SUMMARY
Aerobic glycolysis (Warburg effect) is a core hallmark of cancer, but the molecular mechanisms underlying it remain unclear. Here, we identify an unexpected central role for mTORC2 in cancer metabolic reprogramming where it controls glycolytic metabolism by ultimately regulating the cellular level of c-Myc. We show that mTORC2 promotes inactivating phosphorylation of class IIa histone deacetylases that leads to the acetylation of FoxO1 and FoxO3, and this in turn releases c-Myc from a suppressive miR-34c-dependent network. These central features of activated mTORC2 signaling, acetylated FoxO and c-Myc levels are highly inter-correlated in clinical samples, and with shorter survival of GBM patients. These results identify a specific, Akt-independent, role for mTORC2 in regulating glycolytic metabolism in cancer.
INTRODUCTION
Metabolic reprogramming is a core hallmark of cancer (Ward and Thompson, 2012). Cancer cells convert the majority of their glucose into lactate, providing a supply of glycolytic intermediates as carbon-containing precursors for macromolecular biosynthesis. This biochemical adaptation (the Warburg effect) occurs even in the presence of sufficient oxygen to support oxidative phosphorylation (Dang, 2012b; Koppenol et al., 2011; Vander Heiden et al., 2009; Warburg, 1956), and enables cancer cells to meet the coordinately elevated anabolic and energetic demands imposed by rapid tumor growth (Tong et al., 2009). Uncovering the molecular circuitry by which the Warburg effect is activated and maintained may provide new insights into cancer pathogenesis that might be exploited through identification of new drug targets or detection of drug resistance mechanisms.
C-Myc is a critical regulator of cancer cell metabolism, including the Warburg effect (Dang et al., 2009). Here, we report an unexpected Akt-independent role for mTOR complex 2 (mTORC2) in regulating c-Myc levels and inducing metabolic reprogramming in glioblastoma (GBM), the most common and lethal form of brain cancer. We show that mTORC2 is required for the growth of GBM cells in glucose, but not galactose, and demonstrate that this is mediated by regulating the intracellular level of c-Myc. mTORC2 is shown to control these levels by Akt-independent phosphorylation of class IIa histone deacetylases which leads to the acetylation of FoxO1 and FoxO3, causing release of c-Myc from a suppressive miR-34c-dependent network. The net consequence of this series of events is the conferral of resistance to PI3K and Akt inhibitor in vivo and shorter survival in patients.
RESULTS
mTORC2 Is Required for GBM Growth in Glucose, through Myc-dependent, Akt-independent Signaling
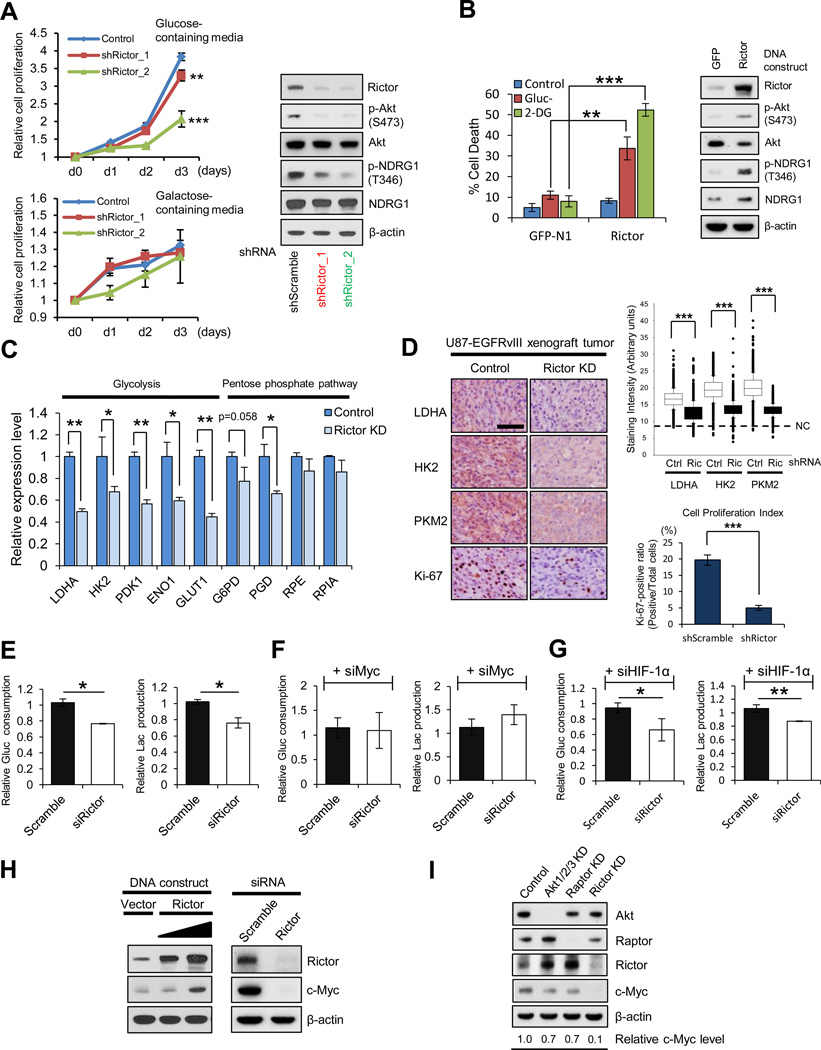
To determine the role of mTORC2 in regulating glycolytic metabolism, we performed genetic depletion of mTORC2 using rictor shRNA in GBM cells expressing EGFRvIII, a commonly mutated oncogene in GBM (Cancer Genome Atlas Research Network, 2008). EGFRvIII potently activates mTORC2 (p-Akt S473 and p-NDRG1 T346; Tanaka et al., 2011) and promotes glycolytic gene expression, tumor cell proliferation and aerobic glycolysis (Babic et al., 2013; Guo et al., 2009) (Figures S1A-S1C). In a dose-dependent fashion, rictor shRNA knockdown suppressed the ability of GBM cells to grow in glucose, the effect of which became apparent by day 2 with increasing magnitude of effect by day 3. In contrast, control and rictor knockdown GBM cells displayed the similar proliferation rate by day 3 grown in galactose, a medium that reduces glycolytic flux and forces cells to rely on mitochondrial oxidative phosphorylation (Finley et al., 2011; Marroquin et al., 2007) (Figure 1A). Further, rictor overexpression rendered GBM cells exquisitely vulnerable to glucose-deprivation or treatment with the glycolytic inhibitor, 2-Deoxy-D-glucose (2-DG) (Figure 1B). Rictor shRNA knockdown also suppressed glycolytic gene expression (Figures 1C and 1D), significantly inhibited glucose consumption, lactate production, glutamine uptake and glutamate secretion (Figures 1E and S1E) and limited tumor cell proliferation in an in vivo GBM xenograft model (Figure 1D). These results demonstrate that mTORC2 promotes glycolysis, enhancing the ability of GBM cells to grow in glucose, but also making them more dependent on glycolysis for survival.
Figure 1. mTORC2 Is Required for GBM Growth in Glucose through c-Myc.
(A) Growth curves of scramble or Rictor knockdown (KD) U87-EGFRvIII cells, cultured in media containing glucose or galactose. Error bars, ± SD. Immunoblot showing the verification of Rictor KD in U87-EGFRvIII cells.
(B) Cell deaths of GFP or Rictor overexpressing U87 cells after 48 h treatment with glucose deprivation (Gluc-) or the glycolytic inhibitor, 2-Deoxy-D-glucose (2-DG, 10 mM). Immunoblot showing the verification of Rictor overexpression in U87 cells.
(C) mRNA levels of glycolysis and pentose phosphate pathway (PPP) enzymes in control or Rictor KD U87-EGFRvIII cells.
(D) Cell-based immunohistochemical analysis for glycolytic enzymes and a proliferative marker Ki-67 in U87-EGFRvIII xenograft tumors with scramble or Rictor shRNA (n = 3). Scale bar, 50 µm. NC denotes the averaged staining intensity obtained by negative control of each sample.
(E-G) Relative glucose consumption and lactate production in control versus Rictor KD U87-EGFRvIII cells (E), combined with c-Myc KD (F) or HIF-1α KD (G).
(H) Biochemical analysis of c-Myc expression for Rictor overexpression in U87 cells and Rictor KD in U87-EGFRvIII cells.
(I) Immunoblot analysis of c-Myc in U87-EGFRvIII cells with indicated siRNAs regarding Akt, mTORC1 (Raptor) and mTORC2 (Rictor).
