Abstract
Insulin is known to be an important regulator of milk secretion in the lactating mammary gland. Here we examine the role of insulin signaling in mammary development in pregnancy using a mouse with a floxed insulin receptor (IR) crossed with a mouse expressing Cre specifically in the mammary gland. In the mammary glands of these IRfl/fl Cre+ mice, expression of IR is significantly diminished throughout development. Glands from these mice had 50% fewer alveoli at midpregnancy; casein and lipid droplets were diminished by 60 and 75%, respectively, indicating a role for IR both in alveolar development and differentiation. In an acinar preparation from mammary epithelial cells (MEC) isolated from pregnant mice, insulin stimulated lumen formation, mammary cell size, acinar size, acinar casein content, and the formation of lipid droplets with a Km of ∼1.7 nM. IGF-I and IGF-II had no effect at concentrations below 50 nM, and a function blocking antibody to the IGF type 1 receptor did not alter the response to insulin. We conclude that insulin interacting with IR is essential for mammary differentiation during murine pregnancy. Using array analysis, we then examined the expression of genes up- or downregulated >1.5-fold in the IRfl/fl Cre+ MECs, finding significant downregulation of differentiation specific genes and upregulation of cell cycle and extracellular matrix genes. We conclude that insulin fosters differentiation and may inhibit cell proliferation in the mammary gland of the midpregnant mouse.
Keywords: mammary development, acinar culture
the hormonal regulation of mammary development and milk secretion has received extensive research attention in the last half-century (14), and the roles of the major sex steroid hormones, estrogen and progesterone (P4), as well as the lactogenic hormones prolactin (PRL) and hydrocortisone (HC) are quite well understood (26, 28). Insulin is another story. It is difficult to ablate the insulin-secreting pancreatic islets, or to increase plasma insulin, without seriously impairing the metabolism of the whole animal; for this reason, alternative approaches have been necessary to elucidate the role of insulin in mammary gland development. The same concern extends to the use of transgenic and knockout mice that have been used to great effect to study steroid hormones. Although genetically modified mice can be used for studying the insulin system, as we will see the experimental design is crucial if complicating systemic effects are to be avoided.
Much early data demonstrated a role for insulin in maintenance of milk secretion in the lactating animal (1, 3, 15, 18, 19, 22, 25, 33, 35). The earliest studies showed that insulin stimulated lipid synthesis in mammary slices from lactating ruminants (13). However, in these experiments, effects on mammary epithelial and adipose tissues were not distinguished. Furthermore, in the dairy cow, raising insulin levels from 0.25 to 1.3 nM in the context of glucose clamp experiments had no effect on milk production (21), suggesting that the lactating gland of the ruminant, which has an acetate-based metabolism, is not sensitive to the type of changes in insulin levels seen after food consumption. However, when lactating rats were treated with alloxan to ablate the endocrine pancreas, pup growth was completely abrogated (51). Replacement of insulin maintained pup growth. These experiments were the earliest to suggest a total dependence of milk synthesis on adequate insulin. Subsequent studies focused mainly on the effects of insulin in the lactating gland, often using high-fat feeding or starvation to alter endogenous levels of insulin. These treatments decreased de novo fatty acid synthesis, and in both circumstances insulin restored fatty acid synthesis almost to normal in part by restoring the activity of acetyl-CoA carboxylase (7, 24). In a nice study (12), Flint et al. showed that labeled insulin bound to mammary acini with a Km of about 1.8 nM consistent with binding to the insulin receptor (IR). Burnol and colleagues (8) examined tyrosine phosphorylation of the IR from the rat mammary gland in response to insulin, finding a Km around 2 nM, again consistent with binding of insulin to its receptor.
The signaling mechanism through which insulin acts in the lactating mammary gland has been elucidated in more recent studies. Hadsell and colleagues (16) showed that infusion of insulin restored AKT phosphorylation following starvation of a lactating mouse for 8 h, but an active form of IGF-I did not, implicating insulin, not IGF-I, in recovery from starvation. AKT phosphorylation was decreased in insulin receptor substrate-deficient (IRS1−/−) mice, providing evidence that insulin acts through its normal signaling pathway in the lactating gland to maintain milk synthesis. Berlato and Doppler (4) and others (36) analyzed the mRNA expression of the A and B isoforms of IR (IRA and IRB), finding that IRB was markedly upregulated in the lactating mammary gland. Notably tyrosine phosphorylation of IRB protein was stimulated only by insulin, not IGF-I or IGF-II (4). Furthermore, IRB was upregulated threefold at lactation day 1. These results could be replicated in HC11 cells, a “normal” mouse mammary cell line derived from Comma 1D cells (9). These experiments provide the strongest possible evidence that insulin signals through IR in lactation, facilitated by the increased expression of IRB. An important question that remains is whether insulin, interacting with IR, is involved in the differentiation of the mammary gland that takes place during pregnancy.
In the studies presented here, we used mammary-specific targeting of a floxed IR (6) by a Cre recombinase driven by the β-lactoglobin (BLG) promoter to show that secretory differentiation in the mammary gland of the pregnant mouse murine is severely compromised when IR expression is abrogated. We were able to replicate secretory differentiation in an in vitro culture of primary mammary epithelium only in the presence of physiological concentrations of insulin, not IGF-I or IGF-II. Finally, examination of gene expression in mammary epithelial cells (MECs) from IRfl/fl Cre+ mice showed that genes associated with differentiated function were downregulated compared with controls and genes associated with the cell cycle; extracellular matrix and the cytoskeleton were upregulated compared with IRfl/fl Cre− controls. Our findings implicate insulin and its receptor as a major regulator of the switch from proliferation to differentiation that occurs in the murine mammary gland in the second half of pregnancy.
MATERIALS AND METHODS
Mice and breeding.
Mouse strains used were the BLG-Cre mouse (C57Bl6 background) (41) obtained as a gift from Trevor Williams (University of Colorado, Anschutz Medical Center); the IRfl/fl mouse (6) was obtained as a kind gift from C. R. Kahn (Harvard University), and the LacZ reporter mouse, Gt(ROSA)26Sor (43), was obtained from Jackson Laboratories (Bar Harbor, ME). To obtain IRfl/fl Cre+ and IRfl/fl Cre− mice, IRfl/fl mice were crossed with heterozygous BLG-Cre mice. The F1 offspring were crossbred to obtain IRfl/fl Cre+ and IRfl/fl Cre− as well as IRfl/+ Cre+ and IRfl/+ Cre− dams. These were mated with the outbred strain CD1 (Taconic) to obtain litters with as few defects as possible. Finding that only litters nursing IRfl/fl Cre+ dams showed a growth defect (see Fig. 3), for further experiments we selected IRfl/fl Cre+ males and IRfl/fl Cre− females, crossbreeding these to obtain IRfl/fl Cre+ and IRfl/fl Cre− littermates for experiments. CD1 females were bred with CD1 males to obtain MECs for acinar preparations. All animal work adheres to the American Physiological Society's Guiding Principles in the Care and Use of Vertebrate Animals in Research and Training and was approved by the Institutional Animal Care and Use Committee at the University of Colorado, Anschutz Medical Campus.
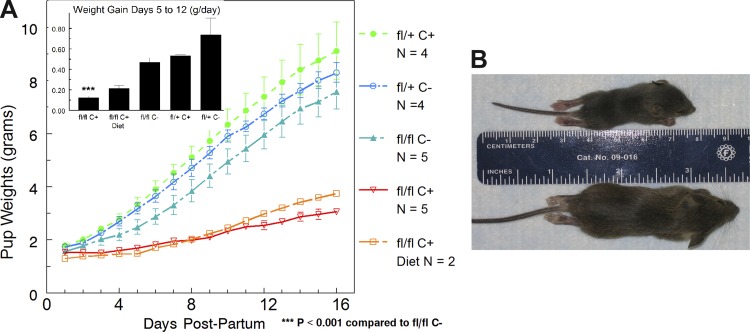
Fig. 3.
Growth of pups nursed by IRfl/fl Cre+ or IRfl/fl Cre− dams. A: litters were weighed daily and divided by the no. of pups per litter, which varied from 4 to 10. Growth curves did not change when weights of pups in largest and smallest litters were excluded. The inset shows the rate of weight gain between days 5 and 12. Two IRfl/fl Cre+ dams were fed a high-fat diet that did not rescue pup growth. B: photographs of pups on day 18. The mouse on top (nursed by an IRfl/fl/Cre+ dam) is fully formed but much shorter and smaller than the pup nursed by a control (bottom).
Detection of Cre recombinase during mammary gland development.
To demonstrate the expression of BLG-Cre transgene throughout mammary development, homozygous LacZ mice were crossed with the BLG-Cre mouse, and mice positive for both transgenes were killed at relevant developmental stages. The mammary glands were dissected, processed for stained whole mounts as described previously (32); the whole mounts were photographed for analysis and then sectioned and stained for histological analysis. Glands were stained for LacZ as previously described (43).
Pup growth.
