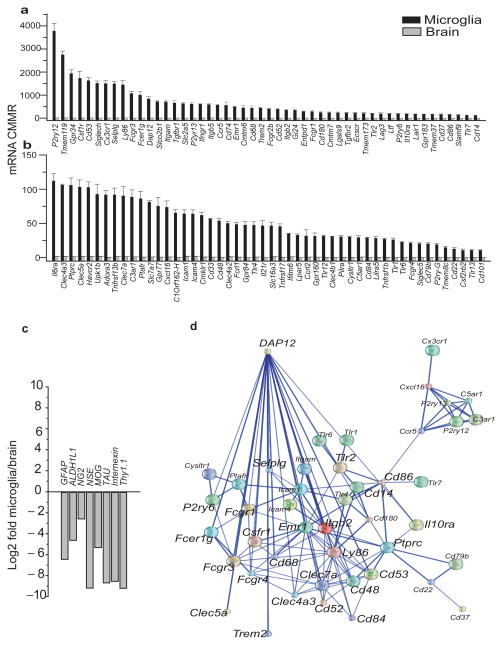
To identify the transcripts that are highly enriched in microglia vs. whole brain, we computed the ratio: and calculated Log2E values, then selected the top 100 transcripts with the highest E values. These transcripts are the most likely transcripts to be microglia specific (Figure 1a, b and Supplementary table 1). The E value for these 100 transcripts ranged from 36 to 292. Of these transcripts 46% have an E value ≥100 indicating a high level of enrichment in microglia (p<0.00001) (Figure 1a, b and supplementary table 1). In contrast, the E values for neuronal genes such as gamma enolase (Neuron-specific enolase NSE), Internexin and Thy1 were 0.0018, 0.0028 and 0.0018, respectively. Similarly the E value for the oligodendrocyte genes myelin-oligodendrocyte glycoprotein (MOG) and chondroitin sulfate proteoglycan 4 (NG2) were 0.026 and 0.17 respectively. Furthermore, the E value for the astrocyte markers glial fibrillary acidic protein (GFAP) and aldehyde dehydrogenase family 1 member L1 (AlDH1L1) were 0.012 and 0.042 respectively (Figure 1c). These data indicate a high level of enrichment of microglia specific sensome genes and a “derichment” of genes expressed by neurons and other non-microglial cells of the neural environment in our purified microglia.
Figure 1. The microglial Sensome identified by direct RNA sequencing.
Of the 21025 transcripts measured, we used gene ontology (GO) analysis and identified 1299 potential sensome genes. Of these, we selected the top 100 transcripts with the highest enrichment of microglia/brain and termed this gene collection as the microglial “Sensome”. a–b. Expression levels of genes of the microglial Sensome in mRNA copies per million reads (CMMR) in microglia and brain. Values are mean ± SD of three different experiments done with microglia pooled from 22, 10 and 20 mice, respectively. and three pools of RNA from 2 brains each. For differences in expression between microglia and brain p<0.00001 for all Sensome genes shown in graph. Data can be found in Supplementary Table 1. c. Log2Fold change (graybars) of non-microglial genes specific for neurons, astrocytes and oligodendrocytes and show “derichment” in microglia compared to whole brain. d. Network analysis of the microglial Sensome by STRING identified a DAP12 centered pathway with 44/100 genes with direct or indirect interaction with DAP12. Of these, 24 have a direct interaction with Dap12 and are highlighted using a largerfont.
We used gene ontology analysis to classify the sensome transcripts into pattern recognition receptors (25%), chemoattractant and chemokine receptors (10%), Fc receptors (7%), purinergic receptors (8%), receptors for extracellular matrix (ECM) proteins (6%), cytokine receptors (10%), receptors involved in cell-cell interaction (10%), other receptors or transporters (13%) and potential sensome proteins with no known ligands (11%) (Supplementary Figure 1e and Supplementary table 1). Some of the receptors identified were expected considering that microglia have macrophage-like functions. These include CD11b, CD14, CD68, TLR2 and TLR7, CXCL16, various Fc receptors, and others. Interestingly, 32% of transcripts of the microglial Sensome have not been previously described in microglia. These include Entpd1, Tgfbr2, Cmtm7, Ly86, CD180, Slco2b1, Gi24, and Clec4a2, a complete list of these transcripts is highlighted in green in Supplementary table 1.
These data comprehensively identify, for the first time, the armamentarium of genes used by microglia in sensing their environment. The diversity of the ligands recognized by the surface proteins encoded by these genes provides strong evidence that support the broad role for microglia in homeostasis, host defense and response to injury. Of note, two of these sensome genes, TREM2 and CD33, were recently identified as risk factors for Alzheimer’s disease30–33, further attesting to the importance and relevance of our dataset.
To identify known and potential protein-protein interactions relevant to the sensome, we used the STRING 9.1 software and database 34. STRING quantitatively integrates protein interaction data from multiple sources for a large number of organisms, and transfers information between these organisms where applicable. The database currently covers 5,214,234 proteins from 1133 organisms. Using STRING we found that 44/100 sensome proteins have direct or indirect association with DAP12, an adaptor that regulates signaling via TREM2 35. Of these, 24 proteins appear to have direct association with DAP12 (Figure 1d). These proteins are Trem2, P2ry6, Fcer1g, Fcgr3, Fcgr1, Fcgr4, Clec4a3, Clec5a, Clec7a, Selplg, Csf1r, Cd14, Cd48, Cd52, Cd53, Cd68, Cd84, Cd86, Ly86, Itgb2, Tlr2, Ptprc, Emr1 and Il10ra. These data indicate that DAP12 may be an important regulator of the microglial sensing function through direct and indirect interaction with other sensome proteins.
Microglia vs. Macrophages
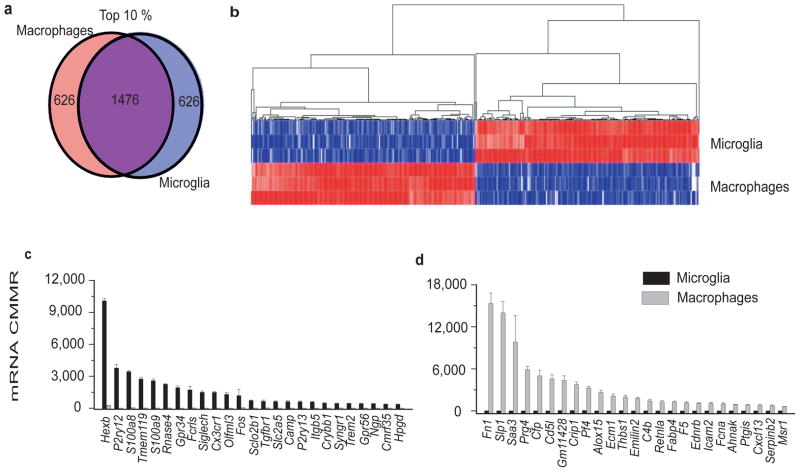
Both microglia and macrophages are resident tissue mononuclear phagocytes and share several functions including phagocytosis, production of reactive oxygen and nitrogen species, response to chemokines and purinergic stimuli 36. To identify similarities and differences between resting microglia and macrophages we used DRS to compare the transcriptome of peritoneal macrophages with that of microglia isolated from the same mice, and analyzed as described in the materials and methods. We compared the top 10% of transcripts with the highest expression on macrophages with those of microglia (Figure 2a). Of these 2102 transcripts, microglia and macrophages share 1476 transcripts indicating significant similarities between the two cell types (Figure 2a). Microglia and macrophages, however, also have significant differences in their transcriptomes and each express 626 transcripts that are not common to the other cell (Figure 2a). The heatmap shown in figure 2b clearly defines the set of transcripts that are unique to microglia and macrophages. Analysis of the top 25 transcripts uniquely expressed in microglia show expression levels ranging from 360–10,088 CMMR (Figure 2c). These transcripts are highly enriched compared to macrophages (p<0.00001, Log2 fold change ranging from 4.8–15.1) (Supplementary Table 2). These genes include several of the Sensome genes discussed above such as P2ry12 and P2ry13, Tmem119, Gpr34, Siglech, Trem2 and Cx3cr1 (Figure 2c and Supplementary Table 2). In addition, microglia highly express several unique transcripts that would not be expected to be expressed only in these cells. These include the enzyme Hexosaminidase B (HexB) and the antimicrobial peptides Camp and Ngp (Figure 2c and Supplementary table 2). The levels of expression of the top 25 transcripts unique to macrophages range from 596–15,327 CMMR (Figure 2d and Supplementary table 2, p<0.00001 for all included transcripts) with a Log2 fold change of 6.1–13.6, indicating a high level of enrichment regardless of the copy number of each transcript (Supplementary table 2). Macrophage-enriched genes include fibronectin, the chemokine Cxcl13, and the endothelin B receptor (Figure 2d and Supplementary Table 2).