All error bars except growth curves (A), SEM. See also Figure S1.
C-Myc siRNA knockdown phenocopied the effect of mTORC2 genetic depletion on glycolytic gene expression (Figure S1D), raising the possibility that mTORC2 controls GBM glycolytic metabolism through c-Myc. Rictor siRNA knockdown suppressed c-Myc expression, whereas rictor overexpression potently enhanced mTORC2 signaling and resulted in elevated c-Myc protein levels (Figure 1H). Most importantly, c-Myc siRNA knockdown completely abrogated the inhibitory effect of mTORC2 genetic depletion on glycolysis (Figures 1F and S1F). HIF-1α is also implicated as a key regulator of the glycolytic phenotype in cancer (Kaelin and Ratcliffe, 2008; Tong et al., 2009) and has been reported to be regulated by mTOR signaling (Hudson et al., 2002). In contrast to c-Myc, genetic depletion of HIF-1α by siRNA knockdown did not alter the inhibitory effect of mTORC2 genetic depletion on glucose consumption, lactate production, glutamine uptake and glutamate secretion (Figures 1G and S1G). Taken together, these results demonstrate that mTORC2 regulates glycolytic metabolism in GBM cells through c-Myc.
mTORC2 is thought to control glycolytic metabolism through Akt (Dang, 2012b; Hagiwara et al., 2012; Plas and Thompson, 2005). However, rictor knockdown almost completely suppressed c-Myc expression, whereas the effect of Akt or raptor (mTORC1) depletion on c-Myc levels was modest (Figure 1I). These results raised the possibility that mTORC2 regulates c-Myc and resultant glycolytic metabolism in GBM cells independent of Akt or mTORC1.
mTORC2 Regulates c-Myc Level and Glycolysis through FoxO Acetylation
To identify an Akt-independent mechanism by which mTORC2 could regulate glycolytic metabolism, we examined two post-translational modifications of the FoxO family of transcription factors. Inactivating phosphorylation by Akt and subsequent nuclear exclusion of FoxOs is a very well-described mechanism by which PI3K-activated tumor cells increase glycolysis (Biggs et al., 1999; Dang, 2012b), potentially by relieving suppression of c-Myc (Dang, 2012a; Ferber et al., 2012; Peck et al., 2013). We reasoned that an alternative post-translational modification might be responsible for the Akt-independent regulation of FoxOs by mTORC2.
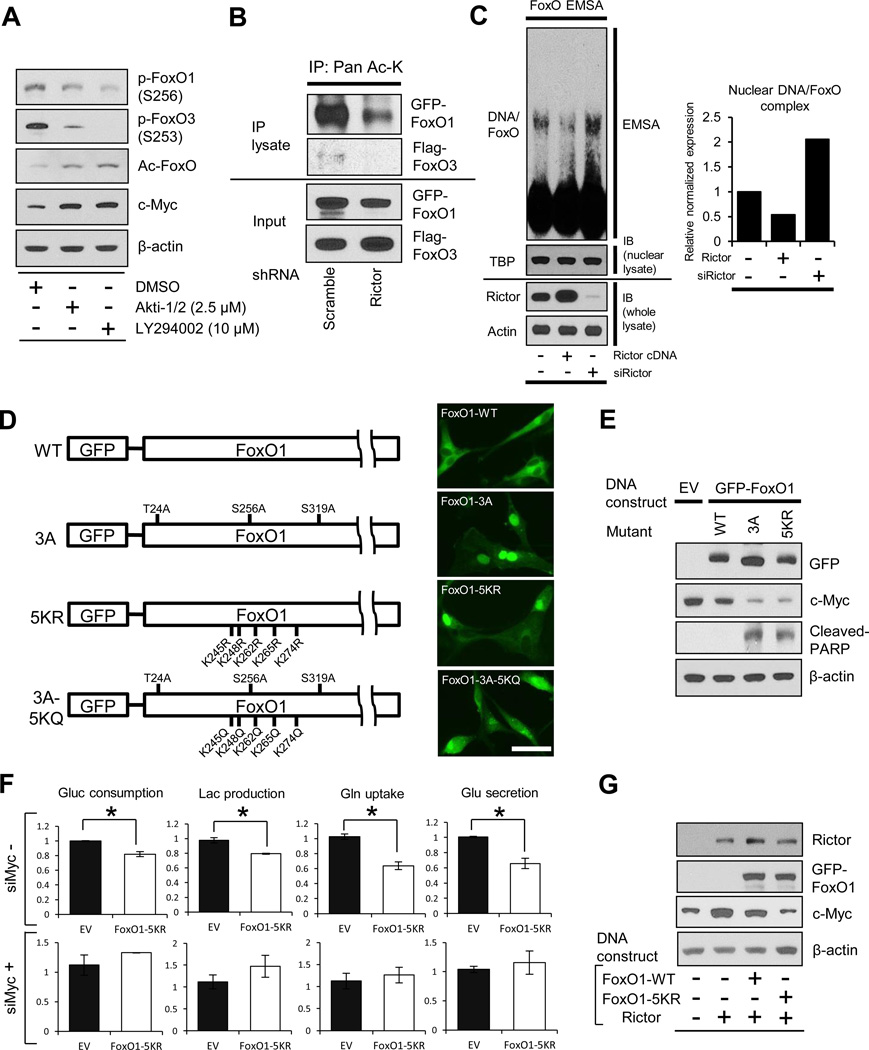
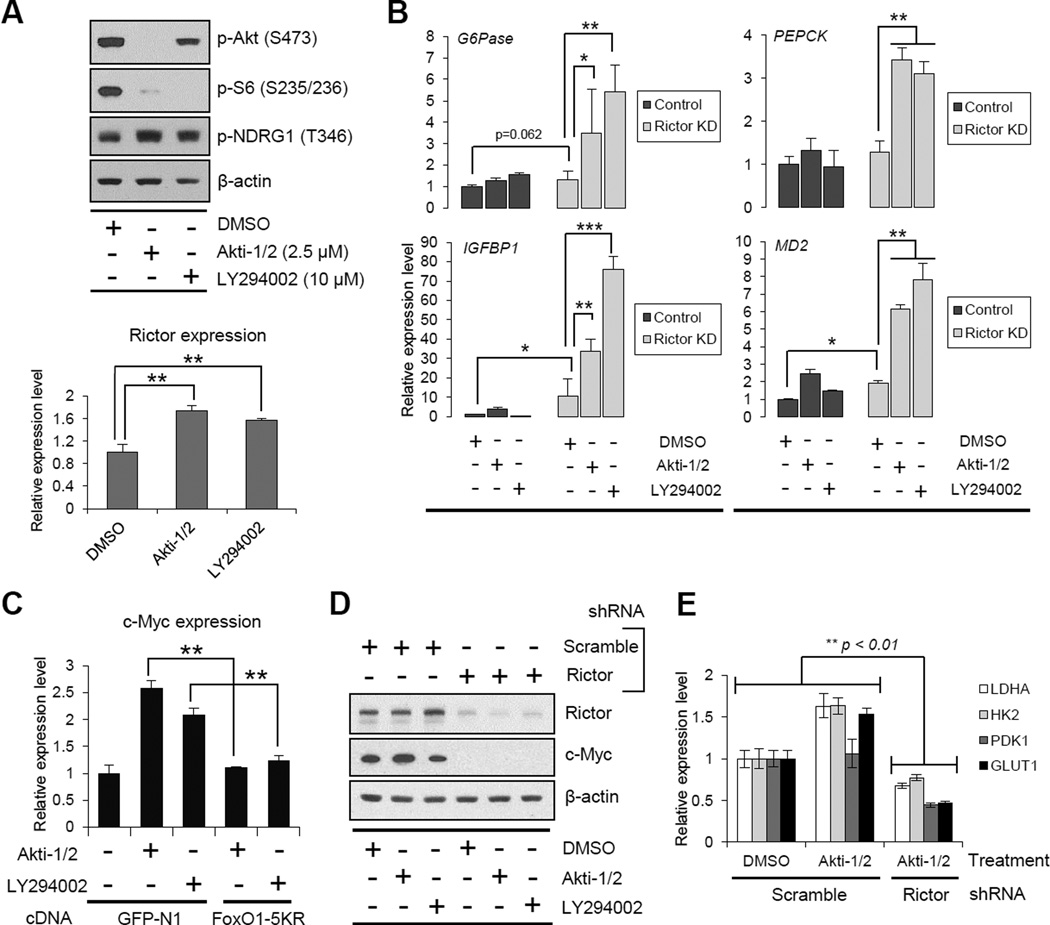
Pharmacologic inhibition of PI3K or Akt potently decreased FoxO phosphorylation, as expected (Figure 2A). However, surprisingly, elevated levels of acetylated FoxO, as well as increased expression of c-Myc, were detected using validated anti-acetyl-FoxO1 antibodies in GBM cells treated with LY294002 (pan-PI3K inhibitor) or Akti-1/2 (Akt inhibitor), raising the possibility of compensatory feedback regulation to maintain c-Myc expression through FoxO acetylation (Figure 2A). By an IP analysis, we determined that genetic depletion of mTORC2 by rictor shRNA knockdown decreased the level of acetylated FoxO1 and 3 (Figure 2B). Rictor knockdown also increased the formation of FoxO-DNA complexes (Figure 2C), enhanced FoxO target gene expression (Figure S2A) and regulated c-Myc and glycolytic gene expression in a FoxO-dependent fashion (Figures S2D and S2E). Conversely, rictor overexpression increased the level of FoxO acetylation (Figure S2B) and suppressed the formation of FoxO-DNA complexes (Figure 2C). Taken together, these results suggest that mTORC2 controls c-Myc expression and glycolytic metabolism in GBM by regulating FoxO acetylation.