To assess pup growth, litters from IRfl/fl Cre+ and IRfl/fl Cre− or IRfl/− Cre+ dams were weighed daily without adjustment for litter size. Weights were divided by the number of pups per litter for averaging.
Immunohistochemistry.
Paraffin-embedded sections were dewaxed in xylene two times for 5 min, rehydrated in graded alcohols at 100, 90, 70, and 30% for 3 min each, and then in PBS for 5 min. Antigen retrieval was performed using Antigen Unmasking Solution no. H-3300 (Vector Laboratories, Burlingame, CA), placing the slides in the solution for 10 rounds of heating to boiling for 10 s followed by cooling for 45 s. Sections were permeabilized with 0.2% glycine in PBS for 30 min and then blocked with 10% normal donkey serum and 0.1 mg/ml saponin in PBS for 60 min. Sections were incubated at 4°C overnight with rabbit anti-casein antibody (Neville 7781) and guinea pig anti-adipophilin (ADPH) antibody (20R-AP002; Fitzgerald, Acton, MA) both at 1:100 in PBS. Sections were rinsed five times in PBS and then incubated with Alexa 488-conjugated donkey anti-rabbit antibody at 1:100 (A21206; Invitrogen, Eugene, OR), Cy3-conjugated donkey anti-guinea pig antibody at 1:100 (706-165-148; Jackson Immunoresearch, West Grove, PA), and 0.5 μg/ml DAPI for 45 min at room temperature. The sections were rinsed five times in PBS and mounted. For proliferating cell nuclear antigen (PCNA) detection, the same method was employed using rabbit anti-PCNA antibody (ab18197; Abcam, Cambridge, MA) at 10 μg/ml and donkey anti-rabbit Cy3 at 1:100 (711-166-152; Jackson Immunoresearch).
Preparation of isolated MECs.
Mammary glands 4 and 5 were dissected from 13.5 day pregnant mice and diced on a glass plate followed by techniques that varied slightly depending on the subsequent use of the MECs.
Isolation of MECs for protein analysis.
The technique was adapted from Rudolph et al. (38). The diced tissue was placed in F-12 medium (Mediatech, Manassas, VA) containing 3 mg/ml collagenase A (Roche, Indianapolis, IN), 1.5 mg/ml trypsin, 50 mM NaF, and 1 mM NaVO4 and agitated at 200 rpm for 30 min at 37°C. The separated cells/organoids were washed four times with ice-cold PBS, spinning 8 min at 2,000 rpm to pellet cells/organoids. Western blots for IR were carried out as follows: 30 μg of protein were resolved on a 10% acrylamide gel and then transferred to a PVDF nylon membrane. The PVDF membrane was blocked with 5% nonfat dry milk and incubated with anti-IR antibody (sc-711, 1:300; Santa Cruz), which detects the 90-kDa β-subunit, and anti-β-actin antibody (8227, 1:20,000; Abcam) overnight at 4°C. Anti-rabbit HRP (1:10,000; GE Healthcare) was used as the secondary antibody, and immunoreactive proteins were detected with ECL Prime Western Detection Reagent (RPN 2232; GE Healthcare). Images were captured on photographic film.
Isolation of MECs for acinar culture.
The diced tissue was placed in DMEM-F-12 medium (Mediatech) containing 1 mg/ml collagenase A (Roche), 50 μg/ml gentamicin, 100 U/ml penicillin, and 100 μg/ml streptomycin and agitated at 100 rpm for 80 min at 37°C. DMEM-F-12 (20 ml) with 5% FBS was added, and the sample was spun at 1,500 rpm for 10 min to pellet the cells/organoids. The pellet was washed four times in 10 ml PBS with calcium and magnesium, spinning only 2 s at 1,500 rpm. The resulting pellet was resuspended in 2 ml of 0.05% trypsin and incubated at 37° for 20 min. DMEM-F-12 (8 ml) with 5% FBS was added to the trypsin mixture and then spun at 1,400 rpm for 3 min. The pellet was resuspended in 10 ml DMEM-F-12 with 5% FBS, passed through a 70-μM cell strainer, and then spun at 1,400 rpm for 3 min. The cell pellet was resuspended in 1 ml PBS for counting and use in acinar culture.
Acinar culture.
The isolated MECs were suspended in 95% Matrigel (no. 356231, growth factor reduced; BD Biosciences) at 6.7 × 105 cells/ml. This cell mixture was plated at 150 μl/chamber of a precooled eight-chamber slide. The Matrigel was allowed to solidify for 30 min in a 37°C incubator, and then 200 μl of growth media [DMEM-F-12 with 5% serum, 1 μg/ml HC, 3 μg/ml PRL, 5 ng/ml epidermal growth factor (EGF), 50 μg/ml gentamicin, and insulin as required] were added over the top. After 7 days incubation, the medium was changed to differentiation medium (same as growth medium only without serum and EGF) for an additional 7 days. The acini in the Matrigel were fixed in 60% methanol, 30% chloroform, and 10% acetic acid for 15 min, placed in Histogel, incubated in 70% ethanol for 24 h, embedded in paraffin, and sectioned. Immunostaining was carried out as described above for whole tissues.
Isolation of MECs for gene expression profiling.
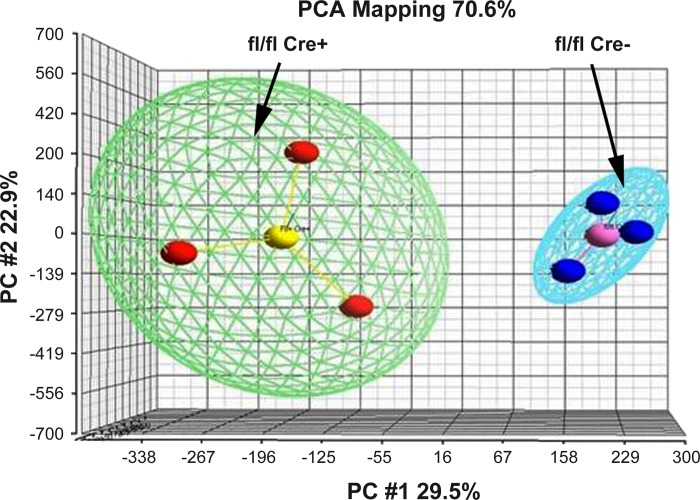
MECs were isolated as above from three or four IRfl/fl Cre+ and IRfl/fl Cre− mice at pregnancy day 13.5 as above. RNA was isolated as previously described (37) and applied to Affymetrix MoGene_1_0-st-v1 chips. Data for both mRNA and miRNA were collected. Only mRNA data are analyzed here, and one sample from the IRfl/fl Cre+ was discarded after principle component analysis (PCA; Fig. 1) as representing incomplete loss of IR. The final analysis was carried out on three IRfl/fl Cre+ and IRfl/fl Cre− mice each. Array data have been deposited with the NCBI Gene Expression Omnibus and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48009.
Fig. 1.
Principle component analysis (PCA) map of IRfl/fl Cre− (blue spheres) and IRfl/fl Cre+ (red spheres). Samples were applied to Affymetrix.ExonExprChip. MoGene-1_0-st-v1 and processed to obtain expression values. These were applied to the PCA function of Partek; the analysis shows that there is distinct separation between experimental (Cre+) and control (Cre−) samples.
Analysis of array results.
Data from the arrays four from IRfl/flCre+ mammary glands and three from MECs from IRfl/flCre− mammary gland were loaded in Genespring X. All entities (28,853) were filtered on variance between samples, eliminating from further analysis genes whose coefficient of variation was >50%. This procedure yielded fewer than 10% of the genes or 2,239 genes. These genes were filtered on expression level in the raw data to give 2,214 genes with expression levels between 20 and 20,000 intensity units. After the use of an unpaired t-test (P < 0.05 with the Benjamini-Hochberg multiple testing correction), 2,014 genes remained. This list was filtered for a fold change of >1.1 between experimental and controls. Of these genes, 890 were down in the Cre+ mice (720 after removal of duplicates and unknowns) and 758 were up in the Cre+ mice (577 after removal of duplicates and unknowns). Most of the analysis in this paper focuses on genes where the fold change between Cre+ and Cre− mice was >1.5 and the P value was <0.01. This filtering produced a list of 218 genes, categorization of which gave a good picture of the physiological processes altered when insulin signaling was abrogated.
Statistics.
For experiments other than the array analysis, we relied on t-tests generated by the appropriate function in either Microsoft Excel or in Slidewrite (see Figs. 5 and 6 and Tables 1 and 2; Advanced Graphics Software, Encinitas CA 91014-2801). A P value of 0.05 or less was considered statistically significant. For Table 2, the best fit to the Michaelis Menton equation was calculated in Slidewrite.
Fig. 5.