Figure 2. Differences between microglia and macrophages revealed by DRS.
a. Venn diagram showing similarities and differences of the top 10% of transcripts expressed in microglia and macrophages. b. Heat map and hierarchal clustering of the transcripts that are unique to microglia or macrophages, showing a distinct signature for each of the cell types. c. The top 25 transcripts with the highest CMMR that are unique to microglia have barely detectable levels in macrophages (p<0.00001 for differences between microglia and macrophage expression). These top 25 transcripts show high level of enrichment (Log2Fold Change >4) over macrophages regardless of the level of expression in microglia. d. The top 25 transcripts unique to macrophages with the highest CMMR have barely detectable levels in microglia (p<0.00001 for differences between macrophages and microglia expression). These top 25 transcripts unique to macrophages show high level of enrichment (Log2Fold Change >5) over microglia regardless of the level of expression in macrophages. (Values in c–d mean ± SD of three different experiments done with microgliapooled from 22, 10and 20 mice, respectively and three pools of macrophages from 10 mice per pool) Data for Figure 1c–d can be found in Supplementary Table 2.
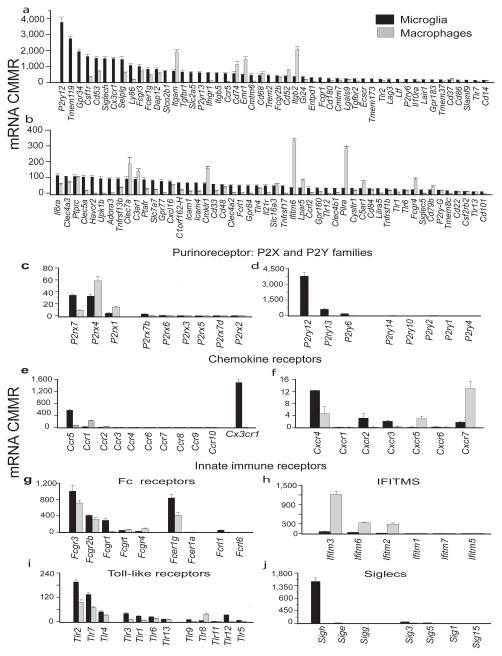
To identify microglial sensome transcripts that are also expressed in macrophages, we compared expression of these genes in the two cell types. Sensome genes that are expressed in both microglia and macrophages include Csfr1, CD53, Selplg, Fcgr3. Some microglial sensome genes have higher expression in macrophages. These include Itgam, CD74, Emr1, Itgb2, CD37, Clec7a, Cmklr1, Ifitm6, Pilra and Fcgr4. Interestingly, of the 22 sensome genes that are exclusively expressed on microglia, 16 genes interact with endogenous ligands rather than with pathogens (Figure 3a, b and Supplementary Table 2). These data imply that microglia express a unique set of genes, distinct from macrophages, that allow them to sense and interact with their local environment.
Figure 3. Comparative expression of the microglial and macrophages genes.
Comparison ofexpression levels of Sensome genes and those involved in regulating the immune response reveal a distinct immune signature for each cell type. a, b. Expression levels of the microglial Sensome in microglia and macrophages show that several genes are differentially expressed. c. Purinergic P2rx receptors. d. Purinergic P2ry receptors. e. Chemokine Ccr and Cx3cr1 receptors. f. Chemokine Cxcr receptors. g. Fc receptors. h. Interferon-inducible transmembrane (Ifitms). i. Toll-like receptors (Tlrs) 1–13. j. Sialic acid binding immunoglobulin lectins (Siglecs). (Values are mean ± SD of three different experiments done with microglia pooled from 22, 10 and 20 mice, respectively and three pools of macrophages from 10 mice per pool.) Data for Figure 3a–j can be found in Supplementary Table 2.
To further characterize the similarities and differences between macrophages and microglia we compared the levels of expression of several specific gene families involved in immune responses, including purinergic receptors (P2y and P2x), chemokine receptors, Fc receptors, Interferon-inducible transmembrane (Ifitms), Toll-like receptors (Tlrs), Sialic acid binding immunoglobulin lectins (Siglecs) and scavenger receptors (Figure 3c-j, Supplementary Figure 2 and Supplementary Table 2). These data show that microglia express significantly higher levels of several sensome genes that include P2rx7, P2ry12, P2ry13, P2ry6, Ccr5, Cx3cr1, Cxcr4, Cxcr2, Tlr2, Siglech and Siglec3 compared to macrophages (all p<0.00001). In contrast, macrophages express significantly higher levels of P2rx4, Ccr1, Cxcr7, Ifitm 2, 3 and 6 and Tlr8 (all p<0.00001). Notably, microglia express negligible levels of all Ifitms compared to macrophages.
Because DRS data is unbiased and quantitative, comparison of the transcriptomes of whole brain, microglia and macrophages allows us to identify a distinct gene signature for microglia and provide a more concrete molecular definition of these cells. Each signature includes a variety of genes with a wide range of functions. For ease of presentation, we have graphed the top 44 of these genes, their microglial and macrophage CMMR values, fold enrichment over brain (Log2FC) in supplementary figure 3. These genes not only reflect unique functional capabilities of microglia, but can also be used as microglial markers to identify these cells in physiologic conditions. Changes in expression levels of these genes under pathologic conditions could also be used as potential biomarkers for such conditions.
Validation of DRS by dual fluorescent in situ hybridization
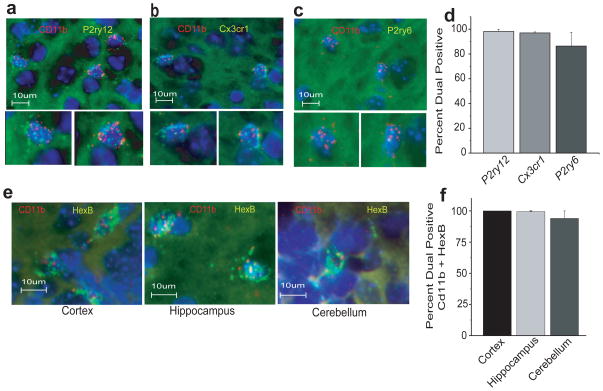
To confirm that microglial sensome genes are only expressed in microglia and not in other brain cells, we performed dual RNAscope, a dual fluorescent in situ hybridization technique 37. We used CD11b as a universal microglial marker and 3 microglial sensome genes P2ry12, Cx3cr1, and P2ry6 with high, intermediate and low expression in microglia, respectively (Figure 1a, b and Supplementary Table 1). P2ry12, Cx3cr1 and P2ry6 mRNA co-localize with CD11b mRNA in the brain parenchyma of young mice (Figure 4a–c). Ninety eight percent of cells expressing CD11b also express P2ry12 and Cx3cr1 and 87% also express P2ry6 (Figure 4d). Cells that do not express CD11b mRNA did not hybridize with probes for P2ry12, Cx3cr1 or P2ry6. These data support our DRS findings that microglial sensome genes are exclusively expressed in microglia in the brain.
Figure 4. RNAscope dual fluorescent in situ hybridization.
Dual RNAscope was performed on brain slices from adult mice for CD11b and sensome genes with high (P2ry12), intermediate (Cx3cr1) and low (P2ry6) levels of expression and for the highly expressed Hexb gene. The results confirm DRS findings and show exclusive expression in microglia and no expression in CD11b negative cells. a. Dual RNAscope for CD11b (red) and P2ry12 (green) probes, nuclei are stained with DAPI (blue). Bottom panels are magnified images of the double positive cells shown in the top panel. b. Dual RNAscope for CD11b (red) and Cx3cr1 (green) probes. Bottom panels are magnified images of the double positive cells shown in the top panel. c. Dual RNAscope for CD11b (red) and P2ry6 (green) probes. Bottom panels are magnified images of the double positive cells shown in the top panel. d. Quantitative image analysis of RNAscope data for CD11b+ cells with P2ry12 and Cx3cr1 and P2ry6. e. Dual RNAscope for CD11b (red) and Hexb (green) probes in the cortex, hippocampus and cerebellum. f. Nearly all of CD11b+ cells in the cortex, hippocampus and cerebellum, respectively, co-express HexB.
