Abstract
Recent studies on microRNA (miRNA) evolution focused mainly on the comparison of miRNA complements between animal clades. However, evolution of miRNAs within such groups is poorly explored despite the availability of comparable data that in some cases lack only a few key taxa. For flatworms (Platyhelminthes), miRNA complements are available for some free-living flatworms and all major parasitic lineages, except for the Monogenea. We present the miRNA complement of the monogenean flatworm Gyrodactylus salaris that facilitates a comprehensive analysis of miRNA evolution in Platyhelminthes. Using the newly designed bioinformatics pipeline miRCandRef, the miRNA complement was disentangled from next-generation sequencing of small RNAs and genomic DNA without a priori genome assembly. It consists of 39 miRNA hairpin loci of conserved miRNA families, and 22 novel miRNAs. A comparison with the miRNA complements of Schmidtea mediterranea (Turbellaria), Schistosoma japonicum (Trematoda), and Echinococcus granulosus (Cestoda) reveals a substantial loss of conserved bilaterian, protostomian, and lophotrochozoan miRNAs. Eight of the 46 expected conserved miRNAs were lost in all flatworms, 16 in Neodermata and 24 conserved miRNAs could not be detected in the cestode and the trematode. Such a gradual loss of miRNAs has not been reported before for other animal phyla. Currently, little is known about miRNAs in Platyhelminthes, and for the majority of the lost miRNAs there is no prediction of function. As suggested earlier they might be related to morphological simplifications. The presence and absence of 153 conserved miRNAs was compared for platyhelminths and 32 other metazoan taxa. Phylogenetic analyses support the monophyly of Platyhelminthes (Turbellaria + Neodermata [Monogenea {Trematoda + Cestoda}]).
Keywords: microRNAs, flatworms, evolution
Introduction
MicroRNAs (miRNAs) are single-stranded, approximately 22-nucleotides long noncoding small RNAs that regulate gene expression in animals by posttranscriptional gene regulation. miRNA families are classified by their target-specific seed regions, usually the positions 2–7 of the mature miRNAs. The binding of the mature miRNA to the mRNA target site can lead to inhibition of the translation and degradation of the respective mRNA (for details see Pasquinelli [2012]). In animals, miRNAs may play key roles in a broad variety of biological processes, such as, for example, cell proliferation and metabolism (Brennecke and Cohen 2003), tissue identity (Christodoulou et al. 2010), developmental timing (Reinhart et al. 2000), cell death (Baehrecke 2003), hematopoiesis (Chen et al. 2004), neuron development (Johnston and Hobert 2003), tumorigenesis (Michael et al. 2003), DNA methylation and chromatin modification (Bao et al. 2004), as well as in immune defense against viruses (Sarnow et al. 2006). By now, more than 19,000 putative miRNAs from animals, plants, and viruses have been described and deposited in public databases such as miRBase (release 19) (Kozomara and Griffiths-Jones 2011). During evolution of most animal lineages, miRNA loci are highly conserved and have been added to genomes rather than being lost (Sperling and Peterson 2009; Wheeler et al. 2009; Tarver et al. 2013). Accordingly, it is primarily the gain of miRNA families that is phylogenetically informative and has led to a number of phylogenetic studies that focus on miRNAs as phylogenetic markers. This seems particularly true for major animal lineages such as, for example, Bilateria, Protostomia, and Deuterostomia, where group-specific gains of miRNA families could be assigned (Wheeler et al. 2009; Christodoulou et al. 2010; Erwin et al. 2011). Only to some extent this has been studied in more derived taxonomic groups (Sperling et al. 2009; Heimberg et al. 2010; Campbell et al. 2011; Rota-Stabelli et al. 2011; Sperling et al. 2011; Wiegmann et al. 2011; Helm et al. 2012), focusing on conserved miRNAs or novel miRNAs that were shared between certain species. In contrast, if losses of conserved miRNAs have been detected, there was usually no correspondence to any obvious phylogenetic pattern of the missing miRNAs (e.g., shared losses). The losses found in, for example, acoelan worms, ascidians, and nematodes were mosaic in nature, leaving enough taxon-specific miRNAs for an assignment to a certain group (for summary see Sperling and Peterson [2009] and Philippe et al. [2011]).