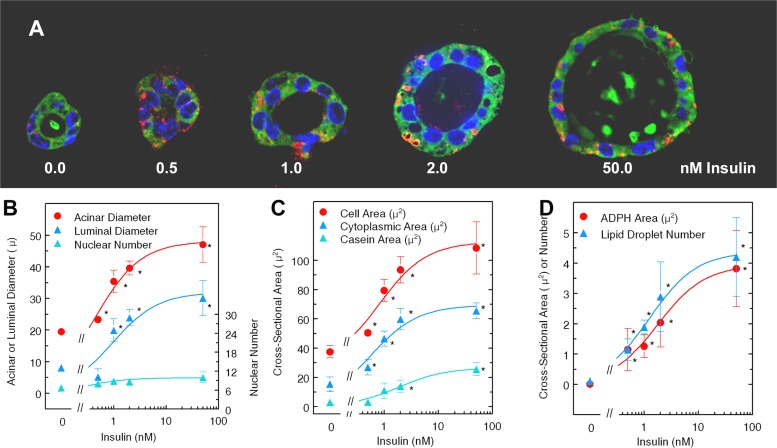
Effect of insulin on properties of acinar cultures. A: representative three-dimensional (3D) acini cultured with designated insulin doses. Acini were fixed and sectioned as in materials and methods and stained with antibodies to adipophilin (ADPH, red), casein (green), and DAPI for nuclei. B, C, and D: Slidebook analysis of acini cultured for 7 days with growth medium followed by culture in differentiation medium (see materials and methods for composition) at concentrations of insulin from 0 to 50 nM. Each point represents the mean parameters from three to five acini. Each parameter was fit to the Michaelis-Menton equation using the curve fit function of SlideWrite. The best-fitting parameters (Table 2) were then used to plot the lines shown. B: acinar and luminal diameters and no. of nuclei. Diameters were measured using the ruler function of Slidebook. Nuclei were counted by hand from the DAPI-stained structures. C: cell, cytoplasmic, and casein areas. Cell areas were calculated from the equation area = {[(DA/2)2 × π] − [(DL/2)2 × π]}/N where DA is the acinar diameter, DL is the luminal diameter, and N is the no. of nuclei in that acinus. Cytoplasmic area was defined as the total area of green stain that did not overlap with DAPI-stained nuclei, and the casein area was defined as the area of high-intensity green stain. Luminal stain was not always present, and, when it was, it was not included in these calculations. D: ADPH area or no. of particles stained for ADPH (red) as calculated by the count and area functions of Slidebook. *Mean value differs from mean at 0 nM insulin, P < 0.05.
Fig. 6.
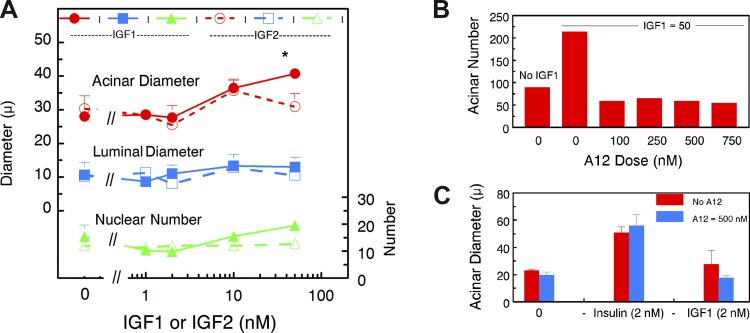
Effect of IGF-I and IGF-II on MEC differentiation in 3D acinar cultures. A: dose-response curves for the effect of IGF-I and IGF-II on acinar and luminal diameter and nuclear numbers in 14-day acinar cultures. No significant difference between any parameters in A were observed with increasing concentrations of IGF-I or IGF-II except that the acinar diameter at 50 nM IGF-I was significantly higher than the diameter in the absence of IGFs (*P < 0.05). B: dose effect of antibody A12, a function blocking antibody for the IGF-I receptor, on acinar no. in the presence of 50 nM IGF-I. C: blocking the IGF-I receptor (IGF-IR) has no effect on the insulin response at 2 nM insulin.
Table 1.
Properties of alveoli in the mammary glands of IRfl/fl Cre+ and IRfl/fl Cre− mice on pregnancy day 13.5
| Parameter | IRfl/fl Cre− | IRfl/fl Cre+ | Probability |
|---|---|---|---|
| Alveolar area, % | 24 ± 4 | 12.3 ± 3.5 | <0.05 |
| Alveolar no./field | 60 ± 2 | 30 ± 2 | <0.001 |
| Alveoli/g gland | 512 ± 109 | 181 ± 51 | <0.03 |
| Nuclei/alveolus | 11.6 ± 0.5 | 12.0 ± 0.3 | NS |
| PCNA-positive cells, % | 0.83 ± 0.2 | 1.8 ± 0.6 | >0.22 |
| Casein area/DAPI | 17.3 ± 1.6 | 5.5 ± 1.4 | <0.002 |
| ADPH area/DAPI | 8.0 ± 1.2 | 2.0 ± 0.6 | <0.005 |
Values are means ± SE; n = 3 mice/strain. PCNA, proliferating cell nuclear antigen; ADPH, adipophilin. Areas occupied by alveoli, as well as casein and lipid droplets, were estimated using the quantitation function of Slidebook, examing 3–6 randomly chosen microscopic fields for each of 3 animals. NS, not significant.
Table 2.
Fit parameters: Michaelis-Menton analysis from curve-fit function in SlideWrite*
| Units | C | Max | Max SE | Km | Km SE | R2 | F-Value | |
|---|---|---|---|---|---|---|---|---|
| Acinar diameter | μ | 19.5 | 29.1 | 3.78 | 1.22 | 0.49 | 0.91 | 30.7 |
| Luminal diameter | μ | 0 | 31.97 | 6.42 | 0.95 | 0.63 | 0.83 | 14.3 |
| Nuclear no./acinus | 6.7 | 3.28 | 0.37 | 0.90 | 0.33 | 0.91 | 34.0 | |
| Cell area | μ2 | 27.4 | 85.3 | 9.27 | 0.81 | 0.31 | 0.94 | 23.1 |
| Cytoplasmic area | μ2 | 0 | 69.4 | 9.95 | 0.56 | 0.32 | 0.90 | 26.0 |
| High-intensity casein | μ2 | 0 | 26.68 | 2.67 | 1.87 | 0.58 | 0.96 | 63.8 |
| ADPH area | μ2 | 0 | 3.94 | 0.21 | 1.77 | 0.3 | 0.98 | 153.9 |
| Lipid droplet no./cell | 4.37 | 0.2 | 1.26 | 0.18 | 0.99 | 246.8 | ||
| 0 | Mean Km 1.17 | |||||||
Equation fitted for analysis in Slidewrite: Y = C + Max + [Ins]/(Km + [Ins]), where, C is the y-intercept when [Ins] = 0, [Ins] is the insulin concentration; Max is the maximum value at infinite insulin, and Km is the dissociation constant for insulin from the insulin receptor (IR).
RESULTS
BLG-Cre deletes floxed sites throughout mammary gland development.
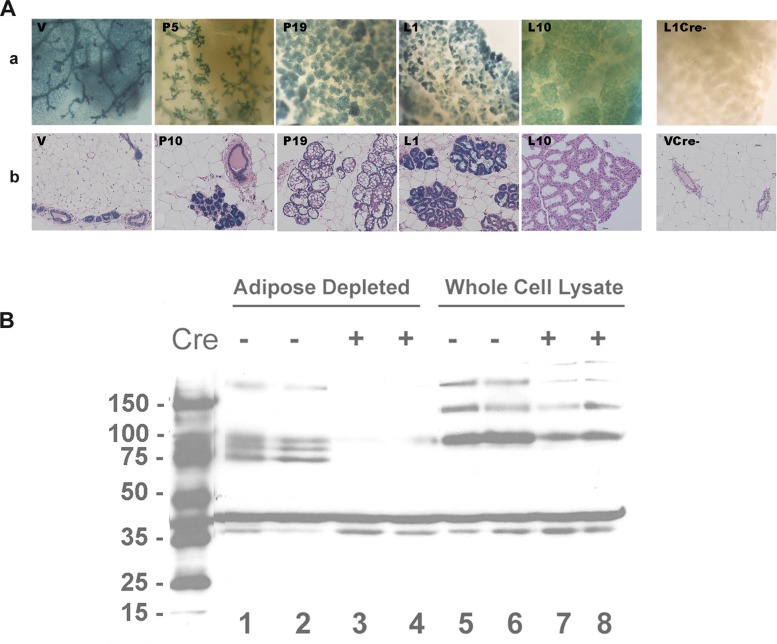
To examine the role of the IR in mammary gland development during pregnancy, we crossed a mouse bearing two floxed alleles of IR, IRfl/fl, with a heterozygous transgenic mouse in which expression of Cre recombinase is driven by the β-lactoglobin (BLG) promoter, a mammary specific gene (41). First, however, to demonstrate when BLG-Cre turns on, we crossed the BLG-Cre mice to the Rosa26R mice (43); expression of Cre recombinase removes stop codons from the Rosa26 constructs and allows expression of lacZ in all Cre−-expressing cells. Expression of lacZ in tissues can be detected with X-gal staining. The blue staining in Fig. 2A shows that LacZ was turned on in the virgin gland and is expressed throughout development. Histological sections (Fig. 2A, bottom) show that Cre is confined to the epithelial compartment with no staining of stroma.