Figure 2. mTORC2 Regulates c-Myc and Glycolysis through FoxO Acetylation.
(A) Phosphorylated FoxO, acetylated FoxO and c-Myc protein levels in U87-EGFRvIII treated with Akt inhibitor (Akti-1/2) or Pan-PI3K inhibitor (LY294002) for 24h.
(B) IP analysis of the association of acetyl-lysine (Ac-K) with FoxO plasmids in U87-EGFRvIII cells which were co-transfected with GFP-FoxO1 and Flag-FoxO3, and depleted with or without Rictor.
(C) EMSA assay with the use of nuclear extracts from U87 with Rictor overexpression or Rictor KD, showing the DNA/FoxO-protein EMSA complex. Immunoblots for TATA-binding protein (TBP) were used to normalize protein loading for nuclear extracts. Quantitative bar graph demonstrated relative DNA/FoxO complex levels in each group.
(D) Schematic illustration of GFP-FoxO1 mutants: 3A, Akt-mediated phosphorylation-resistant; 5KR, acetylation-resistant; 5KQ, constitutively-acetylated form. Immunofluorescent images representing U87-EGFRvIII cells expressing GFP-FoxO1 and mutants. Scale bar, 20 µm.
(E) Immunoblot analysis on the effects of wild type, 3A-, or 5KR-FoxO1 on c-Myc and cleaved PARP.
(F) Relative glucose consumption, lactate production, glutamine uptake and glutamate secretion in empty vector or FoxO1-5KR mutant overexpressing U87-EGFRvIII cells with or without depletion of c-Myc.
(G) Immunoblot assessment of c-Myc in U87 cells co-transfected with Rictor-expressing vector and wild type/mutant FoxO-expressing vector.
Error bars, SEM. See also Figure S2.
To better understand the role of FoxO acetylation in regulating mTORC2-dependent glycolytic metabolism, we employed a panel of FoxO plasmids that are mutated in key phosphorylation and/or acetylation sites (Biggs et al., 1999; Brunet et al., 2004; Nakamura et al., 2000; van der Horst and Burgering, 2007; Zhao et al., 2010) (Figure 2D). These strategic constructs enabled us to compare the relative role of each post-translational modification in regulating glycolytic metabolism. As expected, a phosphorylation-resistant mutant, FoxO1-3A, as well as another phosphorylation resistant mutant (FoxO3-TM, data not shown) localized to the nucleus, suppressed c-Myc expression, and induced tumor cell apoptosis (Figures 2D and 2E). An acetylation resistant mutant (FoxO1-5KR) and an analogous FoxO3 mutant (FoxO3-4KR, data not shown), were also localized to the nucleus, similarly suppressing c-Myc expression and inducing GBM cell apoptosis (Figures 2D and 2E), thus suggesting that either post-translational modification of FoxO could regulate c-Myc expression. Therefore, we introduced a double mutant bearing residues that induce phosphorylation resistance and additional residues that mimic constitutive acetylation. This FoxO1-3A-5KQ construct, which was excluded from the nucleus, partially restored c-Myc levels and suppressed apoptosis induced by the FoxO1-3A mutant (Figure S2C). These results suggest a dominant role for FoxO acetylation relative to phosphorylation in regulating c-Myc and survival in GBM cells and suggest a dual-pronged mechanism of FoxO regulation that is conferred through two different post-translational modifications. To determine whether FoxO acetylation was a critical determinant of the glycolytic phenotype of GBM cells, we assessed the impact of the acetylation resistant mutant on glycolysis. Introduction of the FoxO1-5KR acetylation resistant mutant decreased glycolysis, inhibiting glucose and glutamine uptake and suppressed the secretion of lactate and glutamate (Figure 2F). Importantly, this inhibitory effect of the FoxO1-KR mutant on glycolysis was completely abrogated by c-Myc siRNA (Figure 2F). Further, up-regulation of c-Myc by mTORC2 was also completely blocked by the FoxO1-5KR mutant (Figure 2G). Taken together, these results demonstrate that mTORC2 regulates c-Myc expression and glycolysis in GBM cells through FoxO acetylation.
mTORC2 Controls FoxO Acetylation through Class IIa HDACs, Independent of Akt
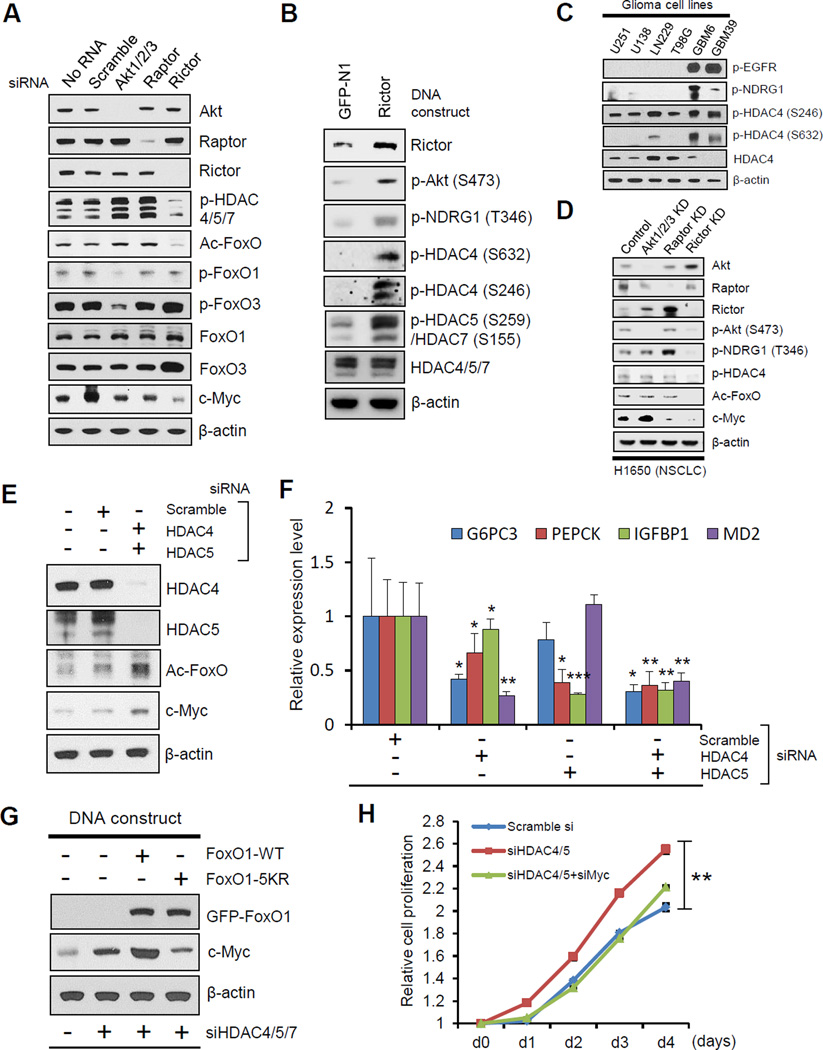
Acetylation of FoxOs is controlled, in part, through the balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs) (van der Horst and Burgering, 2007, Mihaylova et al., 2011; Wang et al., 2011). Gene expression microarray analysis identified only two genes from the HAT and HDAC families that were differentially expressed, HDAC4 and HDAC5 (class IIa HDACs), which were both significantly lower in EGFRvIII-expressing tumors with elevated mTORC2 signaling (Figure S3A). However, their level of down-regulation in our cellular system was modest. Therefore, we focused on other, potentially more GBM-relevant mechanisms of regulation, including post-translational modification (Mihaylova et al., 2011). We tested the possibility that mTORC2-dependent FoxO acetylation was mediated through inactivating phosphorylation of class IIa HDACs. Genetic depletion of mTORC2 with rictor siRNA suppressed class IIa HDAC phosphorylation, concomitant with inhibition of FoxO acetylation and loss of c-Myc expression (Figure 3A). Conversely, rictor overexpression markedly enhanced HDAC4, HDAC5 and HDAC7 phosphorylation (Figure 3B). Importantly, genetic depletion of Akt or raptor (mTORC1) using siRNA knockdowns did not suppress class IIa HDAC phosphorylation or regulate levels of acetylated FoxO and c-Myc. Further, across a panel of GBM cell lines, the level of phosphorylation of HDAC4 was correlated with mTORC2 signaling (Figures 3C, S3B and S3C). Additionally, the expression level of class IIa HDACs is inversely correlated with their phosphorylation status (Figures 3C and S3B), and future studies will be necessary to assess the possibilities that EGFR/mTORC2 signaling controls the level of class IIa HDACs by destabilizing them through phosphorylation (Potthoff et al., 2007) or decreasing their transcription (Figure S3A). These results demonstrate a specific role for mTORC2 in regulating class IIa HDAC phosphorylation in GBM that is independent of Akt. Notably, this mTORC2-HDAC-FoxO-Myc axis was also identified in other cancer types. In EGFR mutant H1650 non-small cell lung cancer cells, rictor knockdown suppressed HDAC4 phosphorylation and abrogated expression of acetylated FoxO and c-Myc (Figure 3D). Further, pharmacological inhibition of Akt, mTORC1 and mTORC2 in H1650 cells, as well as in A549 non-small cell lung cancer cells and HeLa cervical cancer cells confirmed that suppression of mTORC2 signaling correlated with loss of FoxO acetylation and suppression of c-Myc, and that these effects were clearly Akt-independent (Figure S3D). These results do not exclude a role of mTORC1 in controlling c-Myc levels in other cancer types, but rather indicate that the mTORC2-dependent, Akt-independent, Myc-dependent pathway identified here may not be limited to GBM, but may be active in a broader range of cancer subtypes.
