Abstract
We present a spatio-temporal assessment of microRNA expression throughout early human brain development. We assessed the prefrontal cortex, hippocampus, and cerebellum of 18 normal human donor brains spanning infancy through adolescence by RNA-seq. We discovered differentially expressed microRNAs and broad microRNA patterns across both temporal and spatial dimensions, and between male and female prefrontal cortex. Putative target genes of the differentially expressed miRNAs were identified, which demonstrated functional enrichment for transcription regulation, synaptogenesis, and other basic intracellular processes. Sex-biased miRNAs also targeted genes related to Wnt and TGF-β pathways. The differentially expressed miRNA targets were highly enriched for gene sets related to autism, schizophrenia, bipolar disorder, and depression, but not neurodegenerative diseases, epilepsy, or other adult-onset psychiatric diseases. Our results suggest critical roles for the identified miRNAs in transcriptional networks of the developing human brain and neurodevelopmental disorders.
Keywords: microRNA, brain development, gene expression, sex differences
Introduction
Human neurodevelopment requires coordinated expression of thousands of genes, exquisitely regulated in both spatial and temporal dimensions, to achieve the proper specialization and inter-connectivity of brain regions. Consequently, the dysregulation of complex gene networks in the developing brain is thought to underlie many neurodevelopmental and psychiatric disorders (1). In order to understand these pathologic gene expression changes, it is critical to achieve a comprehensive understanding of normal gene expression regulation throughout human neurodevelopment. While broad surveys of gene expression across the developing human brain have recently been described (2), the molecular regulators of this gene expression--most notably microRNAs-- have only been assessed in a few brain regions or developmental periods (3–6). As microRNAs are increasingly recognized in fundamental brain developmental processes and neurologic diseases (7), a comprehensive understanding of their expression dynamics throughout human brain development is important.
Therefore, we analyzed the differential expression of all microRNAs (miRNAs) detected by RNA-sequencing of 82 neurologically-normal post-mortem human brain tissue samples, which derived from 18 individual donor brains spanning 4 months through 19 years of age (see Methods). Donor samples were grouped into four developmental time windows (infancy, early childhood, late childhood, and adolescence, Table 1). Six distinct brain regions were assessed: four regions of the prefrontal cortex, the hippocampus, and the cerebellum. We also assessed for differential miRNA expression between males and females in the prefrontal cortex. Then, we identified putative gene targets of the differentially expressed miRNAs, determined if these gene targets were enriched for particular functional processes, and finally assessed if the identified targets were enriched for genes associated with common neurodevelopmental, psychiatric, and neurodegenerative diseases.
Table 1.
Developmental periods and average number of donor tissue samples assessed.
| Developmental Period | Ages | Avg. Samples per Region |
|---|---|---|
| Infancy | 4 months – 1 year | 3.5 |
| Early Childhood | 2 – 4 years | 3.0 |
| Late Childhood | 8 – 13 years | 2.8 |
| Adolescence | 15 – 23 years | 4.3 |
Methods
miRNA Data and Pre-processing
All data was obtained from the Allen Institute for Brain Science BrainSpan Atlas of the Developing Human Brain (www.brainspan.org). Details of tissue acquisition, processing, and RNA-sequencing can be found on the BrainSpan website. Data was downloaded at: download.alleninstitute.org/brainspan/MicroRNA. The full dataset contained 1620 miRNAs measured across 215 brain samples. Only brain samples originating from the orbitofrontal prefrontal cortex (OFC; Brodmann’s Area (BA) 11), dorsolateral prefrontal cortex (DFC; BA 9, 46), medial prefrontal cortex (MFC; BA 32, 33, 34), ventrolateral prefrontal cortex (VFC; BA 44,45), hippocampus (HIP), or cerebellum (CER) were retained (82 total samples, Supplementary Table 11). For analysis between sexes, brain regions were aggregated from the prefrontal cortex samples. Next, miRNAs with read counts likely to be noise rather than true reads were removed, in order to increase subsequent statistical power; this has been demonstrated not to affect the dispersion model used to calculate differential expression (8). Importantly, we did this prior to any analysis of the data. To do so, the sum total of read counts for each miRNA across all 82 samples was calculated. miRNAs with zero total reads were immediately discarded (58 miRNAs). Next, miRNAs were ordered from most to least reads (range 1 to 41,540,463) and the dispersion of read counts was plotted for visualization. Then, any miRNA with a total read count less than 60 was discarded, resulting in 902 retained miRNAs (Supplementary Table 11).