Fig. 2.
A: LacZ stain of mammary glands from Rosa-β-lactoglobin (BLG)-Cre mice. a, Whole mounts stained with Xgal. b, Whole mounts were embedded in paraffin and sectioned. After Xgal staining, sections were counterstained with hematoxylin and eosin (H&E). The two images on the right were from Cre− ROSA mice. V, virgin; P5, P10, P19, pregnancy days 5, 10, and 19, respectively; L1 and L10, lactation days 1 and 10, respectively. B: effect of BLG-Cre on Western blot for insulin receptor (IR). Lysates of whole tissue and of epithelial tissue depleted of adipose cells [mammary epithelial cells (MECs)] were prepared from mammary glands of IRfl/fl Cre+ and IRfl/fl Cre− mice. In the whole tissue lysates (lanes 5–8), stain for IR (major band at 90 kDa) can be found in samples from both the IRfl/fl Cre+ and IRfl/fl Cre− animals. In adipose-depleted lysates, IR (major band at 90 kDa) can readily be seen in the Cre− lanes although slightly degraded because of the enzymatic digestion necessary to deplete the tissue of adipocytes. There is little or no stain corresponding to these bands in lysates from the Cre+ mammary glands. The band at 45 kDa is the actin loading control, stained with the appropriate antibody in the same Western blot. Two mice were used per treatment as shown.
Having assured ourselves that BLG-Cre produces MEC-specific gene deletion, we crossed the IRfl/fl mouse with BLG-Cre. Figure 2B shows a Western blot for IR; the results from whole tissue lysates on the right hand side demonstrate several bands representing the 90-kDa β-subunit of IR as well as some higher-molecular-weight complexes in both the Cre+ and Cre− lanes. When adipose cells are removed from the tissue before epithelial cell lysis, it can be seen that IR protein has been almost entirely depleted from the remaining epithelial cells by expression of Cre recombinase.
Effect of deletion of IR on pup growth.
To examine the functional effects of deletion of IR, we first bred females of the control strains IRfl/fl Cre−, IRfl/+ Cre+, and IRfl/+ Cre− and the fully targeted strain IRfl/fl Cre+ with CD1 males to obtain litters where IR was not compromised. The growth rate of mouse litters can be used as a test of milk secretion in the dam (30). Figure 3 shows that pup growth is severely compromised only when dams had the IRfl/fl Cre+ genotype. Cre targeting of IR in the homozygous floxed mouse decreased pup growth rate from ∼0.5 to 0.13 g/day (Fig. 3A, inset). Interestingly, most of the litters of this genotype continued to grow, albeit slowly; only one out of six litters failed to survive. By day 18, the pups appeared to have matured with good hair growth and appropriate activity (Fig. 3B), but they were very small. Thus IR appears to be necessary for appropriate milk secretion during lactation. This finding is in accord with previous results (4, 36) showing that IRB is upregulated two- to threefold in the mammary gland of the lactating mouse and plays an important role in lactational function. The next question we asked was whether this receptor is also important in mammary proliferation and differentiation during pregnancy.
Effect of deletion of IR on development of the alveolar epithelium in pregnancy.
To examine the effect of deletion of IR in the pregnant mouse, we examined the histology of mammary glands from 13.5 day pregnant mice. This time point was chosen because it represents a time when alveolar proliferation has slowed at least 75% from its peak at day 5 at pregnancy and the expression of proliferation-related genes has fallen by ∼25% (26). In addition, the growth of cytoplasmic lipid droplets in the epithelium, a reliable marker of differentiation, is substantial (39).
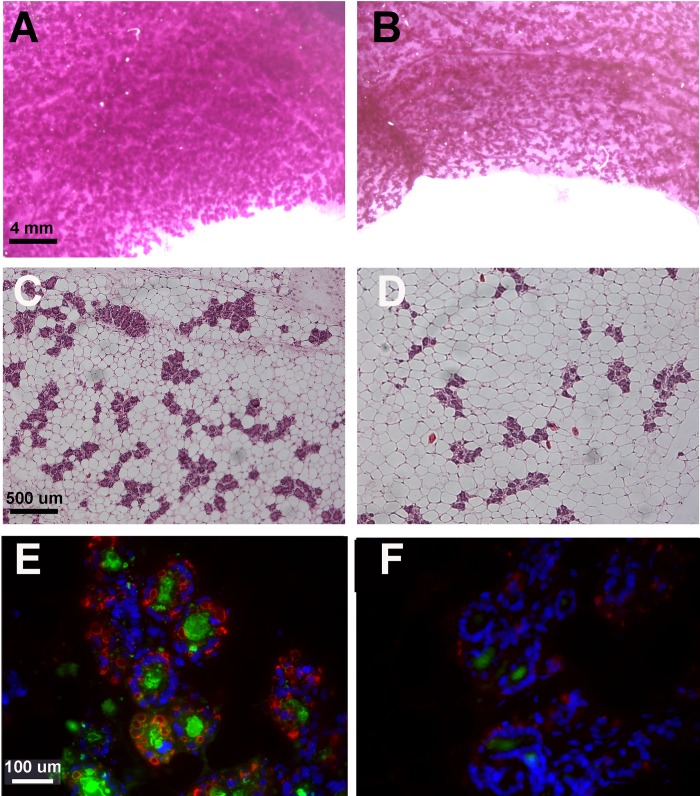
The views of whole mounts (Fig. 4, A and B) suggest wider spacing of alveoli along the ducts, and histological views (Fig. 4, C and D) suggest a severe developmental defect in the gland at pregnancy day 13.5. Consistent with the changes in histology, the area occupied by alveoli as well as the number of alveoli per field was reduced by 50% in sections from the IRfl/fl Cre+ glands (Table 1). When these results are translated to total alveoli per gland by multiplying the volume per section times the weight of the gland in grams, the total number of alveoli appears to be reduced 65%. Although proliferative activity as estimated by PCNA-positive alveolar cells in the gland showed a tendency to be higher in the Cre+ mice (Table 1), the difference was small. These observations suggest that alveolar number is reduced because of wider spacing along the ducts, a process that may have begun in early pregnancy when the floxed IR gene would already have been depleted in the Cre+ strains. The alveoli are also somewhat smaller in the Cre+ glands because of smaller lumens and decreased numbers of fat droplets in the cytoplasm. Using Slidebook software, we were able to determine the area occupied by casein and ADPH, also known as perilipin 2, in sections stained with antibodies to murine β-casein and ADPH (Fig. 4, E and F). Both are reduced >65% (Table 1).
Fig. 4.
Differentiation of MECs is diminished in pregnant mice expressing Cre recombinase. Whole mounts (A and B), histological views (C and D), and immunocytochemistry (E and F) of mammary glands from IRfl/fl Cre− (A, C, and E) and IRfl/fl Cre+ (B, D, and F) mice at pregnancy day 13.5. A and B: whole mounts fixed as described and stained with carmine red. C and D: sections from paraffin-embedded glands stained with H&E. E and F: immunocytochemistry for casein (green), adipophilin (perilipin 2; red), and nuclei (DAPI, blue). Scale bars as shown. See Table 1 for quantitation of the properties of these images.
In summary, these data provide strong evidence that IR is important for normal alveolar growth and differentiation during pregnancy. The next question is whether insulin, IGF-I, or IGF-II is interacting with this receptor to produce these results. Because the effects of deleting IR were more pronounced with respect to differentiation than proliferation, we will focus on differentiation in the remainder of this paper.
Is insulin the ligand signaling through IR?
The formation of acinar cultures from primary MECs placed on a base of Matrigel was first reported by Barcellos-Hoff et al. (2) followed closely by a report on the topography of the structures from the laboratory of Neville et al. (29). Both laboratories were able to demonstrate hollowed acini-like structures that were positive for luminal casein. Similar cultures, variously called mammospheres, three-dimensional (3D) acini, or acinar cultures have found wide utility in many laboratories where they have been used to elucidate the interaction of the mammary epithelium with the extracellular matrix (reviewed in Ref. 52) as well as the effects of oncogenes on acinar development (reviewed in Ref. 42). Recently, similar cultures using matrixes that do not promote attachment of isolated epithelial cells have been used to characterize mammary stem cells (17); the term mammospheres is now restricted to these cultures. To elucidate the ligand responsible for differentiation of MECs in 3D acinar cultures, we started with single cells isolated from the day 13.5 pregnant mammary glands of CD1 mice. Importantly, when these cells are mixed with full-strength Matrigel, they associate to form spheres with a single layer of epithelium. These spheres gradually increase in size mainly because of expansion of the lumen. Unlike cultures where cells are placed on top of Matrigel, very little apoptosis is seen (data not shown). When cells are cultured for 7 days with growth medium containing concentrations of insulin greater than 2 nM, followed by 7 days with differentiation medium again containing >2 nM insulin (see materials and methods for media composition), the cells acquire several characteristics associated with differentiating epithelium, including an increase in cell size, staining for β-casein, and acquisition of cytoplasmic lipid droplets that stain with ADPH. Figure 5 shows several acini incubated in the presence of increasing doses of insulin. The acini are formed as a single layer of epithelial cells that expand around a lumen that is often filled with material staining with the anti-β-casein antibody. Differentiated acini contain numerous lipid droplets identified as hollow-appearing cytoplasmic structures surrounded by red stain for ADPH.