Figure 3. mTORC2 Controls FoxO Acetylation through Class IIa HDACs, Independent of Akt.
(A) Immunoblot analysis of c-Myc, phosphorylated HDAC, and several forms of FoxO in U87-EGFRvIII cells with indicated siRNAs regarding Akt and mTOR complex.
(B) Immunoblot showing change in phosphorylated class IIa HDACs from U87 cells overexpressing GFP or Rictor DNA plasmids.
(C) Immunoblot analysis for the status of p-EGFR, p-NDRG1 and class IIa HDACs in several glioma cell lines.
(D) Immunoblot analysis of phosphorylated HDAC, acetylated FoxO and c-Myc in EGFR-mutated, non-small cell lung cancer (NSCLC) cells (H1650) with indicated KD regarding Akt and mTOR complex.
(E) Immunoblot showing change of acetylated FoxO and c-Myc in U87 cells with indicated siRNAs against class IIa HDACs.
(F) qRT-PCR from U87 cells of FoxO target genes following siRNA-mediated depletion of HDAC4/5.
(G) Immunoblot showing c-Myc amount from U87 cells bearing siRNAs against class IIa HDACs, combined with overexpression of FoxO DNA plasmids.
(H) Cell proliferation assay of scramble or class IIa HDAC KD U87 cells, combined with or without c-Myc depletion. p < 0.01 for comparison between siHDAC cells and scramble siRNA cells, or siHDAC/siMyc cells. Error bars, ± SD.
All error bars except growth curves (H), SEM. See also Figure S3.
We next assessed the dependence of GBM cells on class IIa HDACs for regulation of FoxO acetylation and Myc expression. Knockdown of class IIa HDACs increased FoxO acetylation and c-Myc expression (Figure 3E), impairing FoxO transcriptional activity (Figure 3F). Genetic depletion of class IIa HDACs with siRNA knockdown, or pharmacological inhibition of class IIa HDACs, abrogated the effect of rictor knockdown on c-Myc expression (Figures S3E and S3F), indicating that mTORC2 regulates c-Myc through class IIa HDACs. Further, the acetylation-resistant FoxO1-5KR mutant prevented c-Myc up-regulation in response to knockdown of class IIa HDACs (Figure 3G), confirming that class IIa HDACs regulate c-Myc through FoxO acetylation. More importantly, knockdown of class IIa HDACs promoted GBM cell proliferation, which was reversed by c-Myc co-depletion (Figure 3H). Taken together, the findings demonstrate that inactivating phosphorylation of class IIa HDACs by mTORC2 controls FoxO acetylation and c-Myc levels, promoting GBM proliferation.
Acetylated FoxO Regulates c-Myc through miR-34c
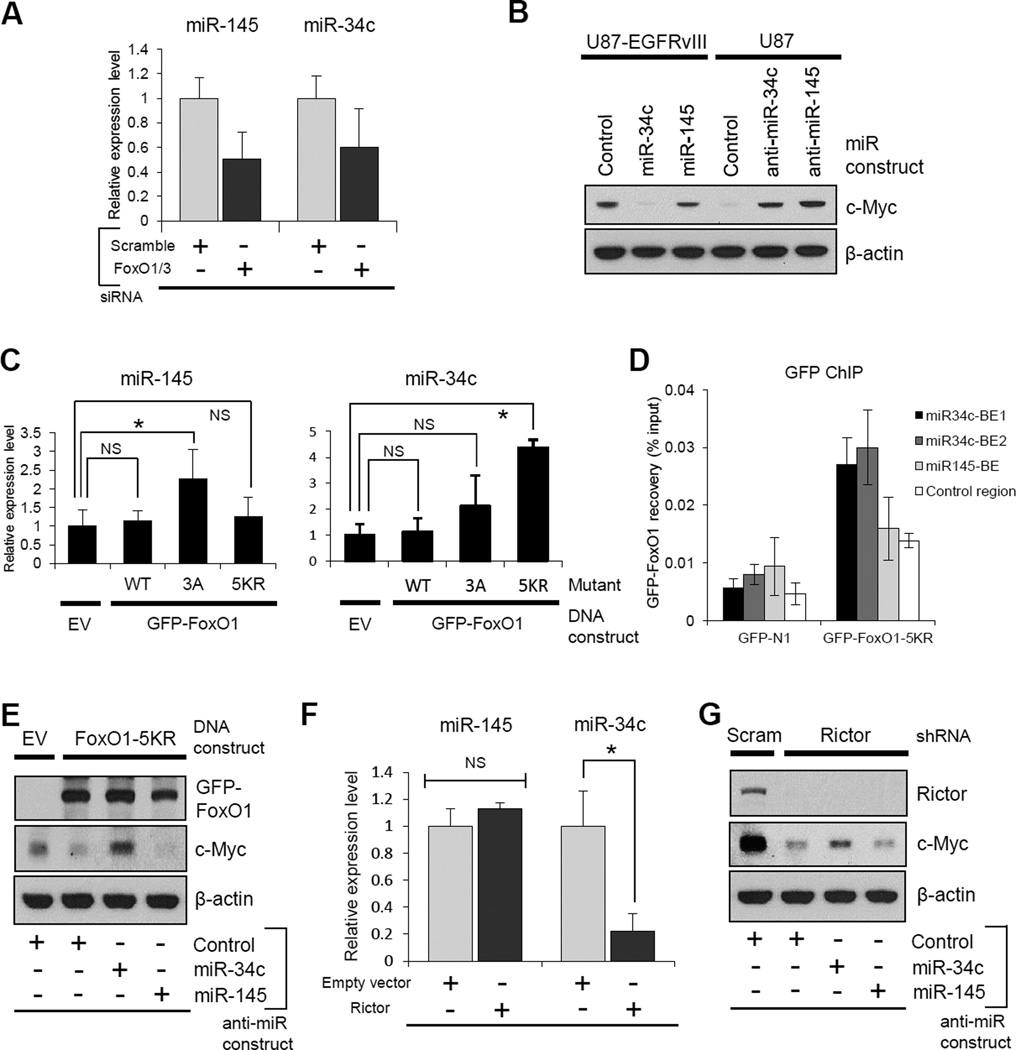
FoxOs antagonize c-Myc (Dang, 2012a, Ferber et al., 2012; Peck et al., 2013), possibly by increasing the expression of miR-145 (Gan et al., 2010) and miR-34c (Kress et al., 2011), limiting c-Myc mRNA stability and translation. In GBM cells, miR-145 and miR-34c levels were both suppressed by FoxO1/FoxO3 knockdown (Figure 4A), and introduction of miR-34c or miR-145 or their anti-miR constructs, demonstrated that these two microRNAs regulate c-Myc levels in GBM cells (Figure 4B). To determine whether these microRNAs are independently regulated by differential post-translational modifications of FoxO, we took advantage of the FoxO phosphorylation and acetylation mutants. Introduction of the FoxO phosphorylation-resistant mutant (FoxO1-3A) into GBM cells up-regulated expression of miR-145, but not miR-34c (Figure 4C). In contrast, introduction of the acetylation resistant mutant (FoxO1-5KR) increased the expression of miR-34c, but not miR-145 (Figure 4C). ChIP analysis further showed that the FoxO1-5KR acetylation resistant mutant was preferentially enriched in the miR-34c promoter regions, but not in the miR-145 promoter region (Figure 4D), and the decrease in c-Myc expression in response to the FoxO1-5KR acetylation resistant mutant was reversed by anti-miR-34c, but not by anti-miR-145 (Figure 4E). Furthermore, mTORC2 activation resulted in a decrease in miR-34c expression, but not miR-145 (Figure 4F), while rictor knockdown partially restored the suppression of c-Myc by anti-miR-34c, but not by anti-miR-145 (Figure 4G). The effect of anti-miR-34c on c-Myc expression in the setting of rictor depletion is limited, suggesting that, in addition to the miRNA regulation, there may exist other functions of rictor in control of c-Myc, considering the fact that the expression and activity of c-Myc is regulated by a variety of factors at the multiple levels (Albihn et al., 2010; Liu and Levens, 2006). Taken together, these results indicate that acetylated FoxO increases c-Myc levels by relieving miR-34c-dependent suppression.