miRNA Differential Expression Analysis
Differentially expressed miRNAs were discovered using the edgeR package (9) run in the R programming environment. The edgeR user guide was followed as detailed in the “classic analysis” section. We chose this method to evaluate differential expression because its performance is intermediately conservative among various RNA-seq analysis packages (10). miRNAs were considered to be significantly differentially expressed between groups only if the false discovery rate (FDR) p-value was < 0.05 and the absolute log2 of Fold Change (FC) was > 1.5. Differentially expressed miRNAs were identified across three dimensions: spatial, temporal, and by sex. Spatial miRNAs were differentially expressed between two anatomic brain regions within one developmental time period. Temporal miRNAs were differentially expressed over developmental time within the one anatomic brain region. Sex-biased miRNAs were differentially expressed between male and female prefrontal cortex samples within one time period (data was combined from all four prefrontal cortex regions).
Downstream analysis of miRNA targets
miRNAs that were determined to be differentially expressed temporally or by sex were further analyzed for putative target genes under their control. To do so, we used the target prediction algorithms of TargetScanHuman 6.2 (11) and miRDB (12). We considered as significant only those targets that were predicted by both algorithms. Gene ontology (GO) enrichment analysis of the target genes was performed using DAVID Bioinformatics Resources 6.7 Functional Annotation Tool (13). Gene ontologies were considered significant only if their Benjamini-Hochberg multiple testing corrected p-value was < 0.05. GO enrichment analysis was performed on lists of aggregated targets (all time periods) that were brain region specific.
Test for enrichment of disease-associated genes
To determine if the target genes of differentially expressed miRNAs may relate to neurological diseases, we assessed for their enrichment into disease-related gene sets. Disease related gene sets were downloaded from the Genotator database (14). Enrichment was tested using the Hypergeometric probability distribution function in Excel. The population universe (i.e. all protein-coding genes in the human genome) was set to 20,687 (15). A success in the Hypergeometric function test was a gene that was both a predicted miRNA target and previously associated with a disorder. P-values were corrected for multiple testing by applying the conservative Bonferorri method. Enrichment was only considered significant if the Bonferorri-corrected p-value was < 0.01.
Results and Discussion
In total, we discovered 75 miRNAs differentially expressed across developmental time within brain regions (absolute log2. fold change > 1.5 and FDR < 0.05, Supplementary Table 1). Similar to previously described changes in gene expression, the greatest differential expression of miRNAs occurred during the transition from infancy to early childhood (Figure 1). The dorsolateral prefrontal cortex exhibited the greatest number of differentially expressed miRNAs (35 miRNAs) and the cerebellum a similar amount (22 miRNAs); the hippocampus and other regions of the prefrontal cortex each displayed less than five differentially expressed miRNAs. In contrast, differential expression of miRNAs between brain regions increased over developmental time (Figure 2, Supplementary Table 2). This finding is opposite previously described patterns of mRNA expression, which has been shown to become more globally similar between brain regions over development (2).
Figure 1. Number of differentially expressed miRNAs within each brain region over development.

Figure 2. Number of differentially expressed miRNAs between brain regions over development.
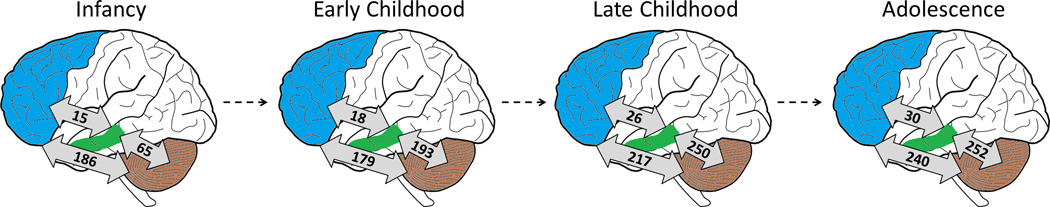
Light blue represents the prefrontal cortex, green represents the hippocampus, and the cerebellum is shaded in brown.
As many neurodevelopmental disorders display a significant sex-bias in their prevalence (16–18), we also assessed for differential miRNA expression by sex in the prefrontal cortex. We discovered 40 miRNAs with significant sex-biased expression differences between the prefrontal cortex of males and females (Table 2, Supplementary Table 3). Strikingly, 93% were more highly expressed in females, again a trend opposite to that of sex-biased gene expression (2). Furthermore, the majority of sex-biased gene expression occurred in adolescence (65%), suggesting that miRNA-targeted gene expression differences in the prefrontal cortex of males versus females becomes most pronounced around puberty.