To determine whether insulin was interacting with the IR in the cultures from CD1 mice, we performed dose-response curves using concentrations of insulin ranging from 0 to 50 nM, well within the physiological range in vivo, measuring the size of the structures and their lumens, the number of nuclei per structure, and the intensity and area occupied by stain for ADPH and casein. Figure 5 shows the effect of increasing insulin doses on acinar morphology (Fig. 5A), acinar and luminal diameters as well as number of nuclei per acinus (Fig. 5B), cell size, calculated as described in the legend, cytoplasmic area, and area of high-intensity casein stain (Fig. 5C), as well as lipid droplet number and area (Fig. 5D). Each point represents the mean and SE from analysis of three to five acini. The lines are theoretically plotted using the parameters calculated from the best fit to the Michaelis-Menton equation, as shown in Table 2. The conclusion is that insulin produces a half-maximal effect at a concentration of ∼1.7 nM for all the differentiation parameters. Nuclear number was increased only ∼33% in these cultures in the presence of 50 nM insulin, but again the Km for this effect was about 1 nM, suggesting that insulin does have a small proliferative effect. In fact, we observed PCNA staining in 5–20% of nuclei of acini cultured for 4 to 7 days in the presence of 2 nM insulin.
We were able to obtain consistent acini in cultures from IRfl/fl Cre+ or IRfl/fl Cre− mice at 100 nM insulin, although not at the lower concentrations; we were able to use MECs from CD1 mice. It is possible that this problem reflects the difficulties Bl6C57 mice (the parent strain) have with lactation in general (30). Fewer acini formed when the MECs were taken from IRfl/fl Cre+; those that did form showed very few of the signs of differentiation in the presence of 2 nM insulin (data not shown).
The conclusion from these experiments is that insulin has a strong effect on alveolar differentiation at concentrations in the physiological range. The next question was “Can either of the IGFs produce these same effects?”
Effect of IGF-I and IGF-II on differentiation parameters of acinar cultures.
Figure 6A shows dose-response curves for IGF-I and IGF-II on acinar and luminal size and nuclear number. IGF-I had a small but significant effect on acinar diameter and number of nuclei only at a concentration of 50 nM. It had no effect on luminal diameter. IGF-II had no effects at the concentrations shown on any of these parameters. At 100 nM, it began to promote differentiation as shown by increases in staining for casein and ADPH (data not shown). Thus neither of these ligands promotes acinar differentiation at concentrations in physiologically relevant ranges. To make certain that insulin is acting solely through IR, we used a function blocking antibody against the IGF-I receptor (IGF-IR). The concentration used, 500 nM, effectively blocks IGF-I action on acinar cultures (Fig. 6B). However, it had no effect on insulin action, as shown by the effect of 2 nM insulin on acinar diameter in the presence and absence of antibody (Fig. 6C). Similar results were obtained for other differentiation parameters. We conclude that insulin acting through IR is essential for alveolar differentiation in the latter part of pregnancy in the mouse.
What are the downstream effects of reduction in signaling from IR?
The images in Figs. 4 and 5 and their analysis in Table 1 suggest that differentiation is diminished when signaling through IR is abrogated. To further test this hypothesis and to begin to identify genes whose expression is affected by insulin signaling in midpregnancy, we prepared MECs from the mammary glands of 13.5 day pregnant IRfl/fl Cre− and IRfl/flCre+ mice, isolated RNA, and, after affirming the RNA quality, applied it to Affymetrix MoGene-1_0-st-v1 arrays. There was a distinct separation between gene expression in the two strains as shown by the PCA analysis in Fig. 1. The signal intensities obtained from processing these arrays, which allow evaluation of the expression of 28,853 genes, were put into GeneSpring X and analyzed as described in materials and methods to give a list of 217 genes of which 93 were upregulated and 124 were downregulated 1.5-fold or more in Cre+ MECs compared with Cre− MECs. Pathway analysis of these genes using standard programs proved unsatisfactory because the literature on pathway analysis in the differentiating mammary gland is not yet reflected in most standard pathway programs (5). For this reason, we used a combination of our own knowledge of synthesis pathways for the production of milk, Gene Ontology analysis from Genespring X, and pathway analysis using Metacore. This combinatorial approach yielded 11 categories of genes that were predominantly downregulated when IR expression is abrogated (Table 3). Affected genes include those for the major milk proteins and milk fat globule proteins, genes involved in both lipid synthesis and lipid degradation, and genes for lactose synthesis as well as for ion transport, immune regulation, and ubiquination. More detailed data on the genes in each of these categories are given in Supplemental Tables S1 and S2 (Supplemental data for this article may be found on the American Journal of Physiology: Endocrinology and Metabolism website.). The same approach yielded five categories of genes that were predominantly upregulated, including cell cycle genes, extracellular matrix and cytoskeleton genes, as well as genes for mRNA transport and processing, and a few signal transduction genes. Again the detailed gene lists can be found in Supplemental Tables S1 and S2. Finally, five categories, the largest of which was small molecule metabolic processes, had more or less equal numbers of up- and downregulated genes, suggesting that they belong to more complex metabolic pathways.
Table 3.
Array analysis of MECs from IRfl/fl Cre− and IRfl/fl Cre+ mice
| Genes Up | Genes Down | Total | |
|---|---|---|---|
| No. of Genes | |||
| Total genes fold Change >1.1, P < 0.05 | 557 | 720 | 1,277 |
| Total genes on chip | 28,817 | ||
| Percent of genes significantly changed in IRfl/fl Cre+, % | 1.93 | 2.50 | 4.43 |
| Total genes fold change >1.5, P < 0.02 | 93 | 124 | 217 |
| Classified genes, fold change >1.5, categories below | 84 | 113 | 197 |
| Time course data available from Refs. 37 and 44 | 33 | 42 | 75 |
| Categories with >75 % downregulated genes in IRfl/fl Cre+ | Genes up | Genes down | Percent down |
| Transport, ion | 2 | 13 | 87 |
| Lipid and triglyceride biosynthesis | 1 | 11 | 92 |
| Immune regulation | 2 | 8 | 80 |
| Transport, organic molecule | 0 | 8 | 100 |
| Receptor | 1 | 6 | 86 |
| Milk proteins | 0 | 7 | 100 |
| Fatty acid oxidation (peroxisomal and mitochondrial) | 0 | 6 | 100 |
| Lactose and glycan synthesis | 0 | 5 | 100 |
| Milk lipid droplet formation | 0 | 5 | 100 |
| Ubiquination | 1 | 4 | 80 |
| Transport, vesicle mediated | 0 | 4 | 100 |
| Categories with >75% upregulated genes in IRfl/fl Cre+ | Percent up | ||
| Cell cycle | 24 | 5 | 83 |
| Cytoskeleton | 18 | 4 | 82 |
| Extracellular matrix | 8 | 1 | 89 |
| mRNA export and processing | 6 | 1 | 86 |
| Signal transduction | 3 | 1 | 75 |
| Categories with mixed regulation in IRfl/fl Cre+ | Percent up | ||
| Small molecule metabolic process | 5 | 13 | 28 |
| Cell-cell adhesion | 5 | 5 | 50 |
| Transcription | 5 | 3 | 63 |
| Apoptosis | 3 | 3 | 50 |
See Supplemental Table 2 for gene identities by category.
Physiologically, what do these findings mean? Because many categories of mammary differentiation specific genes were downregulated with diminished insulin signaling, our findings suggest that secretory differentiation in the mammary gland of the late pregnant mouse is dependent on insulin. In addition, the decrease in cell proliferation that occurs in the second half of pregnancy would appear to be at least partially dependent on insulin signaling as shown by the upregulation of cell cycle genes in the Cre+ mice. Finally, changes in both the cytoskeleton and interactions with the extracellular matrix that prepare the gland for secretory activity may also have a similar dependency on insulin. If these conclusions are correct, it seems likely that the expression trajectories for the downregulated, differentiation-related genes would be upward in the latter half of pregnancy, and, conversely, the expression trajectories for the upregulated genes would be downward in the same period. We are able to use previously published data on the time course of gene expression in the developing mammary gland (37) to test these hypotheses.
Comparison of IRfl/fl Cre+ data with time course data.