Figure 4. Acetylated FoxO Regulates c-Myc through miR-34c.
(A) Relative expression of miR-145 and miR-34c in scramble versus FoxO KD U87 cells.
(B) Myc protein levels in U87-EGFRvIII cells with miRNA mimics, and U87 with miRNA inhibitors.
(C) Relative expression changes of miR-145 and miR-34c in U87-EGFRvIII cells transfected with indicated FoxO plasmids.
(D) ChIP analysis on U87-EGFRvIII cells transfected with control vector or GFP-FoxO1-5KR, and assessed for GFP-FoxO1 recovery on binding elements (BEs) in miR-34c promoter (Kress et al., 2011) and miR-145 promoter (Gan et al., 2010) regions.
(E) 5KR-FoxO1-mediated down-regulation of c-Myc was reverted by the inhibition of miR-34c, but not miR-145 in U87-EGFRvIII cells.
(F) mRNA changes of miR-145 and miR-34c in U87 cells transfected with empty vector or Rictor-expressing vector.
(G) Immunoblot assessment of c-Myc change in U87-EGFRvIII cells co-transfected with shRictor and miRNA inhibitors.
Error bars, SEM.
Resistance to PI3K and Akt Inhibitors Is Mediated by mTORC2-dependent Acetylation of FoxO and Consequent Maintenance of c-Myc Levels
Having shown that FoxO and its downstream regulation of c-Myc are controlled through two independent, and highly specific pathways of post-translational modification and microRNA suppression, we reasoned that therapeutic resistance to PI3K or Akt inhibitors by sustained c-Myc activity could be one critical and potentially clinically actionable consequence. To test this hypothesis, we examined the effect of pharmacologic inhibition of PI3K or Akt on FoxO acetylation, c-Myc expression and tumor cell survival. Treatment of GBM cells with LY294002 or Akti1/2 suppressed PI3K and Akt signaling, respectively, but failed to promote FoxO target gene expression, concomitant with a compensatory elevation in p-NDRG1 and rictor levels (Figures 5A, 5B and S4A). This continued suppression of FoxO activity was mediated through mTORC2, because rictor knockdown markedly elevated FoxO target gene expression in the presence of PI3K or Akt inhibition (Figures 5B and S4A). The restoration of FoxO activities by the combined inhibition of mTORC2 and PI3K/Akt was striking, and could potentially be due to the fact that mTORC2 inhibition could regulate FoxO function both through acetylation and through Akt and SGK1-dependent phosphorylation (Guertin et al., 2006; Zhao et al., 2011). Additionally, this recovery of FoxO activity by combined inhibition of PI3K/Akt and mTORC2 was reversed by class IIa HDACs inhibition to a greater degree than it was by expression of a constitutively active form of Akt (HA-Akt-E17K), suggesting the dominance of acetylation in regulating FoxO transcriptional activity (Figures S4B and S4C). Importantly, treatment of GBM cells with PI3K or Akt inhibitors significantly elevated c-Myc levels, and this was completely reversed by the acetylation-resistant FoxO1-5KR mutant (Figure 5C), and by rictor knockdown (Figure 5D). Further, treatment of GBM cells with Akti-1/2 actually increased the mRNA level of the glycolytic enzymes LDHA, HK2, PDK1 and GLUT1, which was completely abrogated by rictor knockdown (Figure 5E). Taken together, these results strongly indicate that GBM cells treated with PI3K or Akt inhibitors maintain c-Myc levels and enhanced glycolysis through mTORC2 feedback-promoted FoxO acetylation.
Figure 5. Resistance to PI3K and Akt Inhibitors Is Mediated by mTORC2-dependent Acetylation of FoxO and Consequent Maintenance of c-Myc.
(A) mTORC2 activation under Akt/PI3K inhibition in U87-EGFRvIII cells shown by Western blotting for p-NDRG1 protein and qRT-PCR for Rictor mRNA.
(B) qRT-PCR analysis for FoxO target genes in U87-EGFRvIII cells treated by PI3K/Akt inhibitors for 24 h, combined with or without Rictor KD. Targeting both PI3K/Akt and mTORC2 dramatically restores FoxO activity.
(C) qRT-PCR from U87-EGFRvIII cells of c-Myc gene following treatment with PI3K/Akt inhibitor, transfected with or without FoxO1-5KR.
(D) Immunoblot assessment of c-Myc in U87-EGFRvIII cells treated by PI3K/Akt inhibitors, combined with or without Rictor KD.
(E) mRNA levels of Myc-related metabolic enzymes in U87-EGFRvIII cells treated by PI3K/Akt inhibitors, combined with or without Rictor KD.
Error bars, SEM. See also Figure S4.