Table 2.
Differentially expressed miRNAs between male and female prefrontal cortex over development.
| Up-regulated in Males | Up-regulated in Females | Total | |
|---|---|---|---|
| Infancy | 1 | 1 | 2 |
| Early Childhood | 1 | 9 | 10 |
| Late Childhood | 0 | 2 | 2 |
| Adolescence | 1 | 25 | 26 |
| Total | 3 | 37 | 40 |
To explore the potential biologic and pathogenic roles of the differentially expressed miRNAs, we identified putative targets of the temporally and sex-biased differentially expressed miRNAs (see Methods, Supplementary Table 4). We then assessed for enrichment of gene ontology (GO) categories in all lists of putative target genes. Overall, miRNA target genes were highly related to the process of transcription regulation in almost all lists (Supplementary Tables 5–10). This finding is in line with the well-known function of miRNAs as master regulators of gene expression networks (19, 20), and underscores the importance of identifying these key hubs of brain transcriptomes. Additionally, putative gene target lists were enriched for biological processes relating to nervous system development, synaptogenesis, and other basic intracellular processes.
Of particular note was the functional enrichment of miRNA targets that were differentially expressed between male and female prefrontal cortex. In addition to the processes implicated in all lists, the sex-biased targets were further enriched for Wnt signaling and transforming growth factor-beta (TGF-β) pathways. This result suggests these pathways may partially underlie normal behavioral differences in executive functioning between males and females. Furthermore, these two pathways are implicated in neurological disorders with sex-biased differences in prevalence (21, 22), and therefore may relate this sex disparity to underlying miRNA expression differences during normal brain development.