An unexpected finding revealed by our DRS analysis is that HexB is highly enriched in microglia compared to brain (Supplementary Figure 3a). To determine if HexB is predominantly expressed in microglia in situ, we performed dual RNAscope on mouse brains using probes for CD11b and HexB. As observed for P2ry12, Cx3cr1 and P2ry6, HexB mRNA also co-localizes with CD11b in the cortex, hippocampus and cerebellum (Figure 4e). Nearly all cells expressing CD11b also express HexB (Figure 4e, f). Cells that do not express CD11b mRNA did not hybridize with probes for HexB (Figure 4e and not shown). These data support our finding that HexB mRNA is exclusively expressed in microglia in the brain.
Proteomic analysis of microglia and macrophages
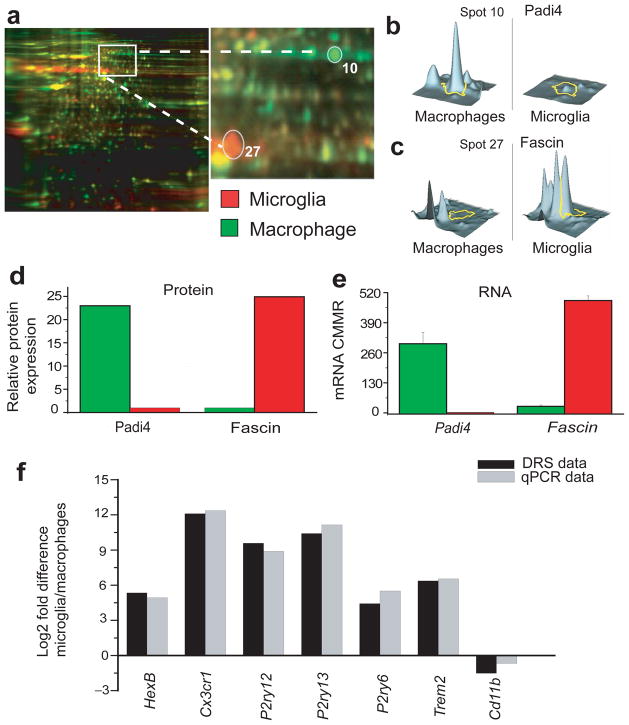
To determine if levels of mRNA transcripts compare with protein expression, we evaluated protein expression differences between microglia and macrophages by two dimensional difference gel electrophoresis (2D-DIGE), as described in materials and methods, and compared the results with DRS data for the proteins identified. Protein samples were extracted from each cell type and labeled with CyDye (microglia in red and macrophages in green), mixed and loaded onto the same 2D-electrophoresis gel. Red protein spots represented microglia-enriched proteins, green spots represented macrophage-enriched proteins and yellow spots represented proteins expressed in both cell types (Figure 5a). To insure unbiased analysis, we randomly selected 30 spots of interest (15 green and 15 red) excised them from the gel and protein ID was determined via mass spectrometry. As an example of such analysis we show one protein labeled in green (Figure 5a, Spot 10, Padi4) and one protein labeled red (Figure 5a Spot 27, Fascin). Three dimensional graphs of the levels of each protein expression in both cell types are shown in Figure 5b–c. The mRNA level for each of these proteins (CMMR) was obtained from our DRS data set. Protein and mRNA levels of both Padi4 and Fascin exhibited similar trends in expression (Figure 5d–e). Of the 30 proteins identified, 22 exhibited similar tends of expression between protein and mRNA levels (Supplementary Figure 4 and not shown). In the remaining 8 of the 30 proteins identified, significant differences were observed in the mRNA levels between microglia and macrophages but not in the protein levels (not shown). An advantage of this approach is that it allows the unbiased comparison between protein and mRNA levels since protein identification is made after the spots are selected. A limitation of this approach is that it only allows assessment of proteins that are intracellular or membrane proteins that are resistant to the proteases collagenase and dispase, the two enzymes we used to purify microglia. Nonetheless, our data show that expression levels of a majority of the proteins identified correlate with their respective mRNA levels, thereby providing added validation for our DRS analysis.
Figure 5. Proteomic analysis of microglia and macrophages.
a. Fluorescent 2D-DIGE of microglia (labeled in red) and macrophages (labeled in green) proteins showing common (labeled in yellow) and unique proteins for each cell type. Right panel shows an enlarged view of the area delineated in left panel. b. quantitative diagram of spot # 10 identified by mass spectrometry as Padi4 showing lower level of expression in microglia compared to macrophages. c. quantitative diagram of spot # 27 identified by mass spectrometry as fascin showing higher level of expression in microglia compared to macrophages. d, e. Comparison of protein levels (measured by mass spectrometry) and mRNA levels (measured by DRS) of Padi4 and fascin in macrophages and microglia. (DRS values are mean ± SD of three different experiments done with microglia pooled from 22, 10 and 20 mice, and from three pools of macrophages from 10 mice per pool, protein values are from pooled microglia and macrophages isolated from 70 and 50 mice, respectively). f. Validation by qPCR of some genes obtained with DRS on new cohorts of mice. The new cohorts comprised 5 sorted microglia pools and 6 macrophage pools from sorting of six sets using five mice per set. Ratio values for qPCR and DRS data represent the ratio of gene of interest to B2-micoglobulin expression. Data are plotted as log2 fold differencebetween microglia/macrophageratio values.
Quantitative PCR of select genes in microglia and macrophages
We further validated our DRS data by quantitative PCR (qPCR) using a new cohort of animals. Microglia and macrophages from thirty 5 months old mice were isolated as described above for DRS and divided into 5 and 6 pools, respectively. We then performed qPCR analysis on 7 transcripts including HexB, CX3CR1, P2ry12, P2ry13, P2ry6, TREM2 and CD11b. As seen in our DRS analysis, HexB, CX3CR1, P2ry12, P2ry13, P2ry6, TREM2 were highly expressed on microglia compared to macrophages (Figures 2a and 3a–b). In contrast CD11b is more expressed on macrophages than microglia (Supplementary Figure 1). We compared qPCR and DRS data by quantifying the Log2 of the ratio of microglia/macrophages. Figure 5f shows that the results obtained from qPCR and DRS are highly comparable indicating that our DRS data is strongly supported by qPCR data on a different set of mice. These results further confirm the validity of our DRS data.