Combined Inhibition of PI3K/Akt and mTORC2 Suppresses Acetylated FoxO-c-Myc Signaling and Promotes Tumor Cell Death
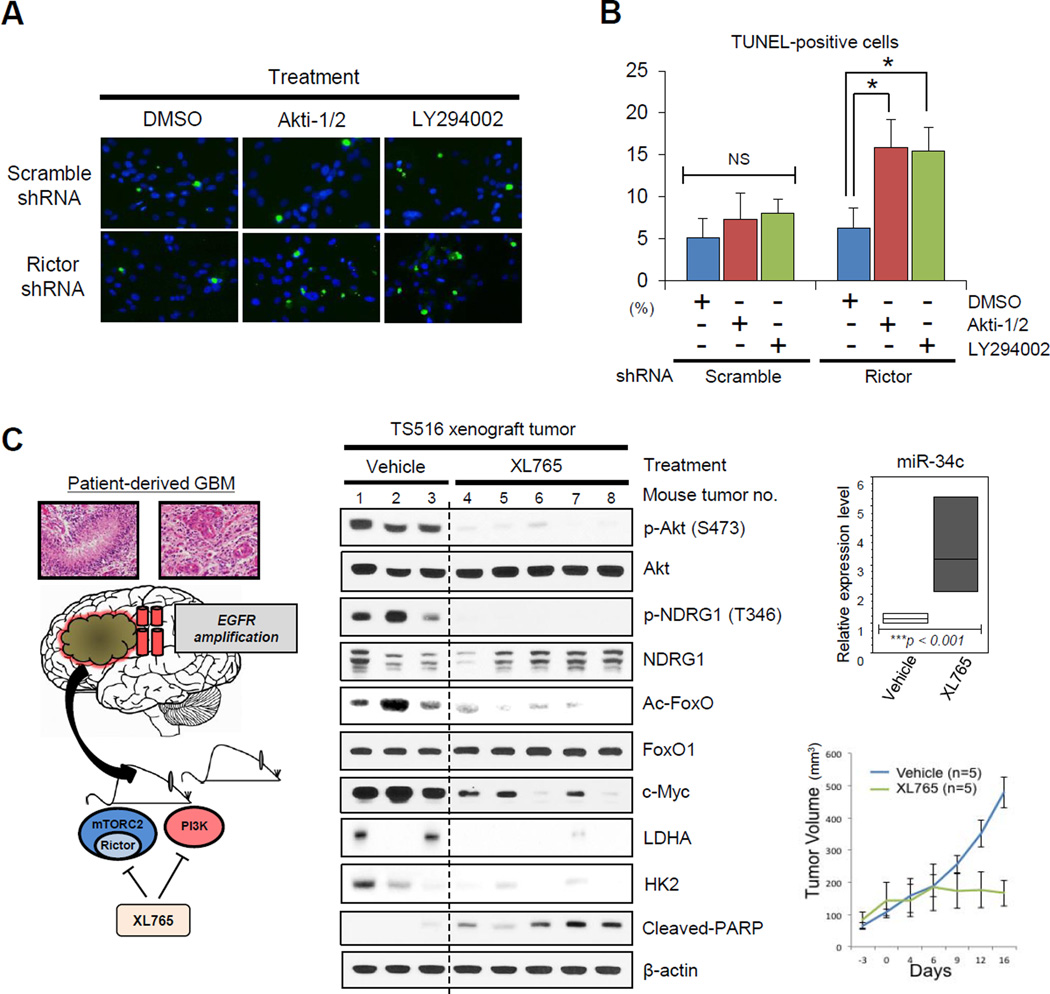
If mTORC2-dependent FoxO acetylation maintains c-Myc levels to drive PI3K or Akt inhibitor resistance, then combined suppression of mTORC2, and PI3K or Akt should decrease cellular levels of c-Myc, potentially causing tumor cell death. Consistent with this hypothesis, rictor shRNA knockdown synergized with LY294002 or Akti-1/2 to cause GBM tumor cell death (Figures 6A and 6B). Therefore, we asked whether dual pharmacologic inhibition of PI3K and mTOR kinase could also suppress FoxO acetylation, decrease c-Myc levels, lower glycolytic enzyme expression and cause tumor cell death. To test this, we treated the patient-derived GBM xenograft model TS516 (Rohle et al., 2013) with the dual PI3K/mTOR inhibitor XL765 (SAR245409) (Figure 6C). XL765 inhibited mTORC2 signaling, blocked FoxO acetylation, increased miR-34c expression, decreased cellular levels of c-Myc and reduced expression of glycolytic enzymes LDHA and HK2. Most importantly, XL765 potently reduced tumor growth, inducing tumor cell apoptosis (Figure 6C). Currently, no specific mTORC2 inhibitors exist, and compounds and ATP-competitive mTOR kinase inhibitors like XL765 also affect mTORC1 signaling. However, in the context of the mTORC2 knockdown experiments (Figures 6A and 6B), these results indicate that pharmacological inhibition of mTORC2 prevents c-Myc-dependent PI3K inhibitor resistance (Dang, 2012a; Ilic et al., 2011; Muellner et al., 2011) through inhibition of FoxO acetylation.
Figure 6. Combined Inhibition of PI3K/Akt and mTORC2 Suppresses Acetylated FoxO-Myc Signaling and Promotes Tumor Cell Death.
(A) TUNEL staining in U87-EGFRvIII cells treated by PI3K/Akt inhibitors for 24 h, combined with or without Rictor KD. Green, TUNEL staining; blue, DAPI staining.
(B) Quantified TUNEL-positive U87-EGFRvIII cells treated by PI3K/Akt inhibitors, combined with or without Rictor KD in the bar graph.
(C) An EGFR-amplified patient-derived TS516 GBM tumor sphere was implanted into immunodeficient mice which were subsequently treated with the dual PI3K/mTOR inhibitor XL765. Representative immunoblots displaying the status of FoxO acetylation, c-Myc and glycolytic enzyme expression, and apoptotic tumor cell death (cleaved PARP). Relative expression of miR-34c is shown in the box graph. Tumor volumes were measured by using length and width for vehicle-treated (n = 5) and XL765-treated (n = 5) groups.
Error bars, SEM.
mTORC2/Acetylated FoxO/c-Myc Alterations in Clinical Human GBMs
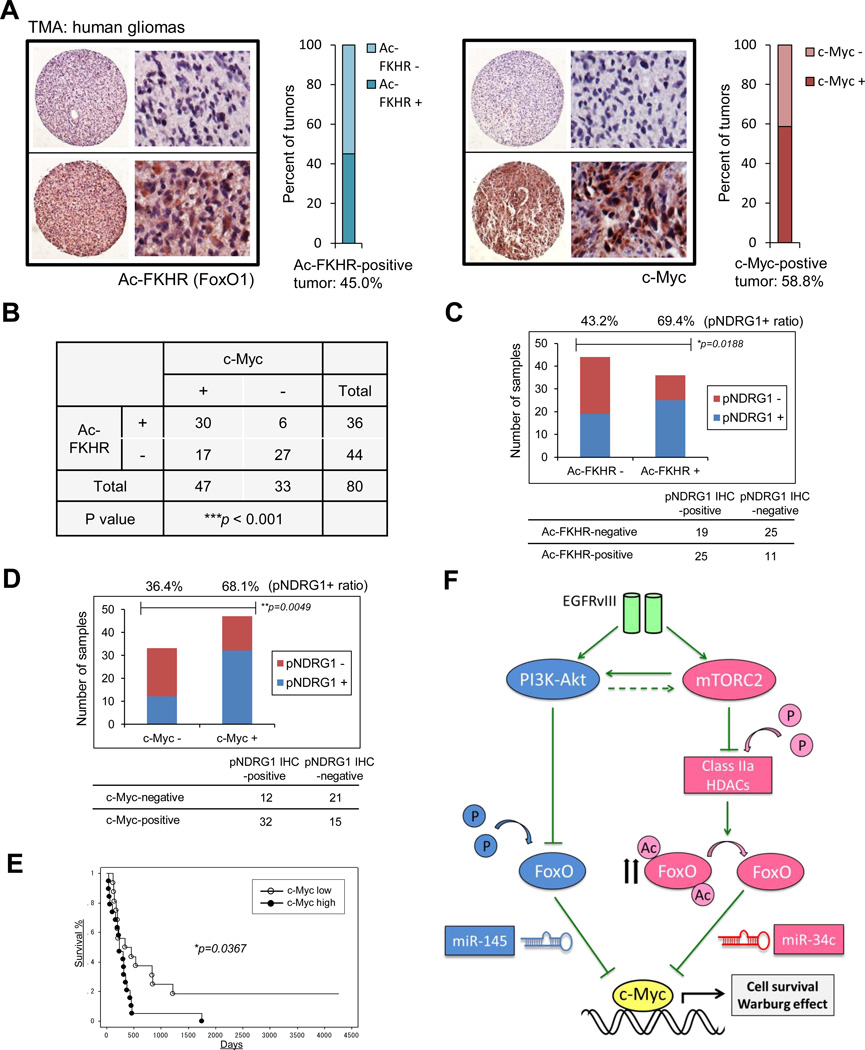
To explore the clinical implications of mTORC2-acetylated FoxO-Myc signaling and assess its impact on prognosis, we performed immunohistochemical analysis of a tissue microarray (TMA) that contained 26 normal brain samples and 80 GBM tumor samples (Figure 7A). Acetylated FoxO and c-Myc were both highly elevated in GBMs relative to normal brain (Ac-FoxO: p = 0.013883, c-Myc: p = 9.58832E-05), being individually up-regulated in 45.0% and 58.8% of tumors, respectively (Figure 7A). mTORC2 signaling (as measured by pNDRG1; Tanaka et al., 2011), acetylated FoxO and c-Myc were also highly correlated with each other (Figures 7B-7D, S5A and S5B). Immunoblot analysis of GBM autopsy samples confirmed coordinate increases in the mTORC2-acetylated FoxO-Myc axis, as well as glycolytic enzymes, in tumor tissue relative to normal brain (Figures S5C and S5D), and elevated acetylated FoxO and c-Myc were found to be highly correlated (Spearman’s rank correlation coefficient, r2 = 0.8288) in this limited number of autopsy samples (n = 8). Finally, elevated acetylated FoxO and c-Myc on TMA were significantly associated with shorter overall survival in GBM patients (p = 0.0367 for c-Myc, p = 0.0799 for Ac-FoxO; Figures 7E and S5E). These results demonstrate that mTORC2 signaling, acetylated FoxO and c-Myc expression are coordinately up-regulated in GBM patients with worse prognoses (Figure 7F).
