Abstract
Objectives
To evaluate the biologic validity of ovarian cancer (OVCA) screening and early detection efforts and to characterize signaling pathways associated with human cancer metastasis and patient survival.
Study Design
Using genome-wide expression profiling and DNA sequencing, we compared pelvic and matched extra-pelvic implants from 30 patients with advanced-stage OVCA for expression of molecular signaling pathways and p53 gene mutations. Differentially expressed pathways were further evaluated in a series of primary or early-stage versus metastatic or recurrent cancer samples from 389 ovarian, prostate, and oral cancer patients. Metastasis pathways were also evaluated for associations with survival in nine independent clinico-genomic datasets from 1,691 ovarian, breast, colon, brain, and lung cancer and leukemia patients. The inhibitory effects of one pathway (TGF-WNT) on in-vitro OVCA cell migration were studied.
Results
Pelvic and extra-pelvic OVCA implants demonstrated similar patterns of signaling pathway expression and identical p53 mutations. However, we identified 3 molecular pathways/cellular processes that were differentially expressed between pelvic and extra-pelvic OVCA samples and between primary/early-stage and metastatic/advanced or recurrent ovarian, oral, and prostate cancers. Furthermore, their expression was associated with overall survival from ovarian cancer (P=0.006), colon cancer (1 pathway at P=0.005), and leukemia (P=0.05). Artesunate-induced TGF-WNT pathway inhibition impaired OVCA cell migration.
Conclusions
Advanced-stage OVCA has a unifocal origin in the pelvis, supporting validity of early detection/screening efforts. Molecular pathways associated with extra-pelvic OVCA spread are also associated with metastasis from other human cancers and with overall patient survival. Such pathways represent appealing therapeutic targets for patients with metastatic disease.
Keywords: Gene Expression, p53 mutation, Serous Ovarian Cancer, Unifocal
INTRODUCTION
Ovarian cancer (OVCA) has the highest mortality rate of all gynecologic cancers, with an estimated 22,280 cases of OVCA diagnosed in 2012 and 15,500 women dying from the disease 1. Most OVCA patients are diagnosed at advanced-stage with disseminated intra-peritoneal metastases, such that the majority succumb to the disease within 5 years 2. Although significant efforts have focused on strategies to detect OVCA at an earlier, more curable, stage, these efforts are based on the assumption that advanced-stage OVCA originates from a single focus in the ovarian epithelium and that extra-pelvic, abdominal implants result from disease spread from the primary lesion along peritoneal surfaces, so-called unifocal disease. In contrast, proponents of a multifocal, field-effect phenomenon support a view that advanced-stage epithelial OVCA does not begin as a single focus in the pelvis, but rather appears at multiple sites on the peritoneal surface throughout the pelvis and abdomen, somewhat simultaneously. Advocates of a multifocal theory of OVCA continue to question the value of efforts directed toward development of screening technologies that detect localized (early-stage) OVCA before it has spread outside the pelvis.
In this study, we sought to define the focal origin of OVCA. We evaluated the pathoetiologic relationship between primary pelvic and matched extra-pelvic implants via genome-wide RNA-based expression data and p53 DNA mutational analysis in matched samples from 30 patients with advanced-stage OVCA. Our hypothesis is based on the assumption that, if multiple pelvic and extra-pelvic OVCA lesions develop in a simultaneous fashion (a multifocal origin), then the activation of molecular signaling pathways that occurred during carcinogenesis in each individual lesion (and particularly between pelvic and extra-pelvic disease) is likely unique. Moreover, a multifocal origin would cast doubt on the likely success of screening efforts focused on disease originating in the pelvis. Such conclusions would have significant public health implications.
In contrast, if OVCA has a unifocal origin, then the pelvic and extra-pelvic implants should express similar signaling pathways and have similar p53 mutational patterns. Furthermore, in the face of a unifocal origin, any differences in expression profiles between pelvic and extra-pelvic OVCA implants would represent processes associated with metastatic spread that may be also common to other cancer types, may influence clinical outcome, and, importantly, may serve as valid therapeutic targets for patients with metastatic cancer.
MATERIALS AND METHODS
Pelvic OVCA samples and matched, non-confluent, extra-pelvic implants were obtained from 30 patients who had provided written informed consent to the Moffitt Cancer Center Institutional Total Cancer Care (TCC)™ Protocol (www.moffitt.org), prior to undergoing primary cytoreductive surgery for advanced-stage serous epithelial OVCA. The study was carried out with approval from the University of South Florida Institutional Review Board. A pelvic sample was resected from the ovarian tissue, which, in the opinion of the surgeon, most likely represented the primary site in the pelvis. From each patient, a matched, non-confluent extra-pelvic implant was identified and collected. Samples were flash-frozen in liquid nitrogen within 10 minutes of surgical resection and stored at −80°C. A histopathologic review was performed to confirm diagnosis, and samples were macrodissected to ensure >70% tumor content. Total RNA and genomic DNA were extracted from each sample.
Normal ovarian surface epithelium (NOSE) samples were obtained from patients who had provided written informed consent to the TCC™ Protocol and had undergone oophorectomy at Moffitt Cancer Center for non-malignant disease, not associated with the ovary. Immediately after surgical resection, the surface epithelium was gently scraped from the surface and immediately subjected to RNA isolation. To ensure sufficient quantities of RNA, NOSE RNA samples were pooled in groups of 3 or 4 to produce a minimum RNA quantity of 50 ng. As a result of such pooling, 49 normal ovaries were analyzed on 12 Affymetrix GeneChip assays.