Effects of aging on the microglial biological pathways
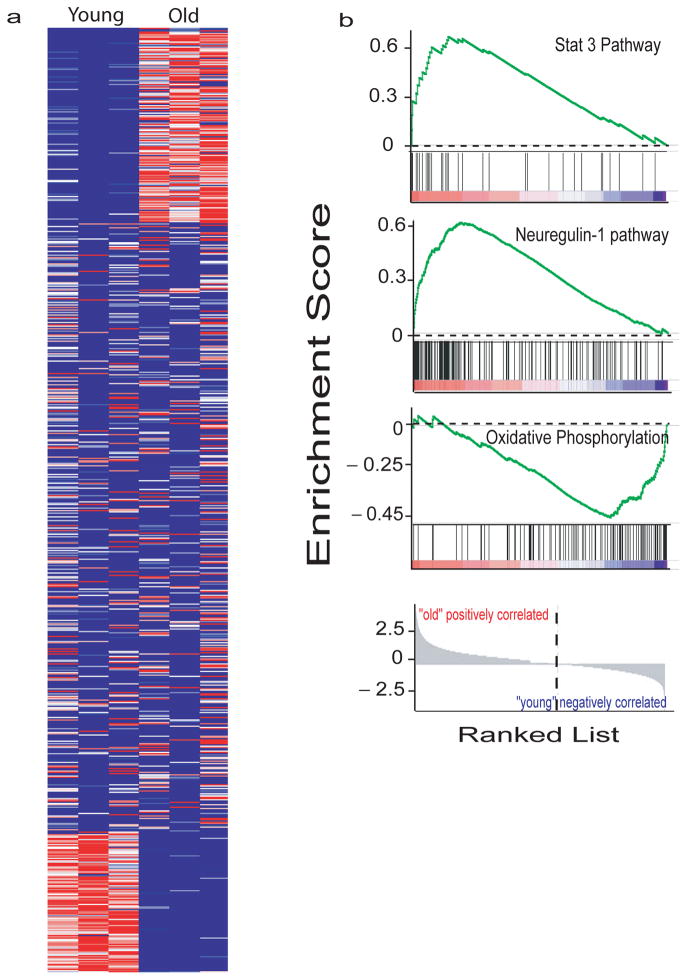
Aging is associated with significant alteration in the gene expression profile of the whole brain 38. To determine the effects of aging on the microglia transcriptome, we used DRS to identify the transcriptome of microglia isolated from 24-month old mice and compared it to that of microglia isolated from 5-month old mice as described above. We limited the analysis to transcripts expressed at ≥ 1CMMR. Of these 10,598 transcripts, 3503 transcripts were significantly different between young and old microglia (p<0.05), 1831 were upregulated, 1672 were downregulated and the remainder were not significantly changed in old vs. young (Figure 6a). We performed Gene set enrichment analysis (GSEA) (see supplementary methods) to identify pathways that are differentially changed in aging microglia. An interesting trend emerged from our analysis. Several pathways that have been described in the literature as promoting neurotoxicity appear to be down-regulated in aging. In contrast, pathways involved in neuroprotection appear to be upregulated. An example of such analysis is shown in figure 6b and supplementary figures 7–8. Genes involved in Stat 339 and Neuregulin-140 pathways are up-regulated in aging mice while those associated with oxidative phosphorylation 41 pathways are down regulated.
Figure 6. Effects of Aging on the microglial mRNA expression profile.
a. Heatmap of the 10598 microglial transcripts expressed at >1CMMR shows that 1831 transcripts were upregulated, 1672 were downregulated and 7095 remained unchanged with aging. b. GSEA pathways analysis showed upregulation of potentially neuroprotective pathways such as Stat 3 and Neuregulin-1 and downregulation of potentially neurotoxic pathways such as oxidative phosphorylation. Each bar at the bottom of each panel represents a member gene of the respective pathway and shows its relative location in the ranked list of genes (lowest panel).
Effects of Aging on the microglial priming state
Microglia exhibit various priming states that determine their responses to subsequent injury or infection of the brain. Interferon-γ induces microglia to assume the classical priming state, which is associated with a neurotoxic phenotype promoting neuronal degeneration. In contrast, the alternative priming state, associated with exposure of these cells to IL4 or IL13, is neuroprotective and promotes axonal sprouting and elongation 8, 12. Each priming state is characterized by increased expression of a defined set of genes that regulate microglial behavior and are considered markers for the corresponding priming state 11.
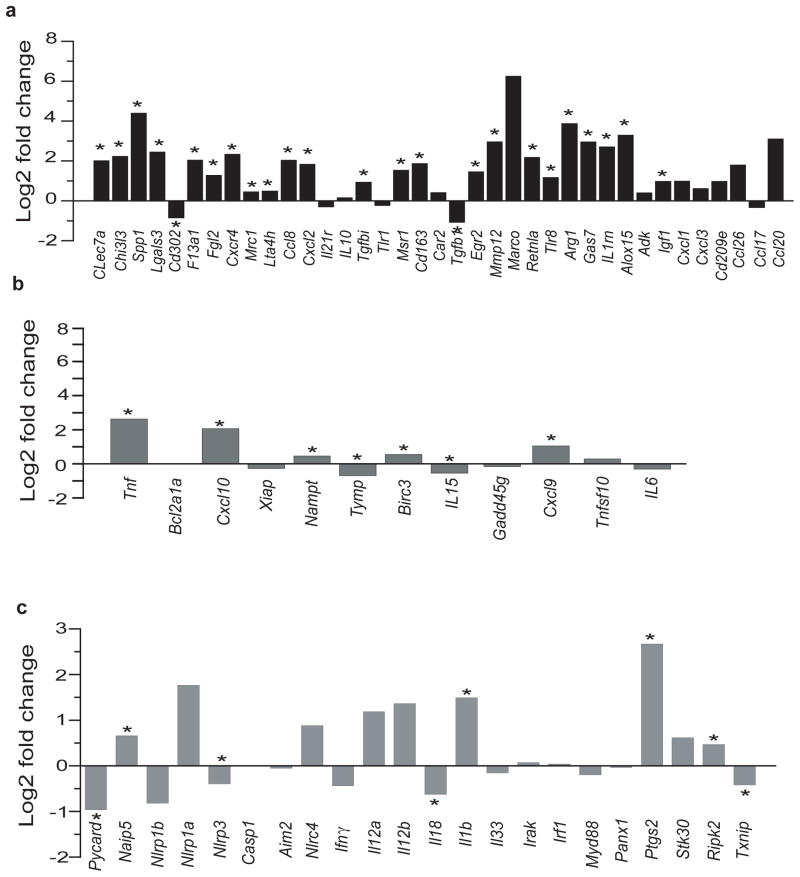
To determine the effect of aging on the microglial priming state we compared expression of classical and alternative priming genes 10, 42 in microglia isolated from 5 months vs. 24 months old mice. We found that in microglia from 24 months old mice, expression of alternative priming markers ranged between 6–348 CMMR and classical priming markers were expressed between 4 and 105 CMMR (Supplementary Table 3). Sixty-two percent (23/37) of alternative priming markers were significantly upregulated in microglia from aged mice (Mean Log2Fold change = 1.97, p<0.012), while 32% (12/37) were not significantly changed and only CD302 and TGFβ1 were down-regulated (Figure 7a and Supplementary table 3). In contrast, aging was associated with downregulation or no change in 7/12 (58%) of the classical priming state markers (Mean Log2FC=0.37) (Figure 7b and Supplementary Table 3). These data suggest that during aging there is a shift of the microglial phenotype towards an alternative neuroprotective priming state.
Figure 7. Upregulation of alternative priming genes in microglia from aged mice.
a,b. Alternative and classical priming genes in microglia from 24 months compared with mice 5 months of age old mice show a wide range of expression levels. a. In old mice, 24 of 37 alternative priming state markers were statistically significantly upregulated (*p<0.016). b. In 24 months old vs. microglia from 5 months old mice, 5 of 12 markers of the classical priming state were significantly upregulated in microglia, while the remaining 7 were down-regulated or not significantly changed. c. Analysis of 22 inflammasome-associated genes shows that 4 are significantly up-regulated in old mice compared with young ones (*p<0.025), while the remaining 18 genes are down-regulated or not significantly changed. Taken together the data suggest a trend toward increased expression of genes involved in resolution of inflammation and neuroprotection. Values are Log2 Fold change of three different experiments done with microglia pooled from 22, 10 and 20 young mice, respectively and from three pools of 10 mice per pool from old mice.. Data for this figure is found in Supplemental Table 3.
We also analyzed changes in expression of 22 inflammasome-associated genes. Interestingly 82% (18/22) of inflammasome genes included in our analysis were either significantly downregulated or not significantly changed (Figure 7c and Supplementary table 3). Because upregulation of some inflammasome genes such as NLRP3 have been implicated in the pathogenesis of Alzheimer’s disease 43 these findings further support that during normal aging there is a shift of the microglial phenotype towards an alternative neuroprotective priming state.