There are two major strategies when using miRNAs as phylogenetic markers: A first group of studies use miRNAs as additional evidence for testing phylogenetic hypothesis inferred with other markers. Frequently, the presence or absence of conserved miRNA families is scored as phylogenetic information (Tarver et al. 2013). Within Diptera, for example, the distribution of miRNAs was shown to be in concordance with phylogenomic and morphological results (Wiegmann et al. 2011). Interestingly, the presence/absence of miRNA loci also supported earlier findings based on the analyses of mitochondrial genomes and expressed sequence tags (EST) markers that acoelan worms, that is, Symsagittifera roscoffensis and Hofstenia miamia, do not belong to Platyhelminthes (Philippe et al. 2011). The second group of studies is using miRNAs as the only phylogenetic marker. Applying this approach, the monophyly of Cyclostomes (hagfish and lampreys as closest relatives) could be confirmed (Heimberg et al. 2010). Similarly, Sperling et al. (2009) found miRNA support for the monophyly for Annelida, and Helm et al. (2012) confirmed the close relationship of the parasitic and morphologically enigmatic Myzostomida and Annelida. However, most studies using miRNAs as phylogenetic markers rely on the miRNA complement of only one species for each group, assuming the relative constancy of the conserved miRNAs within them. This notion is of importance if changes of miRNA complements occurred that have rendered the focus species different than the remainder of the lineage. In particular, losses of conserved miRNA families may occur convergently in distantly related groups of organisms.
Although not being the focus, some recent miRNA-based studies have included flatworms (Platyhelminthes), and by including Schmidtea mediterranea demonstrated that miRNAs support their phylogenetic affiliation to Lophotrochozoans (Erwin et al. 2011; Philippe et al. 2011). A study that included miRNA data of 3 flatworms, Schistosoma mansoni, Schi. japonicum, and S. mediterranea in a presence/absence matrix of 71 conserved miRNA families over 17 species, however, does not support this finding (Helm et al. 2012). In stark contrast to previous studies, they recovered flatworms as paraphyletic and basal to all other Bilaterians. Moreover, it became obvious that the included flatworms had significant differences in their miRNA complements that contradict the common understanding of miRNA evolution. This raises questions about the phylogeny and the evolution of miRNAs in flatworms.
Platyhelminthes (Gegenbauer, 1859) include approximately 20,000 dorsoventrally flattened species. Platyhelminthes lack a true coelom and have traditionally been placed together with Acoela at the root of Bilateria. Today, however, Platyhelminthes are considered Protostomia (Grobben, 1908), either within the Lophotrochozoa (Halanych et al., 1995) or the Platyzoa (Cavalier-Smith, 1998). Acoela are an independent clade that is most likely basal to all bilaterians (Wallberg et al. 2007; Hejnol et al. 2009; Jondelius et al. 2011), or part of deuterostomes (Philippe et al. 2011). Platyhelminthes include the polyphyletic free-living Turbellaria and the monophyletic and strictly parasitic Neodermata Cavalier-Smith (1998) that include almost 75% of all known flatworm species (Littlewood 2006). The divergence time of Neodermata and Turbellaria is difficult to assess because their fossil record is rather poor (Poinar 2003). It might reach 300 My back into the Permian, but recent molecular studies suggest an even older split some 510 Ma (Perkins 2010), which roughly coincides with the occurrence of the early vertebrates (Peterson and Butterfield 2005). Neodermata are strictly dependent on vertebrate hosts and consist of the endoparasitic tapeworms (Cestoda [Rudolphi, 1819]; ∼1,000 species, e.g., Echinococcus, Taenia, and Hymenolepis), the endoparasitic flukes (Trematoda [Rudolphi, 1808]; ∼18,000 species, e.g., Clonorchis, Schistosoma, and Fasciola), and the ectoparasitic Monogenea (Carus, 1863) (e.g., Gyrodactylus). Within Neodermata, the hermaphroditic Monogenea have the simplest life cycle with no intermediate hosts. They—in contrast to tapeworms and flukes—do not cause any human health issues, but some species are severe threats for aquaculture and fishery. In particular, the genus Gyrodactylus v. Nordmann (1832) caught public