Approximately 30 mg of tissue was used for each RNA and DNA extraction. Tissues were pulverized in BioPulverizer H tubes (Bio101) using a Mini-Beadbeater (Biospec Products). Total RNA was collected using the Qiagen RNeasy Mini kit according to manufacturer’s instructions. RNA quality was checked on an Agilent Bioanalyzer to assess quality of RNA via the 28S:18S ribosomal RNAs. Genomic DNA was isolated using the Qiagen QIAamp DNA Mini kit according to manufacturer’s instructions.
For microarray analysis, 10 μg of total RNA was used to develop the targets for Affymetrix microarray analysis, and probes were prepared according to the manufacturer’s instructions. Briefly, biotin-labeled cRNA was produced by in vitro transcription, fragmented, and hybridized to customized Human Affymetrix HuRSTA gene chips (HuRSTA-2a520709). Expression values were calculated using the robust multi-array average algorithm implemented in Bioconductor (http://www.bioconductor.org) extensions to the R statistical programming environment.
Student’s t-test was used to identify differentially expressed genes in comparisons among NOSE, pelvic, and extra-pelvic sample genomic data. For each comparison, the 12 NOSE samples were grouped together. Pelvic and extra-pelvic genomic profiles were analyzed as groups (pelvic as one group, extra-pelvic as another) and as individual pairs (comparisons of matched pelvic/extra-pelvic pairs from same patient). As such, the following comparisons were made: i) grouped NOSE versus grouped pelvic implants, ii) grouped NOSE versus grouped extra-pelvic implants, iii) grouped pelvic versus grouped extra-pelvic implant, iv) grouped NOSE versus individual pelvic implants, v) grouped NOSE versus individual extra-pelvic implants, and vi) individual pelvic versus individual matched extra-pelvic samples from the same patient. For each of the comparisons, differentially expressed genes were analyzed using GeneGO MetaCore™ software to identify represented biologic pathways.
Identified pathways were further evaluated for differential representation in 4 publically available gene expression datasets encompassing 389 patient samples including: 1) OVCA (n=12; 4 early- and 8 advanced-stage), GEO accession number GSE14407, U133Plus genechip; 2) oral cancer (n=27; 22 primary lesions, 5 metastases), GEO accession GSE2280, U133A genechip; 3) prostate cancer (n=271; 196 primary lesions, 75 metastases), GEO accession GSE6919, U95 genechip; and 4) prostate cancer (n=79; 40 non-recurrent, 39 recurrent lesions), GEO accession GSE25136, U133A genechip (by Student’s t-test, gene cutoff P<0.01).
Principal Component Analysis (PCA) was performed using Evince software (evince.umbio.com) for Figure 1. Log-rank tests were used to test associations between pathway expression (using median PCA score value cut-off) and overall survival within nine publically available datasets comprising 1,691 patient samples, including cancers of the ovary, which included 4 datasets [Australian dataset (n=218 GSE9891)3, MCC dataset (n=142)4, MDA dataset (n=53 GSE18520), and TCGA dataset (n=497)], as well as brain (n=182 GSE13041)5, breast (n=187 GSE2990), colon (n=177 GSE17538)6, lung (n=58 TCGA), and blood (leukemia, n=182 TCGA). All survival analysis was performed using R program.
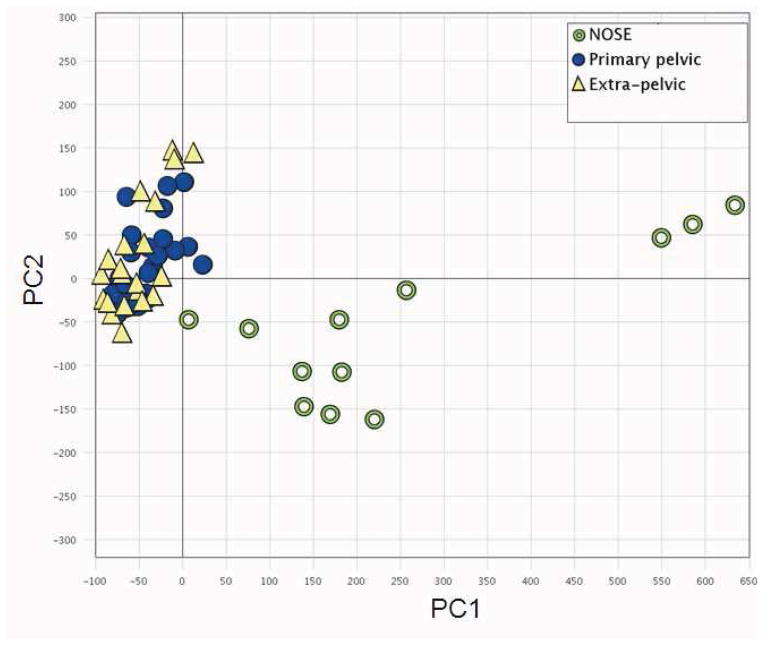
Figure 1.
Comparison of overall expression profiles across all samples. Results for principal component analyses (PCA) of gene expressions in normal ovarian surface epithelium (NOSE; green circles), primary pelvic (filled blue circles), and extra-pelvic (yellow triangles) samples are shown. The first principal component (PC1) explains 35.4% of the variation, whereas the second (PC2) explains 6.3%.