Effects of aging on the microglial sensome
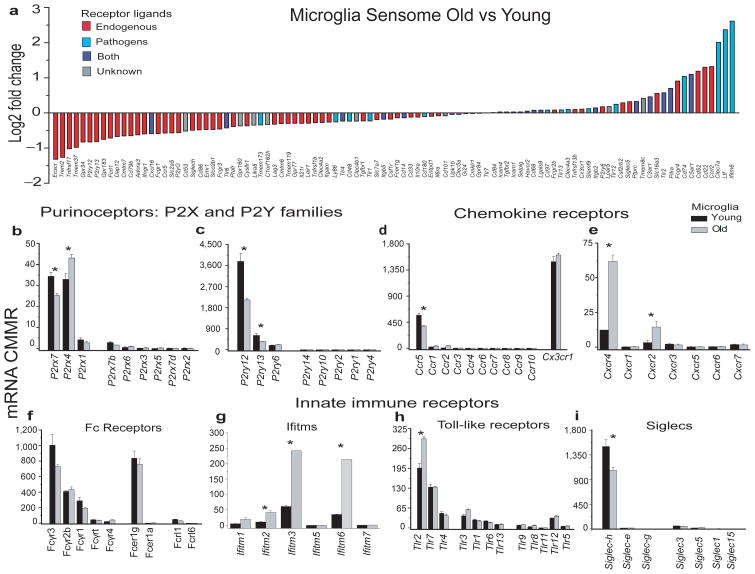
We also assessed any age-related changes in the microglial Sensome genes and in selected families of genes. We found that 31% (31/100) of the Sensome genes are significantly downregulated with aging (p<0.043), and 13% were significantly upregulated with aging (P<0.008) (Figure 8a and Supplementary table 3). Remarkably, 81% of the genes that were significantly downregulated (25/31 genes) encoded proteins involved in sensing endogenous ligands, while 62% (8/13) of the genes that were upregulated encoded proteins involved in sensing infectious microbial ligands (Figure 8a and Supplementary tables 1 and 3). Genes that were significantly downregulated encoded for proteins involved in sensing apoptotic neurons (Trem2, p<0.00005), substances released following neuronal injury such as nucleotides and adenosine and molecules expressed in the cell surface (siglech, p=0.003 and Dap12, p=0.00005) and soluble cytokines (Ccr5, p=0.0006 and Ifngr1, p=0.00033). Genes that were significantly upregulated encoded proteins involved in sensing of bacterial and fungal ligands (Tlr2, CD74, Ltf, Clec7a, Cxcl16 and Ifitm6, all p≤0.0006) or bacterial toxins (C5ar1, p<0.00005) (Figure 8a–i and supplementary tables 1 and 3).
Figure 8. The microglial sensome in aging.
a. measurement of the Log2Fold change of genes encoding the microglial sensome as determined by DRS show that ~81% of the genes are significantly downregulated (Escr1→Tmem173, p<0.043) and encoded proteins involved in sensing endogenous ligands (red bars). Of the 69 genes that are unchanged or upregulated, 45% encoded proteins involved in sensing infectious microbial ligands (blue bars and purple bars). Of the genes that are significantly upregulated ~62% (C3ar1→Ifitm6, p<0.008) encoded proteins involved in pathogen sensing and host defense. b-i. Comparative expression of genes involved in regulating the immune response in old vs. young microglia reveals a selective set of genes that are changed with normal aging. Values are mean ± SD of three different experiments done with microglia pooled from 22, 10 and 20 young mice, respectively and from three pools of microglia from 10 mice per pool., * indicates p<0.03.
Among the chemokine receptors, Cxcr4 and Cxcr2 were significantly (p≤0.0001) increased in aged microglia (Figure 8e and Supplementary table 3). Of note, there were significant changes in expression of members of the Ifitm family of innate immune receptors. Significant increases were seen in Ifitm2, 3 and 6 (Figure 8g, and Supplementary table 3 all p≤0.0001).
DISCUSSION
The data presented in this manuscript fill several major gaps in our understanding of microglia. First, the use of direct RNA sequencing allowed us to generate a quantitative dataset of the normal adult microglia transcriptome, representing an accurate snapshot of these cells’ gene expression profile. This is the only such dataset available at this point for any cell type of the mammalian brain. Our dataset avoids the potential pitfalls associated with other approaches to study gene expression because it does not involve cDNA synthesis or amplification. Our data also differ from semi-quantitative datasets since we provide copy numbers of each transcript rather than relative arbitrary units, allowing the possibility to compare with similar datasets generated in the future. The ability to perform such a comprehensive study using small amounts of unamplified RNA provides proof of concept that this approach can be applied to various cells from different healthy and diseased mammalian tissues. One can envision using DRS on cells obtained from biopsy samples providing a more personalized approach to understand gene expression profiling in the brain. The strength of our data is highlighted by the correlation between the copy number of each transcript obtained by DRS with in situ hybridization, with the relative protein expression of several proteins as determined by unbiased proteomic analysis by 2D-DIGE and mass spectrometry of 30 proteins, and by qPCR of several genes.
Second, we introduce a new area of investigation, namely that of the microglial Sensome. We identified 100 genes that constitute the microglial toolset for sensing changes in the brain’s milieu. Since sensing of the brain environment is a major function of microglia, our data define the apparatus that microglia use to perform these homeostatic functions including sensing of chemokines and cytokines, purinergic molecules, inorganic substances, changes in pH and amino acids. Defining the microglial Sensome under physiological conditions, establishes a baseline to which we can compare and identify changes that occur in this Sensome under pathological conditions. As an example we performed studies to identify changes in the Sensome that occur with aging and identified significant changes as shown above and discussed below. Defining changes in the Sensome that accompany infection, neurodegeneration, traumatic brain injury and inherited disorders is likely to have significant implications for identifying biomarkers, as well as novel therapeutic modalities for such disorders 44. Highlighting the importance of our dataset, two members of the microglial Sensome, TREM2 and CD33, have been found to be independent risk factors for late onset Alzheimer’s disease 30–32. We propose that several additional members of the microglial Sensome are also key players in a number of CNS disorders.
Third, because we compared the copy number of each transcript in microglia, whole brain and macrophages, we were able to identify a molecular signature that defines microglia and distinguishes these cells from other types of resident macrophages and other cells in the brain. Such analyses yielded some expected results such as the high level of expression of the purinergic receptor P2ry12, the chemokine receptor Cx3cr1, and Trem2 the receptor for apoptotic neurons in microglia. In addition, our data showed some very surprising and unexpected findings. For example, we found that the enzyme hexosaminidase B (HexB) is predominantly a microglial enzyme in the brain. The level of expression of this transcript in microglia indicates it is one of the most expressed mRNAs in these cells. HexB mRNA is enriched 164 fold in microglia compared to brain, more than the level of enrichment of the quintessential microglial marker Cx3cr1 (fold enrichment 142.5) and CD11b (fold enrichment 98.8), one of the two markers we used to identify microglia in our studies. These data indicate that HexB is indeed a microglial gene. We further confirmed this observation using dual RNAscope fluorescent in situ hybridization. Since HexB mutations cause the neurodegenerative gangliosidosis Sandhoff disease 45 this finding would have significant implications for our understanding of this disease, suggesting that this inherited disorder is essentially a “microglia” disorder. This suggests that microglia, rather than neurons, may be the better targets for therapeutic intervention in this disorder. This is an example of the usefulness of our data and their potential implications for significantly altering our understanding of microglial biology in sickness and in health. Another likely implication from these findings is the identification of several new microglial markers. In this regard, our data complements recent microarray data 46 and adds significantly to such data. Indeed, 32% of transcripts of the microglial Sensome have not been previously described in microglia.
Another important result in our dataset is the finding that resting microglia express high levels of several antimicrobial peptides not previously known to be expressed on these cells. These include Camp, the cathelin-related antimicrobial peptide (633 CMMR) and Ngp the neutrophilic granule protein (424 CMMR). The high level of expression of these peptides indicates a high level of readiness by the normal “quiescent” resident microglia to perform its innate host defense function in the absence of adaptive immune molecules and cells.
Fourth, we identified several age-related changes in the microglial transcriptome. For example, the microglial Sensome is significantly altered in aging. More than 81% of Sensome genes that were downregulated in aging are involved in sensing endogenous ligands. Of particular interest are purinergic and associated receptors such as P2yr12, P2ry13 and Adora3, and receptors recognizing apoptotic neurons and other cells such as Siglech. Purinergic molecules are released with neuronal injury and cell death, events that are likely to increase with aging. The continuous stimulation of microglia that could result from excessive neuronal injury and death that accompany aging, may initiate a cycle of events that leads to further microglial-mediated neurotoxicity adding “insult to injury”. It is therefore conceptually beneficial to the brain to downregulate the ability of microglia to get activated by dying or injured cells with aging. Interestingly, sensing genes involved in phagocytosis such as CD11b, CD14, ICAMs and CD68 are unchanged suggesting that the ability to clear endogenous debris is not affected. Similarly, genes that mediate the ability of microglia to sense microbial ligands are either not affected or increased. These data contrast with what happens in advanced stages of neurodegenerative disorders like Alzheimer’s disease, multiple sclerosis and amyotrophic lateral sclerosis, where microglia become neurotoxic 7–9. These data also contrast with recent findings of neurotoxic aging associated changes in the peripheral immune system 47. Our data suggest that with aging, there is a shift in the microglial phenotype towards a more neuroprotective type. While the peripheral immune system may become more primed towards a neurotoxic state 47, our data suggest that the microglia maintains its ability to defend against infectious pathogens and clear debris but attempts to “tone down” the stimulatory effects of endogenous debris as if to keep from becoming constantly activated.