Figure 7. mTORC2 Signaling, Acetylated FoxO and c-Myc Expression Are Highly Inter-correlated in Biopsy Samples, and Associated with Poor Prognosis in GBM Patients.
(A) Ac-FoxO and c-Myc immunostaining of GBM tissue microarray (TMA) comprising 80 GBM samples and 26 normal brain tissue. View of the TMA slide and an example of a negative and a positive core at high magnification to show cytoplasmic staining of Ac-FoxO and cytoplasmic/nuclear staining of c-Myc. Ac-FoxO and c-Myc are both individually up-regulated in 45.0% and 58.8 % of tumors, respectively.
(B) Immunohistochemical analysis of TMAs based on correlation of Ac-FoxO with c-Myc.
(C) Bar graph showing differential association of Ac-FoxO-positve or -negative tumors with p-NDRG1 IHC positivity based on TMA.
(D) Differential association of c-Myc +/− tumors with p-NDRG1 immunopositivity based on TMA. P-value was determined by χ2 for independence test (B-D).
(E) Kaplan-Meier survival analysis for overall survival of 36 primary and secondary GBM samples classified by c-Myc expression. Log-rank (Mantel-Cox) test was used to determine p values for Kaplan-Meier survival curve analyses.
(F) mTORC2 inhibits FoxO activity via acetylation which could bypass PI3K/Akt inhibition, leading to the up-regulation of c-Myc, a key downstream effector of cell proliferation and tumor metabolism in GBM.
See also Figure S5.
DISCUSSION
The Warburg effect enables cancer cells to obtain a sufficient supply of macromolecular precursors required for rapid cellular proliferation, while still meeting their energy requirements (Tong et al., 2009). C-Myc plays a central role in regulating this metabolic hallmark of cancer (Cairns et al., 2011; Deberardinis et al., 2008; Koppenol et al., 2011; Levine and Puzio-Kuter, 2010; Vander Heiden et al., 2009; Ward and Thompson, 2012). Although many receptor signaling pathways including Wnt, Shh, Notch, TGF-beta and RTK signaling through the PI3K pathway have been implicated in Myc up-regulation (Dang, 2012a)), the mechanisms by which mutated growth factor receptor signaling pathways engage c-Myc remain incompletely understood.
We recently showed that EGFRvIII regulates glycolytic metabolism and tumor growth through hnRNPA1-dependent alternative splicing of the Myc binding partner Max (Babic et al., 2013). This alternatively spliced product, Delta Max, promotes GBM cell proliferation in glucose and is required for tumor growth in a Myc-dependent fashion (Babic et al., 2013). Taken together with the results presented here, a model is emerging in which aberrant growth factor receptor signaling in GBM engages c-Myc signaling through two complementary and interlacing mechanisms: 1) alternative splicing of Delta Max (Babic et al., 2013) to modulate c-Myc function and 2) control of cellular levels of c-Myc through mTORC2, as demonstrated here. Importantly, both mechanisms are required for GBM growth through their effects of Myc-dependent glycolytic metabolism, because genetic depletion of either Delta Max (Babic et al., 2013) or mTORC2 (Figure 1) blocks the ability of GBM cells to utilize glucose, but not galactose, for tumor cell proliferation, in a c-Myc dependent fashion. These mechanisms also appear to be cooperative, acting at different points of c-Myc regulation, thus highlighting the role for c-Myc in GBM pathogenesis, and suggesting that its pathogenicity, at least in part, may be mediated through up-regulation of glycolytic metabolism.
A surprising implication of this study arises from the observation that GBM cells treated with PI3K or Akt inhibitors maintain c-Myc levels and enhanced glycolysis through mTORC2 feedback-promoted FoxO acetylation. We show that FoxO and its downstream regulation of c-Myc are tightly controlled through two independent, and highly specific pathways of post-translational modification and microRNA suppression. One pathway, inactivating phosphorylation of FoxO by Akt, is a well-characterized mechanism enabling PI3K-activated tumor cells to engage c-Myc (Biggs et al., 1999; Bouchard et al., 2004; Delpuech et al., 2007; Dang, 2012a; Peck et al., 2013), by relieving FoxO-dependent miR-145 suppression of c-Myc. (Gan et al., 2010). In contrast, the results shown here identify a central mechanism of regulation of the FoxO-Myc axis to control glycolysis, as we demonstrate that mTORC2 controls the acetylation of FoxO through class IIa HDACs, independent of Akt.
The pathway identified here complements the previously demonstrated ability of mTORC2 to regulate glycolysis in the liver and in cancer cells through Akt activation (Dang, 2012b; Hagiwara et al., 2012; Plas and Thompson, 2005), providing yet another important signaling mechanism by which growth factor receptor mutations and tumor suppressor losses promote the Warburg effect in cancer (Bensaad et al., 2006; Christofk et al., 2008; Dang, 2012b; Faubert et al., 2013; Tong et al., 2009; Vander Heiden et al, 2009; Ward and Thompson, 2012). Thus, mTORC2 emerges as a particularly critical regulator of cancer cell metabolism through two mechanisms, Akt-dependent and Akt-independent signaling, each one regulating cellular levels of c-Myc by distinct post-translational modifications of FoxO, to relieve suppression of c-Myc through distinct suppressive microRNA networks. FoxO is a key intermediate between growth factor receptor PI3K signaling and c-Myc; thus it is not surprising that the FoxO-Myc axis (Biggs et al., 1999; Bouchard et al., 2004; Delpuech et al., 2007; Dang, 2012a; Peck et al., 2013) has evolved to be regulated by this dual pronged post-translational control. mTORC2 regulates both of mechanisms of FoxO-c-Myc axis regulation and this nominates it as a critical metabolic regulator in cancer that must be suppressed in addition to Akt to abrogate c-Myc-dependent glycolysis and tumor growth.
EXPERIMENTAL PROCEDURES
Image Analysis-Based Scoring of Immunohistochemistry
Quantitative image analysis was performed with Soft Imaging System software (Olympus MicroSuite Analytical Suite). Representative images from each immunostained section were photographed using a Colorview II camera mounted on an Olympus BX61 microscope. Images were captured from representative regions of the tumor with sufficiently high tumor cell content based on H&E staining evaluation. Borders between individual cells were approximated using a filter function. The amount of reaction product per cell was determined by measuring mean saturation per cell in the red-brown hue range. 1000–1500 cells per case (on average) were measured for each marker. Negative control staining was also performed for each section without primary antibodies to determine the threshold for immunopositivity.
Tissue Microarray (TMA)
TMA was constructed as reported previously (Guo et al., 2009), and immunohistochemical staining was performed as described previously (Choe et al., 2003; Lu et al., 2009) to analyze the expression of Ac-FoxO, c-Myc, p-NDRG1 and Rictor in 80 GBM samples and 26 normal brain samples. Cores were scored by a pathologist, and tumor staining intensity was compared to normal brain tissue.
Subcellular Fractionation and Electrophoretic Mobility Shift Assay (EMSA)
Nuclear fractionation was prepared from sub-confluent 10 cm plates using a kit according to the manufacturer’s instructions (Active Motif). EMSA was carried out using EMSA “Gel-Shift” Kit (Panomics). Nuclear extracts were incubated with a biotin-labeled oligonucleotide containing the consensus binding sequence for human FoxO (5'-CAAAACAACAAAACAACAAAACAA-3'), and the transcription factor-bound oligonucleotide was separated from unbound oligonucleotide by electrophoresis on a 6% polyacrylamide gel. After transferred to nylon membrane (GE Healthcare), the biotin-labeled bands were visualized using horseradish peroxidase-based chemiluminescence.
Mutagenesis
To generate the 5KR (using FoxO1) and 5KQ (using FoxO1 and FoxO1-AAA) mutants, we replaced the previously-reported major acetylation sites (van der Horst and Burgering, 2007; Zhao et al., 2010) K245, K248, K262, K265 and K274 with arginine or glutamine, respectively. To generate the 4KR (using FoxO3) mutant, we replaced the acetylation sites (van der Horst and Burgering, 2007) K242, K259, K271 and K290 with arginine. We carried out site-directed mutagenesis using the Quick Change kit (Stratagene).