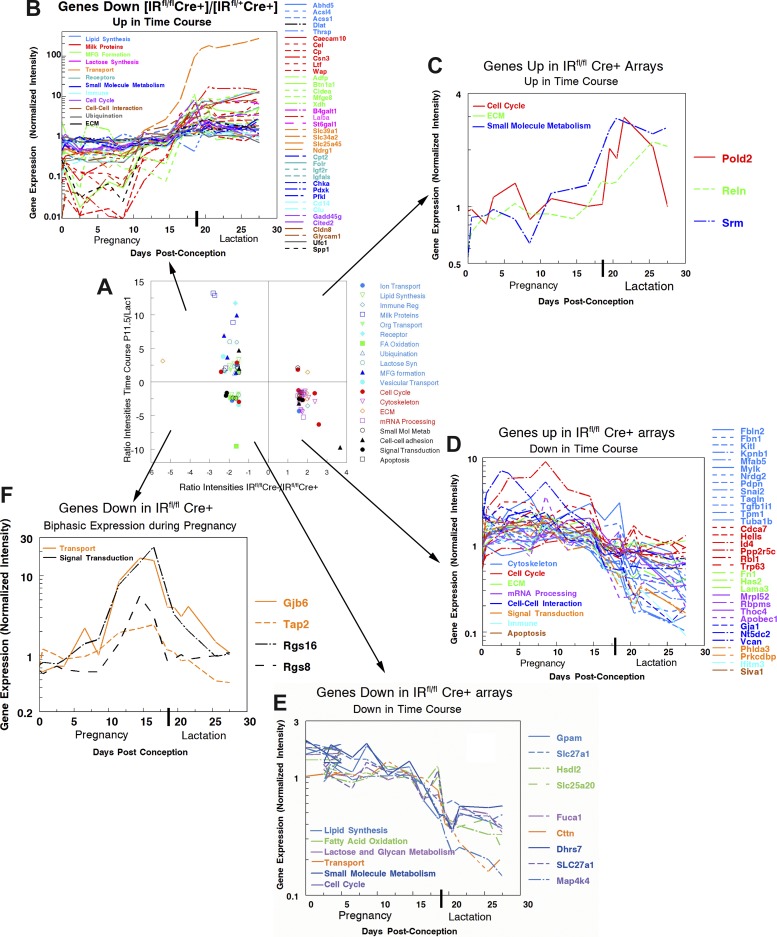
Figure 7A summarizes the relation between changes in intensity in the IRfl/fl Cre+ mice and the observed differences in expression of genes from the time course arrays (37, 44). Most of the downregulated genes in the Cre+ mice are found in Fig. 7A, top left, indicating, as shown in Fig. 7B, that these genes are upregulated in the latter half of pregnancy. Conversely, most of the upregulated genes in the Cre+ mice are found in Fig. 7A, bottom right, indicating that these genes are upregulated in the latter half of pregnancy. Again the detailed time course data are consistent with this interpretation (Fig. 7D). Not all genes fit these predictions. The three genes in Fig. 7A, top right, are increased in the IRfl/fl Cre+ and also increase in the latter half of pregnancy (Fig. 7C). These genes are Pold2, which encodes the 125-kDa catalytic subunit of DNA polymerase delta; Reln, which encodes a large secreted extracellular matrix protein REELIN involved in cell-cell interactions; and Srm, which encodes spermidine synthase, a src-related kinase lacking COOH-terminal regulatory tyrosine and NH2-terminal myristylation sites. The role of these proteins in mammary cell development is unknown although it should be noted that, unlike other genes that are upregulated in the Cre+ MECs, the expression patterns of these genes show a marked tendency to be flat during pregnancy, increasing in expression around secretory activation. Two other categories of genes also do not fit the overall prediction; both are found in Fig. 7A, bottom left. The first category consists of genes that are downregulated in the Cre+ MECs and also downregulated in late pregnancy (Fig. 7E). These genes fall into a number of categories; their major characteristic in the time course is that their expression is mostly level in pregnancy, decreasing only with the onset of secretory activation around pregnancy day 19.
Fig. 7.
Time course of expression of genes changed 1.5-fold or greater in Cre+ mice. A: expression intensity ratios pregnancy day 11.5/lactation day 0.5 from time course as a function of intensity ratios IRfl/fl Cre+/IRfl/fl Cre−. Genes in the upper two quadrants increased 1.5-fold or more between pregnancy day 11.5 and lactation day 0.5, whereas genes in the lower two quadrants decreased 1.5-fold or more over this same period. Genes in the right hand quadrants are down 1.5-fold or more (P < 0.01) in the IRfl/fl Cre+ samples, whereas those in the left hand quadrants were up 1.5-fold or more (P < 0.01) in these samples. Data for each gene plotted is given in Supplemental Table 2 organized into the categories shown in the legend. Note that expression values >20 for only about one-third of the genes in the IRfl/fl Cre experiment were available from the time course experiments that were published in 2003 (37) and 2004 (44) using arrays with many fewer genes. For this reason, the analysis of correlation with the time course of gene expression includes only 42 down-going and 33 up-going genes. In the remaining panels, gene names are shown to the right of the graph, and categories are indicated by color in the list within the graph. B: most of the genes that decreased with decreased IR expression increased on time course arrays during pregnancy. C: three of the genes increased in IRfl/fl Cre+ MECs increased during pregnancy on time course arrays. D: most of the genes increased in IRfl/fl Cre+ MECs decreased during pregnancy on time course arrays. E: a few of the genes decreased in IRfl/fl Cre+ MECs and also decreased during pregnancy on time course arrays. F: time course of expression of genes that increase to pregnancy day 15 and then decrease before secretory activation. These four genes are downregulated in the IRfl/fl Cre+ mouse but show this distinctive time course in data from the Talhouk laboratory (11, 46).
The expression patterns of the most interesting genes in Fig. 7A, bottom left, are shown in Fig. 7F: genes that increase during early pregnancy and then fall rapidly at secretory activation. Interestingly mammary cell expression of Gjb6, gap junction protein 6, also known as connexin 30 has been shown to coincide with the gene expression levels shown in Fig. 7F (47). It has been associated with negative regulation of cell proliferation. The two regulators of G protein signaling, Rgs16 and Rgs8, show a similar pattern of expression as does Tap2, a member of the MDR/TAP subfamily involved in multidrug resistance. One could speculate that this category of molecules is involved in preventing premature secretory activation as the reproductive cycle nears the termination of pregnancy and onset of lactation.
DISCUSSION
The novelty of this work is that we have demonstrated that the IR plays an important role in the development of the mammary gland during gestation. Selective deletion of the IR in the mammary epithelium reduced proliferation and markers of differentiation by midpregnancy and resulted in blunted litter growth rates. In addition, studies in cultured acini from midpregnant MECs provide strong evidence that insulin signaling through its receptor underlies differentiation of the gland in preparation for secretory activation. These observations extend the critical role that insulin and its receptor plays in milk production during lactation to the development of the mammary gland as it prepares for lactation during gestation. These data raise important questions regarding the physiological consequences of perturbed IR signaling in the mammary gland during gestation. To address that question in future work, the role that insulin and its receptor plays must be integrated with the complex set of temporal hormonal signals that prepare the mammary gland and support its function during lactation.
The development of the mammary epithelium during pregnancy takes place in two stages. The first is a proliferative stage that depends on increases in systemic levels of progesterone from the ovary and PRL from the pituitary (27); in this stage the number of cells increases exponentially as alveoli are formed and multiply. By midpregnancy the rate of proliferation has decreased (49) significantly, and differentiation marked by cytoplasmic lipid droplet formation, increasing casein accumulation, and progressive expansion of the lumina takes place (39). The hormonal regulation of this stage is not well understood: in rodents, PRL and its receptor are low, and placental lactogen, present in two forms, likely serves to foster differentiation (48). At the same time, progesterone, secreted by the ovary, inhibits secretory activation (20). Whether progesterone plays a role in differentiation is not known. In rodents, withdrawal of progesterone leads to both parturition and activation of copious milk secretion. Lactation itself depends on the increase in PRL and its receptor that occurs at secretory activation (23) as well as corticosterone and insulin (4, 16). Although strong evidence for a role for the IGF-IR in both proliferation and differentiation during pregnancy has been presented (36, 45), our studies provide the first clear evidence that the IR plays a more important role in late pregnancy. We found that selective deletion of IR using Cre-lox technology in the mouse mammary epithelium reduces the rate of growth of pups suckled by IRfl/fl Cre+ dams by 74%. Insulin and IR are known to support milk production during lactation (4, 16), so our studies cannot definitively attribute the entire reduction in growth rate to impaired mammary gland development. However, the 50% decrease in alveolar number and the 60% decrease in staining for casein and lipid droplets at midpregnancy would suggest that the IR mediates both proliferative and differentiation phases. A decline in IGF-II signaling through the IR could be responsible for the effects on proliferation, since this growth factor is expressed in early pregnancy (34, 36) and does bind to IRA (10), the form of IR most prevalent during the early stages of gestation.