On its face our data appear to conflict with reports that suggest that aging is accompanied by an overall increase in pro-inflammatory status of the brain 48–50. An important distinction between these studies and ours is that they were done using whole brain tissue rather than purified cells. Changes observed in these studies reflect not only the gene expression profiles of microglia but of other glial cells, and possibly circulating blood leukocytes. Furthermore, these studies either included a limited number of genes 48 or involved injecting the brain with a cocktail of cytokines prior to gene expression analysis 50. These technical differences may explain the differences in our results and these published reports.
In support of an aging-associated change in the microglial phenotype towards a more neuroprotective state is our finding that several potential neurotoxic pathways such as the oxidative phosphorylation pathway are downregulated in microglia with aging 41. In contrast pathways that may be neuroprotective such as the STAT3 and Neuregulin-1 pathways are upregulated. Furthermore, the microglial priming state is also altered in aging towards an alternative phenotype. Classical and alternative priming states are not activation states but rather priming states that dictate how the microglia will respond to a certain stimulus. In spinal cord injury, for example, an alternative priming state is associated with improved axonal sprouting and elongation indicating a neuroprotective phenotype 12. Genes that are upregulated in an alternative primed state such as MRC1, Dectin and Lgals3 favor phagocytosis of debris and pathogens. Other genes such as Arg1 and IL1rn downregulate the innate response possibly reducing the damage associated with such a response. Arg1 induces a shift in arginine metabolism from interferon-γ-induced NO production towards production of ornithine and polyamine which are important in wound healing. Similarly, IL1rn antagonizes the effects of IL1 10. Taken together our data suggest that aging is associated with a shift in the microglial gene expression profile towards a more neuroprotective phenotype. We propose that when the brain is faced with an injury or stimulus that acutely activates the microglia, the alternative primed state assumed by these cells with aging may not completely prevent the damages induced by the injurious stimuli, but will likely help reduce the effects of such damage.
Our approach and data presented in this manuscript are a major step towards establishing a definitive quantitative microglia transcriptome under a variety of pathological situations. Understanding the changes that occur in the microglia in aging, neurodegeneration, infection and traumatic injury is the first step in identifying therapeutics that modulate the state of these cells and ultimately alter disease processes.
Supplementary Materials and Methods
Mice
C57Bl/6 mice 5 months and 24 months of age were purchased from the National Institute on Aging and housed in the animal care facility at Massachusetts General Hospital and used within two weeks of arrival. Mice were euthanized according to approved institutional procedures. All protocols were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee and met US National Institutes of Health guidelines for the humane care of animals.
Isolation of peritoneal macrophages and microglia
Peritoneal macrophages
Peritoneal cells were harvested from 5-month old mice by peritoneal lavage with 10ml phosphate buffer saline without Ca++ and Mg++ (PBS=) containing 1 mM EDTA (PBS=/EDTA) and centrifuged at 400×g for 10 mins. The peritoneal cells were then stained for CD11b and CD45for flow cytometry (see below). Each sorting experiment was performed on cells pooled from 5 mice per experiment.
Microglia
Two ages of mice were used for isolation of microglia: 5 months old and 24 months old. Mice were perfused with 50cc PBS=. Brains were then removed, rinsed in PBS= and placed separately into a GentleMacs™ C-tube (Miltenyi Biotech) with RPMI (no phenol red) containing 2mM L-glutamine (Mediatech), Dispase (2U/ml) and 0.2% Collagenase Type 3 (Worthington Biochemicals) according to our previous protocol 7. Brains were processed using the gentleMACS Dissociator (Miltenyi Biotech) on the brain program settings according to manufacturer’s directions. Briefly, the brains were subjected to three rounds of dissociation each followed by a period of incubation at 37°C. After the second round of dissociation, DNase I grade II (Roche Applied Science) was added to a concentration of 40U/ml and incubated for an additional 10 minutes before the final round of dissociation. The digestion enzymes were inactivated by addition of PBS=/EDTA containing 5% fetal bovine serum (FBS) and the digested brain bits were triturated gently, passed over a 100μm filter (Fisher Scientific) and centrifuged. Cell pellets were resuspended in 10.5 ml RPMI/L glutamine, mixed gently with 4.5 ml physiologic Percoll® (Sigma Aldrich), and centrifuged at 850×g for 40 minutes. The resulting cells were rinsed in PBS= and centrifuged. At this time any contaminating red blood cells were lysed using RBC lysis buffer (Sigma) according to manufacturer’s instruction, rinsed twice with PBS= and passed over a 40μm filter (Fisher Scientific). Cells were then stained for CD11b and CD45 for flow cytometry (see below). Each sorting experiment was performed on cells pooled from 5 mice per experiment.
Staining for CD11b and CD45 and Fluorescence Activated Cell Sorting (FACS)
After isolations, cell pellets were resuspended in blocking buffer (PBS=/1mM EDTA/2% donkey serum) containing 1ug/ml Fc block (BD Pharmingen) and incubated on ice for 10 mins. Cells were co-stained for 30 minutes on ice with Alexa-647 labeled anti-CD11b (clone M/170, Biolegend, final conc 5 ug/ml) and Alexa488-labeled CD45 (Biolegend, clone 30-F11, final conc 5 ug/ml). Cells were then rinsed in PBS/EDTA centrifuged, resuspended in PBS/EDTA containing1% fetal bovine serum and filtered into 5 ml polystyrene filter top tubes (BD Falcon) for sorting. Cells were sorted based on CD11b/CD45 expression 24 using FACS ARIA(BD). Sorted cells were centrifuged 400×g for 10 minutes and pellets were either lysed in RLT-Plus buffer (Qiagen) for RNA extraction or stored at −80°C for protein extraction and proteomics analysis.
RNA and DRS
RNA was isolated from flow cytometry-sorted cell populations using RNeasy Plus micro/mini kits (Qiagen) depending on cell number. For RNA from whole brains, 6 mice were euthanized, perfused with PBS=/EDTA and brains were removed and homogenized in Qiazol using the TissueShredder (Qiagen) and RNA isolated with RNeasy Plus Minikit (Qiagen). Three pools of two brains per pool were created for whole brain RNA. Purified RNAs were quantified with a Nandrop 2000 (Thermo Scientific) and the quality of the RNA assessed using an Agilent Bioanalyzer (Agilent), all RNA used had an RNA Integrity Number (RIN)>9.2. RNAs from each population set were pooled as needed to yield samples containing at least 300ng of RNA. There were three pools of microglia RNA from 22, 10, and 20 5-month old mice, three pools of RNA from 24-month old mice comprising 10 mice per pool, and three pools of macrophage RNA that represented 10 mice each. DRS was performed by Helicos (Cambridge, Massachusetts) using Helicos single molecule prototype sequencers 22, 51–53. On average 14,376,408 and 19,007,620 reads were obtained from each microglial and macrophage samples respectively. Of these, 8,480,397 reads and 11,946,274 were annotated for microglia and macrophages respectively. RNA samples that were sent to Helicos were coded such that the identity of the cell types was not known.