For sequence analysis of p53, exons 5–8 of p53 from primary lesions and distal metastases separated by non-involved tissue were analyzed for primary sequence mutation patterns. Genomic DNA (100 ng) was used in PCR amplification reactions essentially as described previously7 using the following primers: exon 5, sense 5′-TTCCTCTTCCTACAGTACTC-3′, anti-sense 5′-GCAACCAGCCCTG-TCGTCTC-3′; exon 6, sense 5′-ACCATGAGCGCTGCTCAGAT-3′, anti-sense 5′-AGTTGCAAACCAGACGTCAG-3′; exon 7, sense 5′-GTGTTGTCTCCTAGGTTCGC-3′, anti-sense 5′-CAAGTGGCTCCTGACCTGGA-3′; and exon 8, sense 5′-CCTATCCTGAGTAGTGGTAA-3′, anti-sense 5′-TGAATCTGAGGCATAACTGC-3′. Amplifications were performed using an Eppendorf Mastercycler thermocycler in 50-μL reaction volumes (100 ng genomic DNA, 1 unit Taq DNA polymerase (Invitrogen, Carlsbad, CA), 1.5 mM MgCl2, 0.2 mM dNTPs, and 0.2 μM primer mix) by standard protocols. Briefly, samples were held at 95°C for 10 minutes followed by 30 cycles of the following: 95°C for 50 seconds, annealing temperature at 56°C or 60°C, depending on the primers, for 90 seconds, and an elongation step at 72°C for 90 seconds. After cycling, samples were held at 72°C for 10 minutes and cooled to 4°C. PCR products were purified using the Purelink PCR purification kit (Qiagen) and evaluated using 4% agarose gels. Sequencing was performed on an Applied Biosystem’s AB3130 Genetic Analysis System using BigDye 3.1 dye terminator chemistry according to the manufacturer’s instructions. Comparative sequence analysis of p53 exons was performed using DNAStar, Lasergene 8 software.
The effects of pathway inhibition on OVCA cell metastatic properties were investigated using the in-vitro scratch assay. HeyA8 OVCA cells were a gift from Dr. Patricia Kruk, Department of Pathology, College of Medicine, University of South Florida, Tampa, FL. Cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Fisher Scientific, Pittsburgh, PA), 1% sodium pyruvate, 1% penicillin/streptomycin (Cellgro, Manassas, VA), and 1% nonessential amino acids (HyClone, Hudson, NH). Monolayers, 75–80% confluent, were cultured in serum-free media for 4 hours and then mechanically disrupted to create a “wound” using a 1-mL pipette tip. Culture plates were washed twice with serum-free media to remove floating cells and then incubated with media containing 10% FBS and either vehicle (DMSO) or drug. The DMSO concentration was maintained below 0.5% so as not to influence cell growth or migration. The underside of the culture plate by the wound area was marked with a Sharpie for reference, and wounds were imaged by phase-contrast microscopy on days 0, 1, and 2.
RESULTS
Comparison of Overall Expression Patterns
PCA modeling was used to assess the overall similarities in gene expression among NOSE, pelvic, and extra-pelvic samples. PCA generates a set of vectors (termed first principal component, PC1, second principal component, PC2, etc.) that summarize the overall genome-wide expression patterns for a sample. Each principal component provides a summary measure for genes that share certain expression characteristics. Comparing PCA values enables a global assessment of how similar or different samples are at a genome-wide level. The two first principal components for all samples are shown in Figure 1. PC1, which explained 35.4% of the variation, separated most of NOSE samples from the primary pelvic and the extra-pelvic samples.
Comparison of Pathway Expression in NOSE, Pelvic, and Extra-pelvic OVCAs
We performed grouped comparisons of NOSE, pelvic, and extra-pelvic genomic data. At a significance of P<0.01 (Bonferroni adjusted), 970 probesets representing 71 signaling pathways (P<0.05) were identified when the grouped NOSE expression data were compared to the grouped primary pelvic sample data, and 1,075 probesets representing 143 signaling pathways were identified when the grouped NOSE expression data were compared to the grouped extra-pelvic implant expression data. Importantly, the 60/71 (85%) signaling pathways present in primary pelvic samples were also represented in extra-pelvic implants. At this level of significance, no probesets were found to be differentially expressed between the grouped primary pelvic and extra-pelvic samples. When the grouped NOSE dataset was analyzed against the individual pelvic primary samples (n=30) and the individual extra-pelvic implants (n=30), an average of 7,392 and 7,772 probesets, respectively, demonstrated differential expression (>2-fold). In contrast, an average of 1,463 probesets were differentially expressed between individual pelvic and matched extra-pelvic implants from the same patient. Consistently, these data suggest significant similarity between primary pelvic and matched extra-pelvic implants (Supplementary Appendix 1).
Mutational Analysis of p53
Exons 5–8 of the p53 gene were examined in primary pelvic and matched extra-pelvic implants (Table 1). A total of 13 nucleotide mutations were found in 11 of 30 primary pelvic samples. A mutation in exon 5 was found in 1 primary pelvic, whereas 3 primary pelvic lesions had a mutation in exon 6, 7 pelvic lesions had a mutation in exon 7, and 2 pelvic lesions had a mutation in exon 8. The majority of identified mutations were missense (9/13); however, one sample showed a frameshift mutation resulting from a deletion in codon 151 of exon 5, one sample showed a nonsense mutation in codon 294 of exon 8, and two samples displayed silent mutations. In every case, the p53 mutation identified in the primary pelvic was also present in the matched extra-pelvic implant.
Table 1.