Metabolite Measurements
Glucose, lactate, glutamine and glutamate in the media of cultured cells were measured using the BioProfile Basic 4 analyzer (Nova Biomedical). Fresh media with 1% FBS were added to a 6-well plate of sub-confluent cells, and metabolite concentration in the media was measured 24 hours later by comparison with blank media without cells, and normalized to the number of cells in each well.
Real-Time RT-PCR and miRNA Studies
Total RNA was extracted by the use of RNeasy Plus Mini Kit (QIAGEN). First-strand cDNA was synthesized by the use of SuperScript III transcriptase (Invitrogen). Real-time RT-PCR was performed with the iQ SYBR Green Supermix (Bio-Rad) on an iCycler (Bio-Rad) following the manufacturer's instructions. Primer sequences are available upon request. microRNAs were extracted by mirVana miRNA Isolation Kit (Applied Biosystems). microRNA reverse transcription was conducted by TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems), and miR-145, miR-34c and RNU19 expression was detected by TaqMan MicroRNA Assays (Applied Biosystems). β-actin was used as an endogenous control for qRT-PCR, and RNU19 was used as an endogenous control for miRNA assays.
Glucose-dependent Cell Proliferation and Cell Death Assay
Cells were placed in 96-well plates at 2.5 × 103 cells/well in 100 µl of growth medium and then incubated in each condition of treatment. For the measurement of glucose-dependent proliferation, DMEM containing glucose (Cellgro) or no-glucose DMEM (Gibco) supplemented with 4.5g/l Galactose (Sigma) was used as previously reported (Finley et al., 2011). Cell proliferation was examined with Cell Proliferation Assay Kit (Millpore) according to the manufacturer’s instructions. The absorbance of the treated and untreated cells was measured with a microplate reader (Bio-Rad) at 420 to 480nm. For the glucose-dependent cell death assay, cells were cultured with no-glucose DMEM + 10%FBS + Galactose (4.5g/l) or DMEM + 10%FBS + 2-Deoxy-D-glucose (2-DG, 10 mM) for 48 h, and live/dead cells were quantified by cell counting with trypan blue exclusion and TC10™ Automated Cell Counter (Bio-Rad). Data represent the mean ± SD of three independent experiments.
Animal Studies
TS516 tumor sphere lines originated from a GBM patient at Memorial Sloan Kettering Cancer Center, or U87 and U87-EGFRvIII cell lines were implanted into immunodeficient SCID/Beige mice for subcutaneous xenograft studies. SCID/Beige mice were bred and kept under defined-flora pathogen-free conditions at the Association for Assessment of Laboratory Animal Care-approved Animal Facility of the Division of Experimental Radiation Oncology, UCLA. For subcutaneous implantation, exponentially growing tumor cells in culture were trypsinized, enumerated by trypan blue exclusion, and resuspended at 3×106 cells/ml in a solution of Dulbecco’s phosphate-buffered saline and Matrigel (BD Biosciences). Tumor growth was monitored with calipers by measuring the perpendicular diameter of each subcutaneous tumor. Tumor was treated with a PI3K/mTOR dual inhibitor (XL765; 60mg/kg) or normal saline every day for 19 days. Mice were euthanized if tumors reached 14mm in maximum diameter or animals showed signs of illness. All experiments were conducted after approval by the Chancellor’s Animal Research Committee of UCLA.
Statistical Analysis
Unpaired Student’s t tests were performed unless otherwise noted. Error bars represented SEM unless otherwise noted, and statistical significance was indicated as *p < 0.05, **p < 0.01, and ***p < 0.001.
Supplementary Material
HIGHLIGHTS.
mTORC2 is required for the growth of GBM cells in glucose
mTORC2 controls glycolytic metabolism by regulating the cellular level c-Myc
mTORC2 signaling acetylates FoxO1 and FoxO3 to up-regulate c-Myc
mTORC2, acetylated FoxO and c-Myc axis confers shorter survival in patients
ACKNOWLEDGEMENTS
We thank Dr. Cameron Brennan and Alicia Pedraza (Memorial Sloan Kettering Cancer Center) for kindly giving us TS516 GBM sphere cell lines, Dr. Ciro Zanca, Tomoyuki Koga, John Anzola and Yuki Ishii (Ludwig Institute for Cancer Research) for GBM6, GBM39, H1650, A549 and HeLa cell lines, and the UCLA Brain Tumor Translational Resource for biospecimen and biorepository support. A PI3K/mTOR dual inhibitor, XL765 (SAR245409) was kindly provided by Exelixis/Sanofi to Dr. Mellinghoff. This work is supported by grants from National Institute for Neurological Diseases and Stroke (NS73831), the National Cancer Institute (CA119347), The Ben and Catherine Ivy Foundation, and generous donations from the Ziering Family Foundation in memory of Sigi Ziering. WKC is a Fellow of the National Foundation for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors report no conflict of interest.
REFERENCES
- Albihn A, Johnsen JI, Henriksson MA. MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res. 2010;107:163–224. doi: 10.1016/S0065-230X(10)07006-5. [DOI] [PubMed] [Google Scholar]
- Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, Zhu S, Gu Y, Villa GR, Akhavan D, et al. EGFR Mutation-Induced Alternative Splicing of Max Contributes to Growth of Glycolytic Tumors in Brain Cancer, Cell Metabolism. Cell Metab. 2013;17:1000–1008. doi: 10.1016/j.cmet.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. 2006. [DOI] [PubMed] [Google Scholar]
- Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. U S A. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Marquardt J, Brás A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23:2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Choe G, Horvath S, Cloughesy TF, Crosby K, Seligson D, Palotie A, Inge L, Smith BL, Sawyers CL, Mischel PS. Analysis of the phosphatidylinositol 3'-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63:2742–2746. [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012a;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Links between metabolism and cancer. Genes Dev. 2012b;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer. Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, Schulze A. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol. Cell. Biol. 2007;27:4917–4930. doi: 10.1128/MCB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber EC, Peck B, Delpuech O, Bell GP, East P, Schulze A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell. Death. Differ. 2012;19:968–979. doi: 10.1038/cdd.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, Hu J, Jiang S, Fletcher-Sananikone E, Zhuang L, et al. FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell. 2010;18:472–484. doi: 10.1016/j.ccr.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Guo D, Hildebrandt IJ, Prins RM, Soto H, Mazzotta MM, Dang J, Czernin J, Shyy JY, Watson AD, Phelps M, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc. Natl. Acad. Sci. U S A. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Rüegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol. Cell. Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc. Natl. Acad. Sci. USA. 2011;108:E699–E708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Kress TR, Cannell IG, Brenkman AB, Samans B, Gaestel M, Roepman P, Burgering BM, Bushell M, Rosenwald A, Eilers M. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol. Cell. 2011;41:445–457. doi: 10.1016/j.molcel.2011.01.023. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- Liu J, Levens D. Making myc. Curr. Top. Microbiol. Immunol. 2006;302:1–32. doi: 10.1007/3-540-32952-8_1. [DOI] [PubMed] [Google Scholar]
- Lu KV, Zhu S, Cvrljevic A, Huang TT, Sarkaria S, Ahkavan D, Dang J, Dinca EB, Plaisier SB, Oderberg I, et al. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009;69:6889–6898. doi: 10.1158/0008-5472.CAN-09-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner MK, Uras IZ, Gapp BV, Kerzendorfer C, Smida M, Lechtermann H, Craig-Mueller N, Colinge J, Duernberger G, Nijman SM. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat. Chem. Biol. 2011;7:787–793. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Ramaswamy S, Vazquez F, Signoretti S, Loda M, Sellers WR. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 2000;20:8969–8982. doi: 10.1128/mcb.20.23.8969-8982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck B, Ferber EC, Schulze A. Antagonism between FOXO and MYC Regulates Cellular Powerhouse. Front. Oncol. 2013;3:96. doi: 10.3389/fonc.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas DR, Thompson CB. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, Gini B, Dang J, Zhu S, Yang H, Jesus JD, et al. Oncogenic EGFR signaling activates an mTORC2-NF-kB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–538. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell. Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, Yates JR, 3rd, Fischer WH, Thomas JB, Montminy M. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang Y, Zhu WG. Applications of post-translational modifications of FoxO family proteins in biological functions. J. Mol. Cell. Biol. 2011;3:276–282. doi: 10.1093/jmcb/mjr013. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell. Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.