RNAscope
RNA scope is a novel RNA in-situ hybridization technique that allows visualization of two RNA biomarkers within single cells. RNAscope uses a unique probe design strategy that results in signal amplification and background suppression 37. RNA scope was performed at Advanced Cell Diagnostics where the technique was developed. Briefly, frozen brains from C57Bl6 mice were cut horizontally and slices were hybridized with several dual probes sets using CD11b conjugated to Alexa546 as the common probe in each set. The companion probes to CD11bAlexa546were HexB, Cx3cr1, P2ry6, and P2ry12 (all conjugated to Alexa-488) and nuclei were visualized using DAPI. The resulting hybridized brain slices were imaged using the Mirax MIDI slide scanner to visualize the entire slice. To quantify cells that expressed both CD11b and other genes of interest, first CD11b positive cells were identified and annotated using Panoramic Viewer 1.15.2 software (3D Histech Limited); for each brain slice 100 cells were counted in cortex, 100 cells in the hippocampus, and 50 cells in the cerebellum. The number of double positive cells was then quantified. Each data point represents mean ± SD of 2–3 brain slices for each probe set. Dual RNAscope experiments were repeated twice on different slide sets with similar results.
Bioinformatics Analysis
Raw reads were filtered to remove low-quality reads and those that do not meet a filter length. For each sample, Helicos provided the alignment files which also contain the query and reference sequences. Each sample alignment file was processed individually by a program written in MolBioLib 25 to determine which gene(s) (if any) were hit by each alignment. The number of hits was recorded both in raw units as well as in copies per million mapped reads [CMMR]-normalized units 54. Only the hits to the sense of the genes are kept. The raw counts were analyzed using EdgeR 27, 55, 56 to generate p-values. EdgeR estimates the common negative binomial dispersion by conditional maximum likelihood. The log2 of the CMMR-normalized values were used as input to discover signatures for class types by using the gene expression monitoring scoring system 57. Scores are sorted with the largest score indicating genes that are most enriched in the class in question and least enriched outside of that class. The gene set enrichment analysis was performed using the GSEA software 26, 58 using the complete set of normalized input values, the c2.all.v3.0.symbols.gmt curated gene set, 1000 gene permutations, and using the classes (e.g. old versus young) as the phenotype. (The gene symbol “chip” was selected as the chip platform.) All processing unless otherwise noted was done in the MolBioLib framework. For all statistical analysis, differences were considered statistically significant if p values calculated by EdgeR as well as student T-test were <0.05.
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those generally employed in the field.
Proteomics
2-D DIGE and Mass spectrometry protein identification were performed by Applied Biomics (Hayward, CA). Proteomic analysis was done on microglial and macrophage lysates using pooled cells isolated from 70 mice and 50 mice, respectively.
Cell lysate preparation
Frozen cell pellets were lysed in 200 ul 2-D cell lysis buffer (30 mM Tris-HCl, pH 8.8, containing 7 M urea, 2 M thiourea and 4% CHAPS). The cells were sonicated at 4° C, incubated on a shaker for 30 minutes at room temperature, followed by centrifugation for 30 min at 14,000 rpm. Supernatants were collected and protein concentrations determined by the Bio-Rad Protein Assay (Biorad). Sample lysates were diluted with the sample 2-D cell lysis buffer to the same protein concentration of 3 to 8 mg/ml.
Minimal CyDye labeling
To 30 ug of cell lysate, 1.0 ul of diluted CyDye (Cy2, Cy3 or Cy5) (1:5 diluted with DMF from 1 nmol/ul stock) was added, incubated on ice for 30 min in the dark, followed by addition of 1.0 ul of 10 mM Lysine to each of the samples and incubation on ice in the dark for additional 15 min. Cy2, Cy3 and Cy5labeled samples were mixed together followed by addition of 2X 2-D Sample buffer (8 M urea, 4% CHAPS, 20 mg/ml DTT, 2% pharmalytes and trace amount of bromophenol blue), and loaded onto a 13 cm IPG strip.
IEF and SDS-PAGE
After loading the labeled samples into the strip holder, IEF was run in the dark at 20 °Caccording to protocol provided by Amersham BioSciences. Upon finishing the IEF, the IPG strips were incubated in equilibration buffer 1 (50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue and 10 mg/ml DTT) for 15 minutes with slow shaking. Then the strips were rinsed in equilibration buffer 2 (50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% glycerol, 2% SDS, trace amount of bromophenol blue and 45 mg/ml Iodacetamide) for 10 minutes with slow shaking. The IPG strips were then rinsed once in the SDS-gel running buffer before being transferred into the SDS-Gel (12% SDS-gel prepared using low florescent glass plates) and sealed with 0.5% (w/v) agarose solution (in SDS-gel running buffer). The SDS-gels were run at 15 °C and until the dye front reached the end of the gel.
Image scan and data analysis
Image scans were carried out immediately following the SDS-PAGE using Typhoon TRIO (Amersham BioSciences) following the protocols provided. The scanned images were then analyzed by Image QuantTL software (GE-Healthcare), and then subjected to in-gel analysis and cross-gel analysis using DeCyder software version 6.5 (GE-Healthcare). The ratio change of the protein differential expression was obtained from in-gel DeCyder software analysis.
Spot picking and Trypsin digestion
The spots of interest were picked up by Ettan Spot Picker (GE Healthcare) based on the in-gel analysis and spot picking design by DeCyder software. The gel spots were washed, followed byin-gel digestion with modified porcine trypsin protease (Trypsin Gold, Promega). The digested tryptic peptides were desalted by Zip-tip C18 (Millipore). Peptides were eluted from the Zip-tip with 0.5 ul of matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/ml in 50% acetonitrile, 0.1% trifluoroacetic acid, 25 mM ammonium bicarbonate) and spotted on the MALDI plate.
Mass Spectrometry
MALDI-TOF (MS) and TOF/TOF (tandem MS/MS) were performed on a 5800 mass spectrometer (AB Sciex). MALDI-TOF mass spectra were acquired in reflectron positive ion mode, averaging 2000 laser shots per spectrum. TOF/TOF tandem MS fragmentation spectra were acquired for each sample, averaging 2000 laser shots per fragmentation spectrum on each of the 5–10 most abundant ions present in each sample (excluding trypsin autolytic peptides and other known background ions).
Database search
Both the resulting peptide mass and the associated fragmentation spectra were submitted to GPS Explorer version 3.5 equipped with MASCOT search engine (Matrix science) to search the database of National Center for Biotechnology Information non-redundant (NCBInr). Searches were performed without constraining protein molecular weight or isoelectric point, with variable carbamidomethylation of cysteine and oxidation of methionine residues, and with one missed cleavage allowed in the search parameters. Candidates with either protein score C.I.% or Ion C.I.% greater than 95 were considered significant.
Quantitative Real Time PCR
Microglia and peritoneal macrophages were isolated from a different cohort of 5-month old mice than the ones used for DRS comprising 6 sets of 5 mice per set. The sorts resulted in 6 pools of macrophages and 5 pools of microglia. Total RNA from each sample of cells (3.0–8.0 ×105 cells) was isolated using the RNeasy® Plus micro kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions and reverse transcribed using Multiscribe™ reverse transcriptase (Applied Biosystems, Foster City, CA). Dilutions of each cDNA prep were used to assess β2-microglobulin RNA levels and samples were then adjusted to give equivalent levels of β2-microglobulin per well in subsequent qPCR reactions for other genes. The qPCR was performed in a Roche 480 Lightcycler qPCR machine (Indianapolis, IN) in duplicates using SYBR Green to detect the amplification products. The following cycles were performed: initial denaturation cycle 95°C for 10 min, followed by 40 amplification cycles of 95°C for 15 secs and 60°C for one min and ending with one cycle at 25°C for 15 secs. Relative quantification of mRNA expression was calculated by the comparative cycle method to obtain the ratio of gene interest/B2M.
The following are genes and primers used in these experiments:
Cd11b (GTGTGACTACAGCACAAGCCG, CCCAAGGACATATTCACAGCCT)
Cx3cr1 (ACCGGTACCTTGCCATCGT, ACACCGTGCTGCACTGTCC)
HexB (ACTCCAAGATTATGGCCTCGAGCA, AGCTATTCCACGGCTGACCATTCT)
B2M (CCGAACATACTGAACTGCTACG, CCCGTTCTTCAGCATTTGGA)
P2ry6 (CTGCGTCTACCGTGAGGATT, GCAATGACGCAGATGTTCAG)
P2ry12 (CACGGATTCCCTACACCCTG, GGGTGCTCTCCTTCACGTAG)
P2ry13 (AACAAAGCTGATGCTCGGGA, GTGTCATCCGAGTGTCCCTG)
Trem2 (GCCTTCCTGAAGAAGCGGAA, GAGTGATGGTGACGGTTCCA
Supplementary Material
Supplementary Figure 1. Overview of cell characterization and experimental flow.