Primary sequence mutations in p53 exons 5–8.
| Histological Type | Grade | Stage | p53 Mutation in Primary Ovarian Cancer | p53 Mutation in Extra-pelvic Implants | Amino Acid Change | ||||
|---|---|---|---|---|---|---|---|---|---|
| Primary Site | Exon | Codon | Extra-pelvic Site | Exon | Codon | ||||
| Serous adenocarcinoma | High | 3C | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Serous adenocarcinoma | High | 4 | Rt Ovary | 6 | 220, TAT to TGT | Omentum | 6 | 220, TAT to TGT | Tyr to Cys |
| 7 | 225, GTT to GTG | 7 | 225, GTT to GTG | Val to Val | |||||
| Serous adenocarcinoma | High | 3C | Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Serous adenocarcinoma | High | 3C | Rt Ovary | 8 | 294, GAG to TAG | Soft tissue, Pelvis | 8 | 294, GAG to TAG | Glu to STOP |
| Serous adenocarcinoma | High | 3C | Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma w/papillary features | High | 4 | Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma w/papillary features | High | 3C | Lt Ovary | 7 | 248, CGG to CAG | Omentum | 7 | 248, CGG to CAG | Arg to Gln |
| Serous adenocarcinoma | High | 3C | Rt Ovary | 7 | 248, CGG to CAG | Omentum | 7 | 248, CGG to CAG | Arg to Gln |
| Serous adenocarcinoma | High | 4 | Rt Ovary | WT | WT | Colon | WT | WT | WT |
| Adenocarcinoma | High | 3C | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma w/papillary features | Not given | 3C | Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Serous adenocarcinoma | High | 3C | Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma w/papillary features | High | 2B | Rt Ovary | 6 | 220, TAT to TGT | Cul-de-sac | 6 | 220, TAT to TGT | Tyr to Cys |
| Clear cell carcinoma | High | 3C | Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma w/papillary features | High | 3C | Lt Ovary | 7 | 245, GGC to GAC | Omentum | 7 | 245, GGC to GAC | Gly to Asp |
| Adenocarcinoma w/papillary features | High | 3C | Rt Ovary | 7 | 248, CGG to CAG | Omentum | 7 | 248, CGG to CAG | Arg to Gln |
| Adenocarcinoma | High | 3C | Lt Ovary | WT | WT | Colon | WT | WT | WT |
| Serous adenocarcinoma | High | 3C | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Serous adenocarcinoma | High | 3C | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Serous adenocarcinoma | High | 4 | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma | High | 2C | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma | High | 3C | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma w/papillary features | High | 4 | Rt Ovary | WT | WT | Omentum | WT | WT | WT |
| Serous adenocarcinoma | High | 3C | Rt Ovary | 7 | 234, TAC to TGC | Omentum | 7 | 234, TAC to TGC | Tyr to Cys |
| Serous adenocarcinoma | High | 3C | Rt, Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Adenocarcinoma | High | 3C | Rt Ovary | 5 | 151, CCC to ^CC | Omentum | 5 | 151, CCC to ^CC | Pro to Frame shift |
| Adenocarcinoma | High | 3B | Rt, Lt Ovary | WT | WT | Omentum | WT | WT | WT |
| Serous adenocarcinoma | Moderate | 3C | Rt Ovary | WT | WT | Soft tissue (periaortic) | WT | WT | WT |
| Serous adenocarcinoma | High | 3C | Rt, Lt Ovary | 8 | 282, CGG to TGG | Omentum | 8 | 282, CGG to TGG | Asp to Trp |
| Clear cell carcinoma | High | 3C | Lt Ovary | 6 | 213, CGA to CGG | Omentum | 6 | 213, CGA to CGG | Arg to Arg |
| 7 | 245, GGC to GAC | 7 | 245, GGC to GAC | Gly to Asp | |||||
Abbreviations: Lt, left; Rt, right; WT, wild-type.
Pathways Associated with Metastasis Influence Clinical Outcome
We also sought to identify pathways present in extra-pelvic samples that were not present in pelvic samples (termed, candidate metastasis pathways, CMPs). We adopted two statistical approaches to this: comparisons of data grouped together and of individual patient matched samples. Five CMPs demonstrated differential expression using both approaches; that is, they were present in extra-pelvic samples but not in pelvic samples when data were compared both in grouped analyses (81 total pathways, Supplementary Appendix 2) and in ≥15/30 (50%) of the patients for whom individual comparisons were made between matched pelvic and extra-pelvic samples (24 pathways total; Supplementary Appendix 3). These 5 CMPs included 1) chemokines and cell adhesion (chemokines/cell adhesion pathway), 2) transforming growth factor-beta and cytoskeletal remodeling (TGF-WNT/cytoskeleton remodeling pathway), 3) histamine signaling in dendritic cells and immune response (histamine signaling/immune response pathway), 4) TLR signaling pathways and immune response (TLR pathway), and 5) protein folding, membrane trafficking, and signal transduction of G-alpha (i) heterotrimeric G-protein (G-alpha pathway).
To further explore the validity of these 5 CMPs, we evaluated each in 4 publically available external gene expression datasets from primary or early-stage cancers versus metastatic/advanced or recurrent cancer. Pathways associated with metastatic, advanced-stage, or recurrent disease included 1) TGF-WNT/cytoskeleton remodeling pathway (P<0.0001) and chemokines/cell adhesion pathway (P<0.001) for ovarian cancer (GSE14407); 2) TGF-WNT/cytoskeleton remodeling (P<0.001) for oral cavity cavity (GSE2280); and 3) TGF-WNT/cytoskeleton remodeling (GSE6919; P<0.001), chemokines/cell adhesion (GSE6919; P<0.001), histamine signaling/immune response (GSE6919; P=0.016), TGF-WNT/cytoskeleton remodeling (GSE6919; P<0.001), and chemokines/cell adhesion (GSE6919; P<0.001) for prostate cancer. Based on their representation in the external datasets, TGF-WNT/cytoskeleton remodeling, chemokines/cell adhesion, and histamine signaling/immune response pathways were defined as metastasis pathways from our initial list of 5 CMPs.