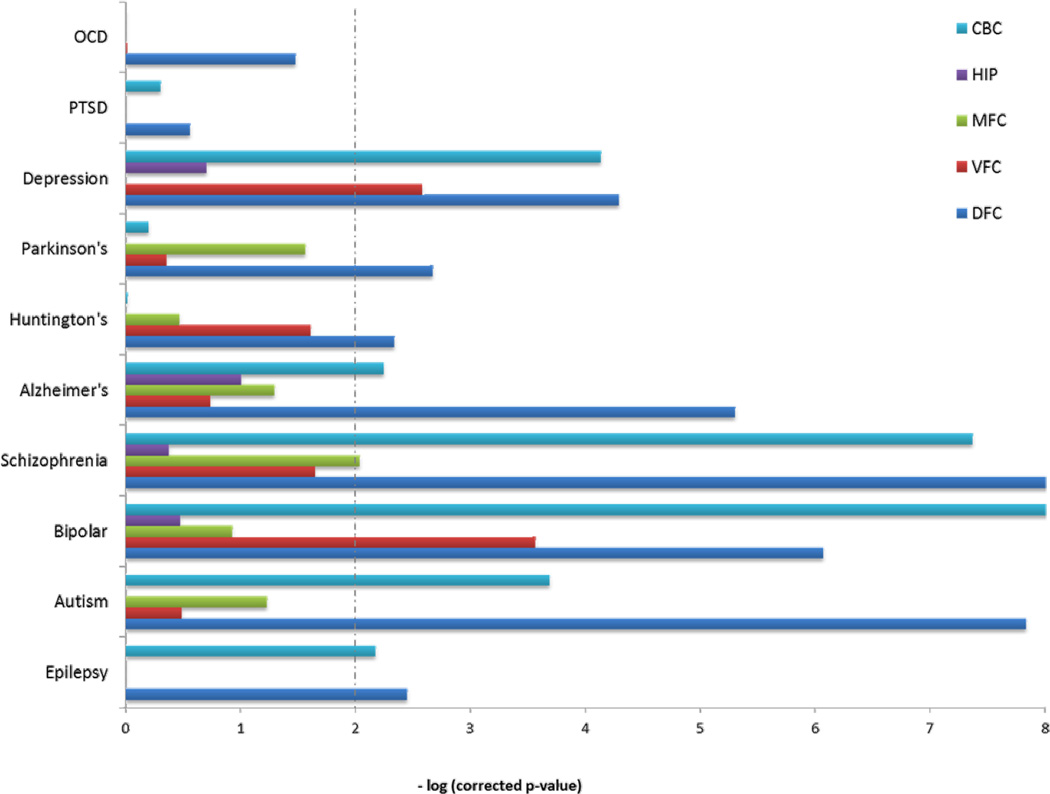
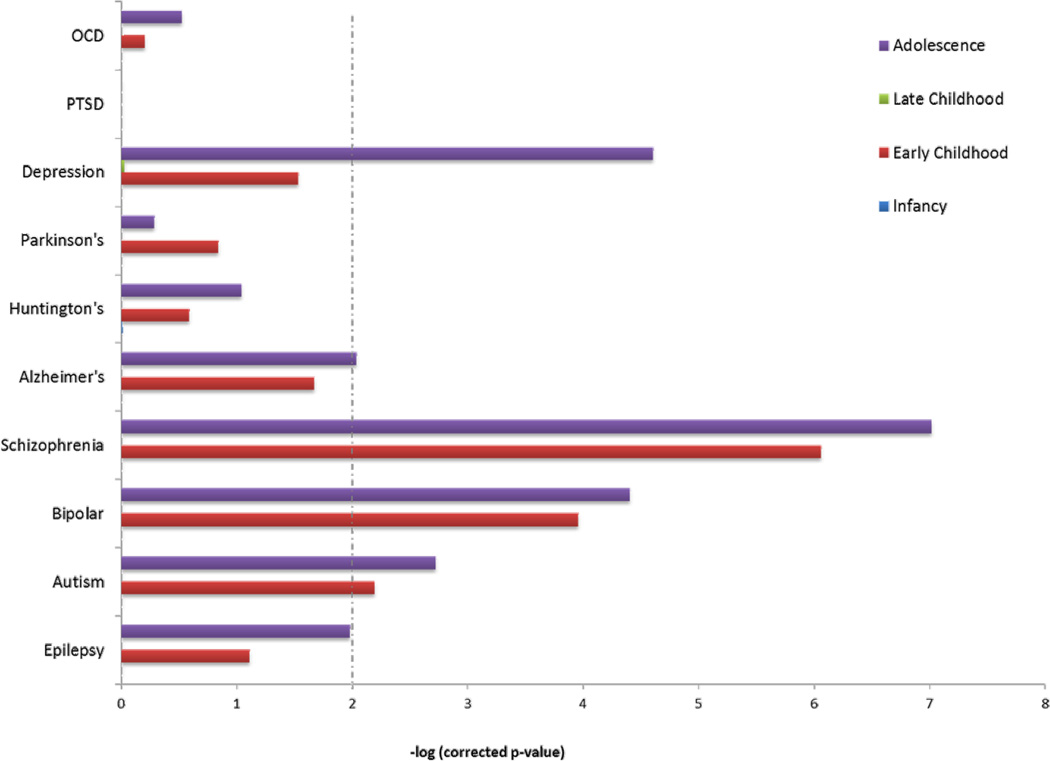
Last, we assessed for enrichment of miRNA targets among genes previously implicated in various neurological and psychiatric disorders that have significant genetic etiology (see Methods). We tested for enrichment of genes involved in epilepsy, three neurodevelopmental disorders (autism, schizophrenia, and bipolar disease), three neurodegenerative disorders (Alzheimer’s, Huntington’s, and Parkinson’s diseases), and three psychiatric diseases (major depressive disorder, post-traumatic stress disorder, and obsessive-compulsive disorder). The enrichment of all gene lists significant for various disorders is shown in Figures 3 and 4. The three neurodevelopmental disorders (ASD, Schizophrenia, and Bipolar) showed a nearly identical enrichment pattern, among many categories. In contrast, there was almost no enrichment for neurodegenerative disease lists. Similarly, the neuropsychiatric disorders showed no enrichment for miRNA target genes, except for major depressive disorder, where the pattern was similar to the neurodevelopmental disorders.
Figure 3. Enrichment of differentially expressed miRNA target genes by brain region for disease associated genes.
Dashed line indicates significance (corrected p-value < 0.01).
Figure 4. Enrichment of differentially expressed miRNA target genes among male versus female sets for disease associated genes.
Dashed line indicates significance (corrected p-value < 0.01).