Supplementary Figure 2. Comparative expression of scavenger receptor genes in microglia (black) and macrophages (red).
Supplementary Figure 3. Molecular signature of microglia
Supplementary Figure 4. Comparison of expression of proteins identified by 2D DIGE with mRNA levels in microglia and macrophages
Supplementary Figure 5. NRG1 and Stat3 are examples of neuroprotective pathways upregulated in microglia from old mice.
Supplementary Figure 6. Oxidative phosphorylation is an example of a potential neurotoxic pathway downregulated in microglia from old mice.
Supplementary Table 1. The Microglia Sensome
Supplementary Table 2. Microglia Sensome Gene Expression in Macrophages
Supplementary Table 3. Young vs. Old Microglia
Acknowledgments
This work was supported by grants NS059005 from NINDS and AG032349 from NIA to JEK. We thank Dr. Fatih Ozsolak at Helicos for supervising the sequencing experiments and Dr. John Liao at Applied Biomics for analysis of the proteomic data.
Footnotes
Note: Supplementary Information is linked to the online version of the paper.
ACCESSION CODES
The Direct RNA sequencing data has been deposited in the NCBI BioProject database under the BioProject ID code PRJNA219501 (URL: http://www.ncbi.nlm.nih.gov/bioproject/219501).
AUTHOR CONTRIBUTIONS
Suzanne Hickman co-designed the experiments performed all cell isolations from peritoneum and brain, RNA extractions and quality analysis and co-wrote the manuscript. Nathan Kingery assisted in all cell isolations and sorting for DRS and proteomics studies. Toshi Ohshumi and Mark Borowsky performed bioinformatics analyses, including development of programs in MolBioLib to annotate sequence information obtained from Helicos. Li-chong Wang performed the RNAscope experiments. Terry Means was involved in designing experiments and data analysis. Joseph El Khoury, co-designed the experiments, analyzed the data and co-wrote the manuscript.
References
- 1.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–70. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- 2.El Khoury J, et al. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13:432–8. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- 3.Rezaie P, Male D. Mesoglia & microglia--a historical review of the concept of mononuclear phagocytes within the central nervous system. J Hist Neurosci. 2002;11:325–74. doi: 10.1076/jhin.11.4.325.8531. [DOI] [PubMed] [Google Scholar]
- 4.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–8. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 5.Block ML, Zecca L, Hong JS. Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat Rev Neurosci. 2007;8:57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 6.Gomes-Leal W. Microglial physiopathology: how to explain the dual role of microglia after acute neural disorders? Brain Behav. 2:345–56. doi: 10.1002/brb3.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J Neurosci. 2008;28:8354–60. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao B, Zhao W, Beers DR, Henkel JS, Appel SH. Transformation from a neuroprotective to a neurotoxic microglial phenotype in a mouse model of ALS. Exp Neurol. 2012;237:147–152. doi: 10.1016/j.expneurol.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muzio L, Martino G, Furlan R. Multifaceted aspects of inflammation in multiple sclerosis: the role of microglia. J Neuroimmunol. 2007;191:39–44. doi: 10.1016/j.jneuroim.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Colton CA. Heterogeneity of microglial activation in the innate immune response in the brain. J Neuroimmune Pharmacol. 2009;4:399–418. doi: 10.1007/s11481-009-9164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kigerl KA, et al. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J Neurosci. 2009;29:13435–44. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berchtold NC, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–10. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremsky I, Morgan TE, Hou X, Li L, Finch CE. Age-changes in gene expression in primary mixed glia cultures from young vs. old rat cerebral cortex are modified by interactions with neurons. Brain Behav Immun. 26:797–802. doi: 10.1016/j.bbi.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–26. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 17.Schena M, et al. Microarrays: biotechnology’s discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–6. doi: 10.1016/s0167-7799(98)01219-0. [DOI] [PubMed] [Google Scholar]
- 18.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 19.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 20.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagalakshmi U, Waern K, Snyder M. RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol. Chapter 4(Unit 4):11, 1–13. doi: 10.1002/0471142727.mb0411s89. [DOI] [PubMed] [Google Scholar]
- 22.Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–8. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 23.El Khoury JB, et al. CD36 mediates the innate host response to beta-amyloid. J Exp Med. 2003;197:1657–66. doi: 10.1084/jem.20021546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sedgwick JD, et al. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc Natl Acad Sci U S A. 1991;88:7438–42. doi: 10.1073/pnas.88.16.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohsumi TK, Borowsky ML. MolBioLib: A C++11 Framework for Rapid Development and Deployment of Bioinformatics Tasks. Bioinformatics. 2012;28:2412–6. doi: 10.1093/bioinformatics/bts458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Khoury J, et al. Scavenger receptor-mediated adhesion of microglia to beta-amyloid fibrils. Nature. 1996;382:716–9. doi: 10.1038/382716a0. [DOI] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2012;368:117–27. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2012;368:107–16. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradshaw EM, et al. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–50. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griciuc A, et al. Alzheimer’s Disease Risk Gene CD33 Inhibits Microglial Uptake of Amyloid Beta. Neuron. 2013 doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Mering C, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–61. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thrash JC, Torbett BE, Carson MJ. Developmental regulation of TREM2 and DAP12 expression in the murine CNS: implications for Nasu-Hakola disease. Neurochem Res. 2009;34:38–45. doi: 10.1007/s11064-008-9657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 37.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–9. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colantuoni C, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–23. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dziennis S, Alkayed NJ. Role of signal transducer and activator of transcription 3 in neuronal survival and regeneration. Rev Neurosci. 2008;19:341–61. doi: 10.1515/revneuro.2008.19.4-5.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Z, et al. Neuroprotection by neuregulin-1 following focal stroke is associated with the attenuation of ischemia-induced pro-inflammatory and stress gene expression. Neurobiol Dis. 2005;19:461–70. doi: 10.1016/j.nbd.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds A, Laurie C, Mosley RL, Gendelman HE. Oxidative stress and the pathogenesis of neurodegenerative disorders. Int Rev Neurobiol. 2007;82:297–325. doi: 10.1016/S0074-7742(07)82016-2. [DOI] [PubMed] [Google Scholar]
- 42.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–83. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 43.Heneka MT, et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2012;493:674–8. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Khoury J. Neurodegeneration and the neuroimmune system. Nat Med. 2010;16:1369–70. doi: 10.1038/nm1210-1369. [DOI] [PubMed] [Google Scholar]
- 45.Sango K, et al. Mouse models of Tay-Sachs and Sandhoff diseases differ in neurologic phenotype and ganglioside metabolism. Nat Genet. 1995;11:170–6. doi: 10.1038/ng1095-170. [DOI] [PubMed] [Google Scholar]
- 46.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–28. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villeda SA, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–4. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cribbs DH, et al. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J Neuroinflammation. 2012;9:179. doi: 10.1186/1742-2094-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–7. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 50.Lee DC, et al. Aging enhances classical activation but mitigates alternative activation in the central nervous system. Neurobiol Aging. 2013;34:1610–20. doi: 10.1016/j.neurobiolaging.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozsolak F, et al. Digital transcriptome profiling from attomole-level RNA samples. Genome Res. 2010;20:519–25. doi: 10.1101/gr.102129.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozsolak F, et al. Amplification-free digital gene expression profiling from minute cell quantities. Nat Methods. 2010;7:619–21. doi: 10.1038/nmeth.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 55.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–7. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 56.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–32. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 57.Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 58.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Overview of cell characterization and experimental flow.
Supplementary Figure 2. Comparative expression of scavenger receptor genes in microglia (black) and macrophages (red).
Supplementary Figure 3. Molecular signature of microglia
Supplementary Figure 4. Comparison of expression of proteins identified by 2D DIGE with mRNA levels in microglia and macrophages
Supplementary Figure 5. NRG1 and Stat3 are examples of neuroprotective pathways upregulated in microglia from old mice.
Supplementary Figure 6. Oxidative phosphorylation is an example of a potential neurotoxic pathway downregulated in microglia from old mice.
Supplementary Table 1. The Microglia Sensome
Supplementary Table 2. Microglia Sensome Gene Expression in Macrophages
Supplementary Table 3. Young vs. Old Microglia