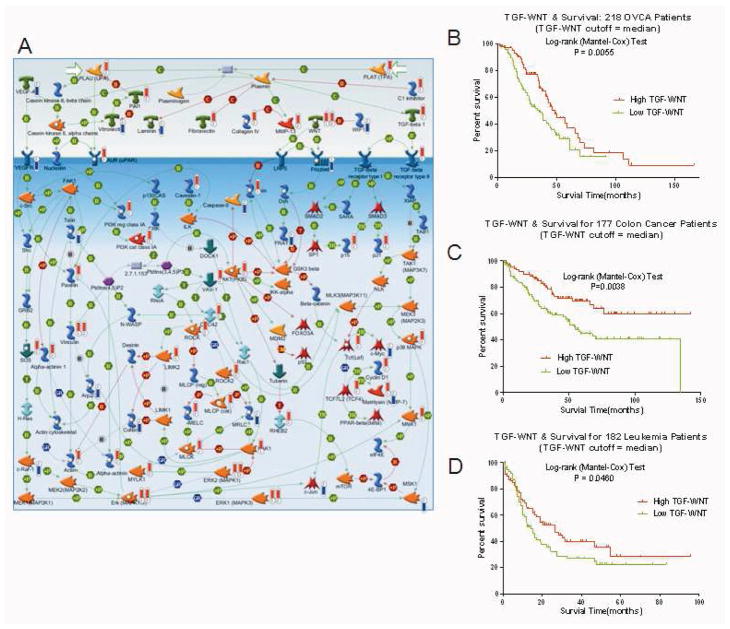
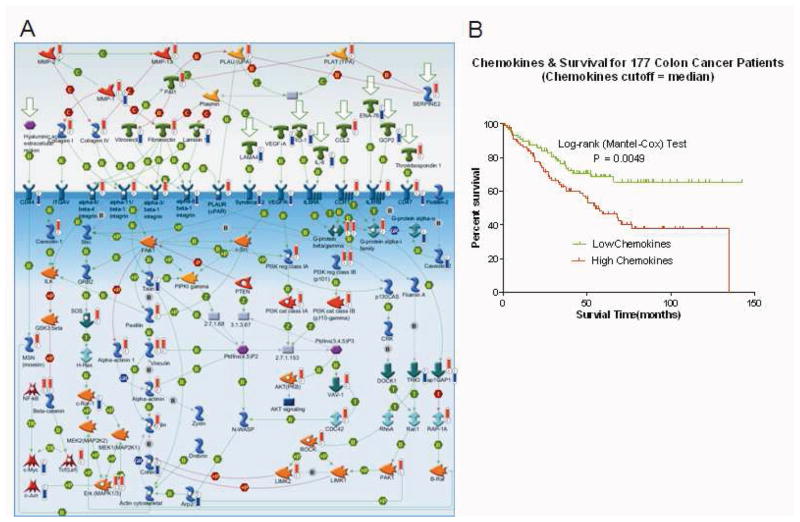
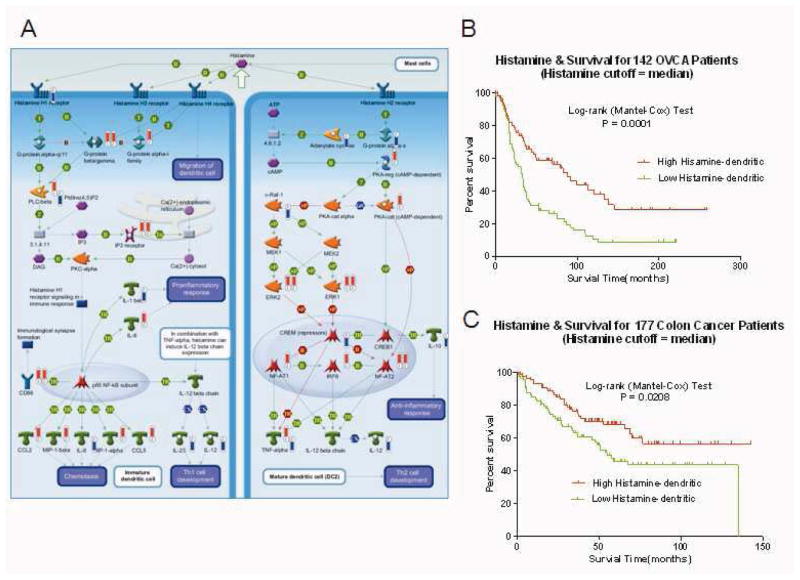
To further explore the clinical relevance of the 3 metastasis pathways, we evaluated associations (log-rank P values) between pathway expression (quantified by PCA modeling) and overall survival in 1,691 patients from a series of 9 external clinico-genomic datasets. Expression of the TGF-WNT/cytoskeleton remodeling pathway (Figure 2A) was associated with survival from OVCA (n=218, P=0.006, Figure 2B), colon cancer (n=177, P=0.004, Figure 2C), and leukemia (n=182, P=0.047, Figure 2D). The chemokines/cell adhesion pathway (Figure 3A) was associated with survival from colon cancer (n=177, P=0.005, Figure 3B), and the histamine signaling/immune response pathway (Figure 4A) was associated with survival from OVCA (n=142, P<0.001, Figure 4B) and colon cancer (n=177, P =0.02, Figure 4C).
Figure 2.
The TGF-WNT/cytoskeleton remodeling pathway is associated with survival from OVCA, colon cancer, and leukemia. (A) TGF-WNT/cytoskeleton remodeling pathway. Thermometers indicate the directional change (upward or downward) in expression of genes associated with extra-pelvic implant samples. Numbers 1–2 at base identify the originating dataset (1 = grouped analysis, unique to NOSE versus extra-pelvic implant; 2 = individual analysis; common to ≥15 paired samples). (B–D) Kaplan-Meier curves depicting the association between the TGF-WNT/cytoskeleton remodeling-pathway PCA score (using median PCA threshold) and overall survival from OVCA (GSE9891, survival information available for 218 of the 220 samples) (B), colon cancer (GSE17538, n=177 (C), and leukemia (TCGA database, n=182 (D). Log-rank test P values indicate significance.