Our studies in acinar cultures of isolated MECs, on the other hand, suggest that the critical ligand for IR-mediated differentiation is insulin. Insulin stimulated luminal expansion, increased cell size, increased high-intensity staining for casein, and increased the number of cytoplasmic lipid droplets in acini. Dose-response curves for insulin action had a Km of ∼1.7 nM, typical of insulin interaction with IR in many physiological systems. Much higher concentrations of both IGF-I and IGF-II had little or no effect on these parameters although our studies do not rule out effects on proliferation, particularly earlier in pregnancy (45). In addition, insulin was still active in the presence of a function-blocking antibody to the IGF-IR, ruling out effects of endogenously secreted IGF-I and IGF-II on differentiation. The inescapable conclusion is that insulin interacting with IR is necessary for the full differentiation of the mammary epithelium in the latter half of pregnancy. The possibility that the IGF-IR is important in proliferation in pregnancy as postulated by Sun et al. (45) is not ruled out by these studies; interestingly, the gene expression studies (Fig. 7 and Table 3) suggest that insulin may downregulate proliferation in late pregnancy, however.
We then began to address a problem that has been somewhat intractable in the mammary gland of pregnancy: what exactly are the downstream molecules involved in secretory differentiation? To do this, we compared microarray data from IRfl/fl Cre+ and IRfl/fl Cre− mice, finding that diminution of signaling through IR decreased expression of many categories of differentiation-specific genes and increased expression of genes encoding cell cycle-, cytoskeletal-, and extracellular matrix-related proteins. Thus the physiological changes in the midpregnant gland leading to a decrease in cell proliferation and an increase in differentiation parameters appear to be dependent on insulin signaling. At midpregnancy, it is also important that mechanisms be present to inhibit secretory activation. The genes downregulated in the IRfl/fl Cre+ mice showed three expression patterns in time course arrays. The predominant pattern was an increase in expression beginning in midpregnancy (Fig. 7B), consistent with an effect on differentiation; most differentiation-specific genes showed this pattern. However, another group of genes tended to be flat during most of pregnancy, decreasing markedly only around pregnancy day 18, when secretory activation begins (Fig. 7E). A final, most interesting group increased up to 10-fold beginning at about day 8 of pregnancy but sharply decreased after day 15 with the onset of secretory activation (Fig. 7F). One of these genes encodes connexin 30, a protein postulated to regulate the function of mammary alveolar cells (46). It is of considerable interest that a microRNA, miR-150, highly expressed in the pregnancy mammary gland shows a similar expression pattern in pregnancy (J. Richer, personal communication). The expression of this microRNA is decreased in the IRfl/fl Cre+ mouse (M. C. Neville, P. Ramanathan, and J. Richer, unpublished observation), suggesting a potential insulin-dependent regulatory mechanism. Is it possible that the two groups of genes, whose expression patterns are shown in Fig. 7, E and F, are related not to differentiation but to the inhibition of secretory activation in pregnancy? If so, one might expect that their expression is related to progesterone signaling. However, there is presently no data that allows evaluation of this hypothesis.
Many questions remain: how is insulin signaling through IR mediated? The evidence is quite clear that insulin signaling through IR is mediated by the IRS2 → AKT pathway during lactation (16). There is only one piece of evidence from cells isolated during pregnancy. In studies of the IGF-IR receptor in the laboratory of T. Wood using MECs isolated from mid- and late-pregnant mammary glands, 50 nM insulin is shown to increase phosphorylation on serine-473 of AKT in both the presence and absence of a dominant-negative form of the IGF-IR (45). The implication of this observation is that insulin is signaling through the canonical pathway IR → phosphatidylinositol 3-kinase → IRS1/2 → AKT in mid- and late gestation.
How does insulin signaling interact with other regulators of differentiation during pregnancy? As mentioned above, it is likely that differentiation is regulated by the systemic placental hormones placental lactogen I and placental lactogen II, and possibly by progesterone and IGF-I. The acinar preparation used in the current studies might be useful in dissecting these pathways about which very little is known.
Finally, from a clinical standpoint, does the sensitivity of differentiation to insulin render women with hyperinsulinemia, in particular obese women or women with gestational diabetes mellitus, susceptible to problems in lactation? Lactation was impaired when the putative downstream mediator of insulin signaling, activated AKT, was overexpressed in the mouse mammary alveolar cell (40). In other studies, diet-induced obese mice, which are hyperinsulinemic, exhibit an impairment in lactation characterized by the production of lipid-poor milk, reduced neonatal growth, and altered neonatal metabolism (50). Lactation is often impaired in obese women for reasons that are currently not understood (31). With the advent of technology to study mammary gene expression in women using RNA isolated from milk fat droplets, it might be possible to test the hypothesis that hyperinsulinemia and/or insulin resistance in the mammary gland leads to impaired glandular development or premature activation of secretory mechanisms in obese women.
In conclusion, insulin interacting with IR is necessary for full mammary differentiation in the latter half of pregnancy. In the absence of IR, lipid droplets and casein staining in lumens are reduced. The mammary alveolar cell shows a normal sensitivity to insulin with a Km for insulin effects around 1.7 nM. Insulin acts both to enhance secretory differentiation and decrease proliferation starting in midpregnancy. An important question is whether the hyperinsulinemia of obesity has deleterious effects on mammary cell differentiation in pregnancy, leading to impaired lactation.
GRANTS
The work in this article was supported by National Institute of Child Health and Human Development Grant PO1-HD-38129, which supported M. C. Neville, P. MacLean, and S. M. Anderson.
DISCLOSURES
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
Author contributions: M.C.N., P.W., P.R., M.P.M., C.P., J.M., S.M.A., and P.S.M. conception and design of research; M.C.N., P.W., P.R., and P.S.M. analyzed data; M.C.N. and P.W. prepared figures; M.C.N. drafted manuscript; M.C.N., P.W., S.M.A., and P.S.M. edited and revised manuscript; M.C.N., P.W., P.R., M.P.M., C.P., J.M., S.M.A., and P.S.M. approved final version of manuscript; P.W., P.R., M.P.M., and C.P. performed experiments; C.P., J.M., S.M.A., and P.S.M. interpreted results of experiments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Teresa Wood, Department of Neurology and Neurosciences, New Jersey Medical School for valuable advice throughout the development of the experiments presented here.
REFERENCES
- 1.Balmain JH, French TH, Folley SJ. Stimulation by insulin of in vitro fat synthesis by lactating mammary gland slices. Nature 165: 807–808, 1950. [DOI] [PubMed] [Google Scholar]
- 2.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphologenesis of primary mammary epithelial cells cultured on a reconstituted basement membrane. Development 105: 223–235, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter MA, Coore HG. The mode of regulation of pyruvate dehydrogenase of lactating rat mammary gland. Effects of starvation and insulin. Biochem J 174: 553–561, 1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlato C, Doppler W. Selective response to insulin versus IGF-I and IGF-II and upregulation of insulin-receptor splice variant B in the differentiated mouse mammary epithelium. Endocrinology 150: 2924–2933, 2009. [DOI] [PubMed] [Google Scholar]
- 5.Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle (Abstract). BMC Genomics 9: 366, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruning JC, Michael MD, Winnay JN, Hayashi T, Horsch D, Accili D, Goodyear LJ, Kahn CR. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell 2: 559–569, 1998. [DOI] [PubMed] [Google Scholar]
- 7.Burnol AF, Ebner S, Ferre' P, Girard J. Regulation by insulin of glucose metabolism in mammary gland of anaesthetized lactating rats. Stimulation of phosphofructokinase-1 by fructose 2,6-bisphosphate and activation of acetyl CoA carboxylase. Biochem J 254: 11–14, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnol AF, Loizeau M, Girard J. Insulin receptor activity and insulin sensitivity in mammary gland of lactating rats. Am J Physiol Endocrinol Metab 259: E828–E834, 1990. [DOI] [PubMed] [Google Scholar]
- 9.Danielson KG, Oborn CJ, Durban EM, Butel JS, Medina D. Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci USA 81: 3756–3760, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denley A, Brierley GV, Carroll JM, Lindenberg A, Booker GW, Cosgrove LJ, Wallace JC, Forbes BE, Roberts CT., Jr Differential activation of insulin receptor isoforms by insulin-like growth factors is determined by the C domain. Endocrinology 147: 1029–1036, 2006. [DOI] [PubMed] [Google Scholar]