In summary, we describe the most comprehensive assessment to date of spatio-temporal miRNA expression in the developing human brain. Our results identified miRNAs differentially expressed both within and between brain regions, and demonstrated that the greatest shifts in miRNA expression occur shortly after birth. However, unlike global gene expression patterns, miRNAs become more differentially expressed between brain regions over time, potentially driving regional specialization as the brain matures. Target genes under putative control by region-specific differentially expressed miRNAs are most related to the processes of transcription regulation and neurodevelopment, highlighting the central function of these miRNAs to brain transcription networks. Additionally, sex-biased expression of miRNAs increases in the prefrontal cortex around puberty, and the pathways related to sex-biased target genes are further enriched for Wnt signaling and TGF-β pathways. Common neurodevelopmental disorders with complex genetic etiologies are highly related to genes targeted by these miRNAs, but this was not found for genes related to neurodegenerative or other neuropsychiatric diseases with adult onset.
This study has a number of important limitations. First, the total sample size is 18 donor brains, potentially limiting the statistical power. Unfortunately, this problem is prevalent throughout human neurosciences research owing to the lack of large repositories of human post mortem brain tissue (23). Therefore, it will be important for future studies to replicate and aggregate the data presented here with larger datasets when they become available. Additionally, while computational prediction of miRNA targets based on sequence homology is an effective discovery tool, individual miRNAs of interest will require in vitro or in vivo experimental validation of their targets.
In conclusion, while thousands of genes are differentially expressed throughout human neurodevelopment, we have identified a set of miRNAs with differential spatio-temporal and sex-biased expression patterns that may regulate these expression changes. The targets of these differentially expressed miRNAs are highly enriched for genes related to transcriptional regulation, neurodevelopmental processes, and common neurodevelopmental disorders. Furthermore, inter-regional expression differences of miRNAs appear to increase over development. These results suggest the identified miRNAs are likely hubs of critical brain developmental and pathologic transcriptional processes.
Supplementary Material
Acknowledgements
We would like to thank the Allen Institute for Brain Science.
Sources of Support
This work was supported by the Intramural Research Program (IRP) of the National Institute of Child Health and Human Development, NIH. MNZ was also supported by the Baylor College of Medicine Medical Scientist Training Program (MSTP) and the NIH-Oxford/Cambridge Biomedical Scholars Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
Supplementary Information
Supplementary Tables.xls Contains all supplementary tables.
References
- 1.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008 Nov;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011 Oct 27;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shao NY, Hu HY, Yan Z, Xu Y, Hu H, Menzel C, et al. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics. 2010;11:409. doi: 10.1186/1471-2164-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu HY, Guo S, Xi J, Yan Z, Fu N, Zhang X, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011 Oct;7(10):e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010 Sep;20(9):1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011 Dec;9(12):e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012 Aug;13(8):528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Jan 1;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robles JA, Qureshi SE, Stephen SJ, Wilson SR, Burden CJ, Taylor JM. Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing. BMC Genomics. 2012;13:484. doi: 10.1186/1471-2164-13-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. Rna. 2008 Jun;14(6):1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Wall DP, Pivovarov R, Tong M, Jung JY, Fusaro VA, DeLuca TF, et al. Genotator: a disease-agnostic tool for genetic annotation of disease. BMC Med Genomics. 2010;3:50. doi: 10.1186/1755-8794-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sep 6;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jazin E, Cahill L. Sex differences in molecular neuroscience: from fruit flies to humans. Nat Rev Neurosci. 2010 Jan;11(1):9–17. doi: 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- 17.Abel KM, Drake R, Goldstein JM. Sex differences in schizophrenia. Int Rev Psychiatry. 2010;22(5):417–428. doi: 10.3109/09540261.2010.515205. [DOI] [PubMed] [Google Scholar]
- 18.Werling DM, Geschwind DH. Sex differences in autism spectrum disorders. Curr Opin Neurol. 2013 Apr;26(2):146–153. doi: 10.1097/WCO.0b013e32835ee548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007 Feb;8(2):93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 20.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008 Mar 28;319(5871):1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 21.Freese JL, Pino D, Pleasure SJ. Wnt signaling in development and disease. Neurobiol Dis. 2010 May;38(2):148–153. doi: 10.1016/j.nbd.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krieglstein K, Zheng F, Unsicker K, Alzheimer C. More than being protective: functional roles for TGF-beta/activin signaling pathways at central synapses. Trends Neurosci. 2011 Aug;34(8):421–429. doi: 10.1016/j.tins.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, Munafò MR. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 2013 May;14(5):365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.