Figure 3.
The chemokines/cell adhesion pathway is associated with survival from colon cancer. A, Chemokines/cell adhesion pathway. Thermometers indicate the directional change (upward or downward) in expression of genes associated with extra-pelvic implant samples. Numbers 1–2 at base identify the originating dataset (1 = grouped analysis, unique to NOSE versus extra-pelvic implant; 2 = individual analysis; common to ≥15 paired samples). B, Kaplan-Meier curves depicting the association between the chemokines/cell adhesion pathway PCA score (using median PCA threshold) and overall survival from colon cancer (GSE17538, n=177). Log-rank test P values indicate significance.
Figure 4.
The Histamine signaling/immune response pathway is associated with survival from OVCA and colon cancer. A, Histamine signaling/immune response pathway. Thermometers indicate the directional change (upward or downward) in expression of genes associated with extra-pelvic implant samples. Numbers 1–2 at base identify the originating dataset (1 = grouped analysis, unique to NOSE versus extra-pelvic implant; 2 = individual analysis, common to ≥15 paired samples). B and C, Kaplan-Meier curves depicting the association between the chemokines/cell adhesion pathway PCA score (using median PCA threshold) and overall survival from OVCA (MCC dataset, n=142) and colon cancer (GSE17538, n=177), respectively. Log-rank test P values indicate significance.
Inhibition of the TGF-WNT/Cytoskeleton Remodeling Pathway Prevents Cell Migration
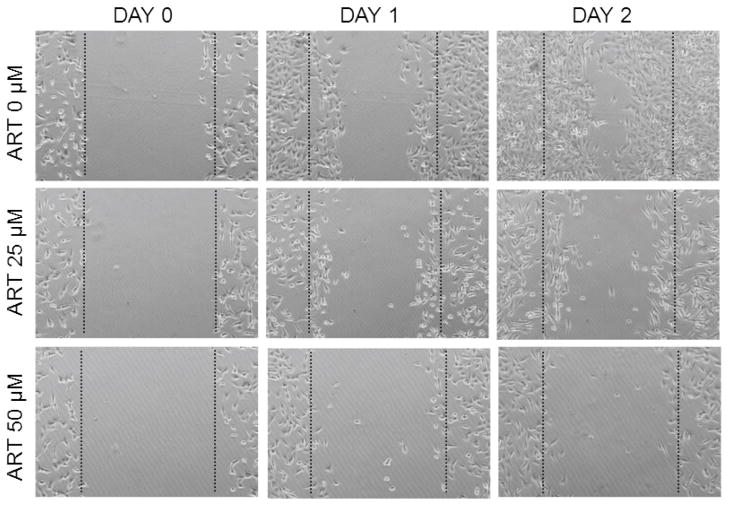
In light of the TFG-WNT/cytoskeleton remodeling pathway expression associations identified above and reports of its influence on metastatic activity in other cancer types,8–11 we performed functional studies to evaluate the effect of this pathway on OVCA cellular metastatic characteristics, specifically, the influence of inhibition of this pathway using artesunate 12, 13 on OVCA cell migratory ability. Inhibition of TGF-WNT signaling using 25 μM or 50 μM artesunate impaired the ability of HeyA8 OVCA cells to fill the gap (Figure 5). In contrast, cells cultured in media containing DMSO vehicle completely filled in the gap within 2 days (Figure 5).
Figure 5.
TGF-WNT/cytoskeleton remodeling pathway inhibition prevents OVCA cell migration. HeyA8 cells treated with 25 μM and 50 μM artesunate (ART) were impaired in their ability to fill in the gap of a scratch test. In contrast, cells cultured in the presence of DMSO vehicle completely closed the gap within 2 days.
DISCUSSION
Our findings support the view that advanced-stage OVCA has a unifocal origin in the pelvis, thus validating efforts dedicated to the identification of early detection/screening strategies. We also identified what we believe are pathways associated with metastasis of OVCA, as well as metastasis/recurrence and overall survival from multiple human cancers. Our functional studies suggest that such pathways may represent appealing therapeutic targets for patients with metastatic disease.
The p53 gene is known to be mutated in 30–80% of OVCAs 14, 15. Because there is a strong selection for these mutations to be distributed over the conserved regions of the gene, we compared the sequence of p53, exons 5–8. Of 30 primary pelvic lesions tested, we found 11 (37%) containing DNA mutations. In every case, the matched extra-pelvic implant contained an identical mutation. Our findings are consistent with early work in this area by Mok et al, who described identical p53 mutations in samples collected from multiple sites in 9 patients with advanced-stage epithelial OVCA 16. Subsequently, analysis of allele loss on chromosome 17 in 16 OVCA samples revealed identical patterns of allelic deletions in all samples resected from the same patient, irrespective of the collection site 17. In 4 of 16 informative samples, analysis of the hypoxanthine phosphoribosyl transferase gene showed that the same parental allele was methylated in samples collected from the primary and metastatic sites 17. Similar findings were reported by Kupryjanczyk et al., who performed p53 sequence analysis to determine the clonality of disseminated serous carcinoma 18. Although this group also concluded a monoclonal origin of OVCA, they found additional mutations in the metastatic implant, suggesting a continual divergence during disease progression 18. Other studies have suggested that, although the extra-pelvic implant may be clonally derived, in some cases, the primary mass may be polyclonal 19, 20. Jacobs et al. evaluated p53 mutation patterns, X-chromosome inactivation, and loss of heterozygosity in 17 cases of advanced epithelial OVCA 19. Although 15 of these cases were confirmed to be unifocal, 2 cases showed evidence of the presence of more than one primary tumor 19. Similar results were reported by Khalique et al. who performed loss of heterozygosity analysis on multiple sites of 22 primary serous OVCAs, each with multiple matched extra-pelvic implants 20. While also concluding that the metastatic implants were clonally derived from the primary pelvic mass, these researchers also suggested a genetic divergence of the primary tumor 20.