attention after Gyrodactylus salaris (Malmberg, 1957) was first reported as a pest of Atlantic salmon (Salmo salar) in Norway in the 1970s (Bakke et al. 2007). Most gyrodactylids are viviparous and highly progenetic ectoparasites, and it is expected that literally all teleost fishes are hosts for at least one Gyrodactylus species. Despite the great variety of forms, the monophyly of the Neodermata is strongly supported by the name-giving Neodermis (Littlewood 2006), but the phylogenetic relationships of the three main lineages within Neodermata have never been resolved unambiguously (Baguna and Riutort 2004). Historically, the Monogenea have been considered the sister group to Cestoda (Rohde 1994; Littlewood et al. 1999). In contrast, Perkins et al. (2010) found the Monogenea basal to a Trematoda + Cestoda clade when analyzing 32 platyhelminth mitochondrial genomes. Their results supported earlier findings based on sequence analyses of ribosomal DNA (Lockyer et al. 2003), as well as individual mitochondrial genes (Park et al. 2007). However, phylogenetic trees based on sequences from a single molecular marker or solely on mitochondrial DNA might be inappropriate (Hurst and Jiggins 2005; Galtier et al. 2009). Direct and indirect selection on the mitochondrial genome as well as its maternal mode of inheritance might confound the inference of evolutionary history (Hurst and Jiggins 2005; Galtier et al. 2009). Furthermore, Platyhelminthes are also a fast-evolving group and contrasting phylogenetic trees were shown to be due to long-branch attraction (Lartillot et al. 2007). Novel phylogenetic markers like miRNAs have not yet been used to study their phylogeny.
Until now, platyhelminth miRNA data have been available for the planarians S. mediterranea (Palakodeti et al. 2006; Lu et al. 2009; Friedländer et al. 2012) and Dugesia japonica (Xu et al. 2013), the trematodes Schi. japonicum (Xue et al. 2008; Hao et al. 2010; Wang et al. 2010), Schi. mansoni (de Souza Gomes et al. 2011), Orientobilharzia turkestanicum (Wang et al. 2012), Fasciola gigantica and F. hepatica (Xu et al. 2012), and Clonorchis sinensis (Xu et al. 2010) as well as the cestodes Echinococcus granulosus, E. multilocularis (Cucher et al. 2011), and Taenia saginata (Ai et al. 2012). Only the miRNA complements of S. mediterranea, Schi. japonicum, and E. granulosus—representatives of all major flatworm groups except Monogenea—are based on either high-throughput sequencing or cloned libraries of small RNAs, along with the respective reference genomes. For all other studies, there is either no reference genome available for the studied species or no small RNA libraries, which limits the significance of their results considerably. The corresponding miRNA complements are very likely incomplete and thus not suitable for comparative analyses.
This study therefore aimed at providing a miRNA complement of the monogenean flatworm G. salaris. With data for at least one species for all major flatworm lineages at hand, we here analyze miRNA evolution in platyhelminths in more general terms.
New Approaches
The bioinformatics pipeline miRCandRef (miRNA–candidate–reference) was developed to enable miRNA predictions for nongenome organisms like G. salaris. Briefly, miRCandRef uses genomic and small RNA data from next-generation sequencing (NGS) and locally assembles genomic reads that match miRNA candidates from small RNA-reads into relatively short genomic contigs (crystal-contigs). MiRCandRef delivers a multi-fasta file of assembled and clustered miRNA crystal-contigs that can be used as a genomic reference for miRNA prediction software rather than a properly assembled genome. Using this approach allows for a comparably fast miRNA prediction directly from NGS reads and circumvents the need for costly and sometimes patchy draft genome-assemblies. In its current version, the pipeline includes a modified version of miRDeep2 (Friedländer et al. 2012) that can handle the miRCandRef-fasta-output files and small RNA reads for miRNA prediction. To simplify the usage of the pipeline, miRCandRef has been implemented at the latest version of the hyperbrowser (http://hyperbrowser.uio.no, last accessed September 26, 2013) (Sandve et al. 2013), which is based on the Galaxy system (Giardine et al. 2005) and allows for a fully automated analysis based on small RNA and DNA-reads uploaded by the user.