- 11.El-Saghir JA, El-Habre ET, El-Sabban ME, Talhouk RS. Connexins: a junctional crossroad to breast cancer. Int J Dev Biol 55: 773–780, 2011. [DOI] [PubMed] [Google Scholar]
- 12.Flint DJ, Clegg RA, Knight CH. Effects of prolactin, progesterone and ovariectomy on metabolic activities and insulin receptors in the mammary gland and adipose tissue during extended lactation in the rat. J Endocrinol 102: 231–236, 1984. [DOI] [PubMed] [Google Scholar]
- 13.Folley SJ, French TH. The intermediary metabolism of the mammary gland. 3. Acetate metabolism of lactating mammary gland slices with special reference to milk fat synthesis. Biochem J 46: 465–473, 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsyth IA, Neville MC. Introduction: hormonal regulation of mammary development and milk protein gene expression at the whole animal and molecular levels. J Mammary Gland Biol Neoplasia 14: 317–319, 2009. [DOI] [PubMed] [Google Scholar]
- 15.Forsyth IA, Strong CR, Dils R. Interactions of insulin, corticosterone and prolactin in promoting milk-fat synthesis by mammary explants from pregnant rabbits. Biochem J 129: 929–935, 1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadsell DL, Olea W, Lawrence N, George J, Torres D, Kadowaki T, Lee AV. Decreased lactation capacity and altered milk composition in insulin receptor substrate null mice is associated with decreased maternal body mass and reduced insulin-dependent phosphorylation of mammary Akt. J Endocrinol 194: 327–336, 2007. [DOI] [PubMed] [Google Scholar]
- 17.Jechlinger M, Podsypanina K, Varmus H. Regulation of transgenes in three-dimensional cultures of primary mouse mammary cells demonstrates oncogene dependence and identifies cells that survive deinduction. Genes Dev 23: 1677–1688, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones RG, Ilic V, Williamson DH. Physiological significance of altered insulin metabolism in the conscious rat during lactation. Biochem J 220: 455–460, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones RG, Ilic V, Williamson DH. Regulation of lactating-rat mammary-gland lipogenesis by insulin and glucagon in vivo. The role and site of action of insulin in the transition to the starved state. Biochem J 223: 345–351, 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn NJ. Progesterone withdrawal as the lactogenic trigger in the rat. J Endocrinol 44: 39–54, 1969. [DOI] [PubMed] [Google Scholar]
- 21.McGuire MA, Grinari JM, Dwyer DA, Bauman DE. Role of insulin in the regulation of mammary synthesis of fat and protein. J Dairy Sci 78: 816–824, 1995. [DOI] [PubMed] [Google Scholar]
- 22.McNeillie EM, Zammit VA. Regulation of acetyl-CoA carboxylase in rat mammary gland: Effects of starvation and of insulin and prolactin deficiency on the fraction of the enzyme in the active form in vivo. Biochem J 204: 273–280, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizoguchi Y, Yamaguchi H, Aoki F, Enami J, Sakai S. Corticosterone is required for the prolactin receptor gene expression in the late pregnant mouse mammary gland. Mol Cell Endocrinol 132: 177–183, 1997. [DOI] [PubMed] [Google Scholar]
- 24.Munday MR, Williamson DH. Insulin activation of lipogenesis in isolated mammary acini from lactating rats fed a high-fat diet: evidence that acetyl-CoA carboxylase is a site of action. Biochem J 242: 905–911, 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munday MR, Williamson DH. Role of pyruvate dehydrogenase and insulin in the regulation of lipogenesis in the lactating mammary gland of the rat during the starved-refed transition. Biochem J 196: 831–837, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neville MC. Lactation and its Hormonal Control. In: Physiology of Reproduction, edited by Neill JD. San Diego, CA: Elsevier, 2006, p. 2993–3054. [Google Scholar]
- 27.Neville MC, McFadden TB, Forsyth I. Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia 7: 49–66, 2002. [DOI] [PubMed] [Google Scholar]
- 28.Neville MC, McFadden TB, Forsyth I. Hormonal regulation of mammary differentiation and milk secretion. J Mammary Gland Biol Neoplasia 7: 49–66, 2002. [DOI] [PubMed] [Google Scholar]
- 29.Neville MC, Stahl L, Brozo LA, Lowe-Lieber J. Morphogenesis and secretory activity of mouse mammary cultures on EHS biomatrix. Protoplasma 160: 110–123, 1991. [Google Scholar]
- 30.Ramanathan P, Martin IC, Gardiner-Garden M, Thomson PC, Taylor RM, Ormandy CJ, Moran C, Williamson P. Transcriptome analysis identifies pathways associated with enhanced maternal performance in QSi5 mice (Abstract). BMC Genomics 9: 197, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen KM, Kjolhede CL. Maternal obesity: a problem for both mother and child. Obesity (Silver Spring) 16: 929–931, 2008. [DOI] [PubMed] [Google Scholar]
- 32.Rasmussen SB, Young LJT, Smith GH. Preparing mammary gland whole mounts from mice. In: Methods in Mammary Gland biology and Beast Cancer Research, edited by Ip MM, Asch BB. New York, NY: Kluwer, 2000, p. 75–86. [Google Scholar]
- 33.Ray DB, Horst IA, Jansen RW, Mills NC, Kowal J. Normal mammary cells in long term culture. II. Prolactin, corticosterone, insulin, and triiodothyronine effects on alpha-lactalbumin production. Endocrinology 108: 584–590, 1981. [DOI] [PubMed] [Google Scholar]
- 34.Richert MM, Wood TL. The insulin-like growth factors (IGF) and IGF type I receptor during postnatal growth of the murine mammary gland: Sites of messenger ribonucleic acid expression and potential functions. Endocrinology 140: 454–461, 1999. [DOI] [PubMed] [Google Scholar]
- 35.Robinson AM, Williamson DH. Comparison of glucose metabolism in the lactating mammary gland of the rat in vivo and in vitro. Effects of starvation, prolactin or insulin deficiency. Biochem J 164: 153–159, 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowzee AM, Ludwig DL, Wood TL. Insulin-like growth factor type 1 receptor and insulin receptor isoform expression and signaling in mammary epithelial cells. Endocrinology 150 3611–3619, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia 8: 287–307, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Rudolph MC, Wellberg EA, Anderson SM. Adipose-depleted mammary epithelial cells and organoids. J Mammary Gland Biol Neoplasia 14: 381–386, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell TD, Palmer CA, Orlicky DJ, Fischer A, Rudolph MC, Neville MC, McManaman JL. Cytoplasmic lipid droplet accumulation in developing mammary epithelial cells: roles of adipophilin and lipid metabolism. J Lipid Res 48: 1463–1475, 2007. [DOI] [PubMed] [Google Scholar]
- 40.Schwertfeger KL, McManaman JL, Palmer CA, Neville MC, Anderson SM. Expression of constitutively activated Akt in the mammary gland leads to excess lipid synthesis during pregnancy and lactation. J Lipid Res 44: 1100–1112, 2003. [DOI] [PubMed] [Google Scholar]
- 41.Selbert S, Bentley DJ, Melton DW, Rannie D, Lourenco P, Watson CJ, Clarke AR. Efficient BLG-Cre mediated gene deletion in the mammary gland. Transgenic Res 7: 387–396, 1998. [DOI] [PubMed] [Google Scholar]
- 42.Shaw KR, Wrobel CN, Brugge JS. Use of three-dimensional basement membrane cultures to model oncogene-induced changes in mammary epithelial morphogenesis. J Mammary Gland Biol Neoplasia 9: 297–310, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71, 1999. [DOI] [PubMed] [Google Scholar]
- 44.Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy M, Heath VJ, Bell AK, Ferrier RK, Sandilands GP, Gusterson B. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res 6: 75–91, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Z, Shushanov S, LeRoith D, Wood TL. Decreased IGF type 1 receptor signaling in mammary epithelium during pregnancy leads to reduced proliferation, alveolar differentiation, and expression of insulin receptor substrate IRS-1 and IRS-2. Endocrinology 152: 3233–3245, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talhouk RS, Elble RC, Bassam R, Daher M, Sfeir A, Mosleh LA, El-Khoury H, Hamoui S, Pauli BU, El-Sabban ME. Developmental expression patterns and regulation of connexins in the mouse mammary gland: expression of connexin30 in lactogenesis. Cell Tissue Res 319: 49–59, 2005. [DOI] [PubMed] [Google Scholar]
- 47.Talhouk RS, Mroue R, Mokalled M, Abi-Mosleh L, Nehme R, Ismail A, Khalil A, Zaatari M, El-Sabban ME. Heterocellular interaction enhances recruitment of alpha and beta-catenins and ZO-2 into functional gap-junction complexes and induces gap junction-dependant differentiation of mammary epithelial cells. Exp Cell Res 314: 3275–3291, 2008. [DOI] [PubMed] [Google Scholar]
- 48.Thordarson G, Fielder P, Lee C, Hom YK, Robleto D, Ogren L, Talamantes F. Mammary gland differentiation in hypophysectomized, pregnant mice treated with corticosterone and thyroxine. Biol Reprod 47: 676–682, 1992. [DOI] [PubMed] [Google Scholar]
- 49.Traurig HH. A radioautographic study of cell proliferation in the mammary gland of the pregnant mouse. Anat Rec 159: 239–248, 1967. [DOI] [PubMed] [Google Scholar]
- 50.Wahlig JL, Bales ES, Jackman MR, Johnson GC, McManaman JL, Maclean PS. Impact of high-fat diet and obesity on energy balance and fuel utilization during the metabolic challenge of lactation. Obesity 20: 65–75, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walters E, McLean P. Effect of alloxan-diabetes and treatment with anti-insulin serum on pathways of glucose metabolism in lactating rat mammary gland. Biochem J 109: 407–417, 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu R, Boudreau A, Bissell MJ. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer Metastasis Rev 28: 167–176, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.