The data generated here support a unifocal origin of advanced-stage OVCA. Moreover, we identified 3 pathways (TGF-WNT/cytoskeleton remodeling, chemokines/cell adhesion, and histamine signaling/immune response) that are not only associated with advanced, metastatic, or recurrent disease, but also with overall survival from a range of cancers. The TGF-WNT/cytoskeleton signaling pathway is known to influence multiple cellular processes including growth, differentiation, and apoptosis in both the adult and embryo 21–23. TGF binding to serine/threonine kinase receptors induces phosphorylation of receptor-regulated SMADs, which enter the nucleus as transcriptional regulators 24, 25. WNT protein receptor binding inhibits the axin/GSK-3/APC complex, increasing nuclear entry of β-catenin where transcription is influenced 26, 27.
Our findings are consistent with other reports of TGF-beta and WNT associations with carcinogenesis, proliferation, and invasiveness 8–10, 28. Similarly, several members of the chemokines/cell adhesion pathway, including PI3K, PAK1, and ERK, may work in conjunction to increase the invasiveness of a variety of cancer cells 29, 30. Finally, histamine signaling has been reported to increase calcium mobilization and proliferation in OVCA 31, maintain cholangiocarcinoma growth 32, 33, and increase the proliferation of lung, breast, and pancreatic cancer cells 34–36.
We conclude that advanced-stage OVCA has a unifocal origin in the pelvis, supporting the validity of early detection/screening efforts aimed at reducing disease mortality. Furthermore, our analysis has also provided insight into the biologic basis of human cancer metastasis, identifying pathways such as TGF-WNT/cytoskeleton remodeling that represent appealing therapeutic targets for patients with metastatic disease from a range of primary sites.
Supplementary Material
Acknowledgments
Source of funding: This work was supported by Jacquie Liggett Fellowship, H.O.W, Hearing the Ovarian Cancer Whisper, and Palm Healthcare Foundation. The research was also supported in part by the National Cancer Institute, part of the National Institutes of Health, through grant number 2 P30-CA76292-14.
We would like to thank the Total Cancer Care Protocol at the Moffitt Cancer Center for their contributions and Rasa Hamilton (Moffitt Cancer Center) for editorial assistance.
Footnotes
DISCLOSURE: The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Baker VV. Salvage therapy for recurrent epithelial ovarian cancer. Hematol Oncol Clin North Am. 2003;17:977–88. doi: 10.1016/s0889-8588(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 3.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 4.Marchion DC, Cottrill HM, Xiong Y, Chen N, Bicaku E, Fulp WJ, et al. BAD phosphorylation determines ovarian cancer chemosensitivity and patient survival. Clin Cancer Res. 2011;17:6356–66. doi: 10.1158/1078-0432.CCR-11-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Scheck AC, Cloughesy TF, Lai A, Dong J, Farooqi HK, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–68. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard DG, Travis LB, Addya K, Dores GM, Holowaty EJ, Bergfeldt K, et al. p53 mutations in leukemia and myelodysplastic syndrome after ovarian cancer. Clin Cancer Res. 2002;8:973–85. [PubMed] [Google Scholar]
- 8.Theriault BL, Nachtigal MW. Human ovarian cancer cell morphology, motility, and proliferation are differentially influenced by autocrine TGFbeta superfamily signalling. Cancer Lett. 2011;313:108–21. doi: 10.1016/j.canlet.2011.08.033. [DOI] [PubMed] [Google Scholar]
- 9.Li GC, Ye QH, Dong QZ, Ren N, Jia HL, Qin LX. TGF beta1 and related-Smads contribute to pulmonary metastasis of hepatocellular carcinoma in mice model. J Exp Clin Cancer Res. 2012;31:93. doi: 10.1186/1756-9966-31-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Zhang H, Tang L, Chen H, Wu C, Zhao M, et al. Resveratrol inhibits TGF-beta1-induced epithelial-to-mesenchymal transition and suppresses lung cancer invasion and metastasis. Toxicology. 2013;303C:139–46. doi: 10.1016/j.tox.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 11.Wright LE, Frye JB, Lukefahr AL, Timmermann BN, Mohammad KS, Guise TA, et al. Curcuminoids Block TGF-beta Signaling in Human Breast Cancer Cells and Limit Osteolysis in a Murine Model of Breast Cancer Bone Metastasis. J Nat Prod. 2013;76:316–21. doi: 10.1021/np300663v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akhmetshina A, Palumbo K, Dees C, Bergmann C, Venalis P, Zerr P, et al. Activation of canonical Wnt signalling is required for TGF-beta-mediated fibrosis. Nat Commun. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li PC, Lam E, Roos WP, Zdzienicka MZ, Kaina B, Efferth T. Artesunate derived from traditional Chinese medicine induces DNA damage and repair. Cancer Res. 2008;68:4347–51. doi: 10.1158/0008-5472.CAN-07-2970. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto A, Sameshima Y, Yokoyama S, Terashima Y, Sugimura T, Terada M, et al. Frequent allelic losses and mutations of the p53 gene in human ovarian cancer. Cancer Res. 1991;51:5171–6. [PubMed] [Google Scholar]
- 15.Salani R, Kurman RJ, Giuntoli R, 2nd, Gardner G, Bristow R, Wang TL, et al. Assessment of TP53 mutation using purified tissue samples of ovarian serous carcinomas reveals a higher mutation rate than previously reported and does not correlate with drug resistance. Int J Gynecol Cancer. 2008;18:487–91. doi: 10.1111/j.1525-1438.2007.01039.x. [DOI] [PubMed] [Google Scholar]
- 16.Mok CH, Tsao SW, Knapp RC, Fishbaugh PM, Lau CC. Unifocal origin of advanced human epithelial ovarian cancers. Cancer Res. 1992;52:5119–22. [PubMed] [Google Scholar]
- 17.Tsao SW, Mok CH, Knapp RC, Oike K, Muto MG, Welch WR, et al. Molecular genetic evidence of a unifocal origin for human serous ovarian carcinomas. Gynecol Oncol. 1993;48:5–10. doi: 10.1006/gyno.1993.1002. [DOI] [PubMed] [Google Scholar]
- 18.Kupryjanczyk J, Thor AD, Beauchamp R, Poremba C, Scully RE, Yandell DW. Ovarian, peritoneal, and endometrial serous carcinoma: clonal origin of multifocal disease. Mod Pathol. 1996;9:166–73. [PubMed] [Google Scholar]
- 19.Jacobs IJ, Kohler MF, Wiseman RW, Marks JR, Whitaker R, Kerns BA, et al. Clonal origin of epithelial ovarian carcinoma: analysis by loss of heterozygosity, p53 mutation, and X-chromosome inactivation. J Natl Cancer Inst. 1992;84:1793–8. doi: 10.1093/jnci/84.23.1793. [DOI] [PubMed] [Google Scholar]
- 20.Khalique L, Ayhan A, Whittaker JC, Singh N, Jacobs IJ, Gayther SA, et al. The clonal evolution of metastases from primary serous epithelial ovarian cancers. Int J Cancer. 2009;124:1579–86. doi: 10.1002/ijc.24148. [DOI] [PubMed] [Google Scholar]
- 21.Mulligan KA, Cheyette BN. Wnt signaling in vertebrate neural development and function. J Neuroimmune Pharmacol. 2012;7:774–87. doi: 10.1007/s11481-012-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012;11:790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dooley S, ten Dijke P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012;347:245–56. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulloa L, Tabibzadeh S. Lefty inhibits receptor-regulated Smad phosphorylation induced by the activated transforming growth factor-beta receptor. J Biol Chem. 2001;276:21397–404. doi: 10.1074/jbc.M010783200. [DOI] [PubMed] [Google Scholar]
- 25.Souchelnytskyi S, Ronnstrand L, Heldin CH, ten Dijke P. Phosphorylation of Smad signaling proteins by receptor serine/threonine kinases. Methods Mol Biol. 2001;124:107–20. doi: 10.1385/1-59259-059-4:107. [DOI] [PubMed] [Google Scholar]
- 26.Semba S, Kusumi R, Moriya T, Sasano H. Nuclear Accumulation of B-Catenin in Human Endocrine Tumors: Association with Ki-67 (MIB-1) Proliferative Activity. Endocr Pathol. 2000;11:243–50. doi: 10.1385/ep:11:3:243. [DOI] [PubMed] [Google Scholar]
- 27.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang JD, Seol SY, Leem SH, Kim YH, Sun Z, Lee JS, et al. Genes associated with recurrence of hepatocellular carcinoma: integrated analysis by gene expression and methylation profiling. Journal of Korean medical science. 2011;26:1428–38. doi: 10.3346/jkms.2011.26.11.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du J, Sun C, Hu Z, Yang Y, Zhu Y, Zheng D, et al. Lysophosphatidic acid induces MDA-MB-231 breast cancer cells migration through activation of PI3K/PAK1/ERK signaling. PLoS One. 2010;5:e15940. doi: 10.1371/journal.pone.0015940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rettig M, Trinidad K, Pezeshkpour G, Frost P, Sharma S, Moatamed F, et al. PAK1 kinase promotes cell motility and invasiveness through CRK-II serine phosphorylation in non-small cell lung cancer cells. PLoS One. 2012;7:e42012. doi: 10.1371/journal.pone.0042012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popper L, Batra S. Muscarinic acetylcholine and histamine-receptor mediated calcium mobilization and cell-growth in human ovarian-cancer cells. Int J Oncol. 1994;4:453–9. doi: 10.3892/ijo.4.2.453. [DOI] [PubMed] [Google Scholar]
- 32.Francis H, DeMorrow S, Venter J, Onori P, White M, Gaudio E, et al. Inhibition of histidine decarboxylase ablates the autocrine tumorigenic effects of histamine in human cholangiocarcinoma. Gut. 2012;61:753–64. doi: 10.1136/gutjnl-2011-300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng F, Han Y, Staloch D, Francis T, Stokes A, Francis H. The H4 histamine receptor agonist, clobenpropit, suppresses human cholangiocarcinoma progression by disruption of epithelial mesenchymal transition and tumor metastasis. Hepatology. 2011;54:1718–28. doi: 10.1002/hep.24573. [DOI] [PubMed] [Google Scholar]
- 34.Stoyanov E, Uddin M, Mankuta D, Dubinett SM, Levi-Schaffer F. Mast cells and histamine enhance the proliferation of non-small cell lung cancer cells. Lung Cancer. 2012;75:38–44. doi: 10.1016/j.lungcan.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 35.Medina VA, Brenzoni PG, Lamas DJ, Massari N, Mondillo C, Nunez MA, et al. Role of histamine H4 receptor in breast cancer cell proliferation. Front Biosci (Elite Ed) 2011;3:1042–60. doi: 10.2741/e310. [DOI] [PubMed] [Google Scholar]
- 36.Wang ZY, Ding Y, Miki T, Warita K, Matsumoto Y, Takeuchi Y, et al. Nerve growth factor and receptors are significantly affected by histamine stimulus through H1 receptor in pancreatic carcinoma cells. Mol Med Report. 2010;3:103–9. doi: 10.3892/mmr_00000225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.