Results
The miRNA Complement of G. salaris
The miRCandRef analyses of NGS data sets of small RNAs and genomic DNA of G. salaris identified initially some 300,000 of the roughly 3 Mio unique small RNA NGS reads with perfect matches in one of the about 90 Mio genomic DNA reads. All genomic DNA subsets, including the respective mate sequences, were assembled into 401,731 contigs, all representing miRNA-candidate-loci. Subsequently, this miRNA-candidate reference assembly and the original small RNA data set were used in miRDeep2*, a modified version of miRDeep2. All predicted miRNA loci were manually checked for appropriate structure and offset of mature and star sequences required for Dicer processing, and thus all accepted miRNA loci had to be represented by at least one mature and one star read (Tarver et al. 2012). Accordingly, 39 G. salaris miRNAs (gsa-mir) with high sequence similarities to previously described miRNAs and 22 miRNAs with yet undescribed seeds were identified (see supplementary file S1, Supplementary Material online, for read-counts, structures including offsets, and alignments). Four highly abundant miRNAs with particularly long hairpins, miR-1 (represented with more than 2 Mio reads in the small RNA NGS read pool), miR-87 (represented with more than 300,000 reads), mir-36 (represented with ∼57,000 reads), and mir-9 (represented with ∼8,000 reads) could only be detected after changing the miRDeep2 algorithmic parameters for the excision of hairpins.
Loss and Gain of miRNAs in Platyhelminth Flatworms
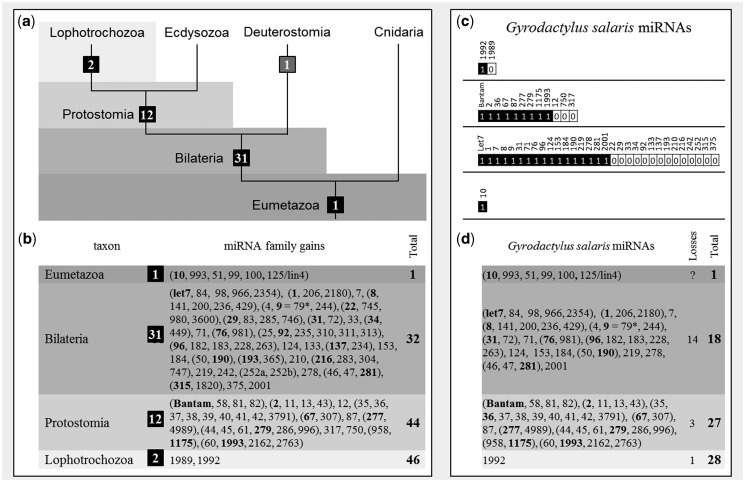
Earlier studies (Wheeler et al. 2009; Christodoulou et al. 2010; Erwin et al. 2011; Tarver et al. 2013) proposed ancestral miRNA complements for several animal lineages, and accordingly 46 conserved miRNAs families were expected in Gyrodactylus, that is, one that is characteristic for Eumetazoa, 31 for Bilateria, 12 for Protostomia, and two for Lophotrochozoa. Twenty-eight of the expected miRNA families were indeed found, but a total of 18 miRNA loci were not represented in the data set (fig. 1). Homologs of 14 bilaterian, 3 protostomian, and 1 lophotrochozoan miRNA families were not recovered. Mir-10, the only conserved miRNA characteristic for Eumetazoa, was found having several representatives in Gyrodactylus. BLAT-Searches (Kent 2002) of missing miRNA families in the genomic reference (crystal-contigs) did not recover additional bona fide miRNAs.
Fig. 1.
The miRNA complement of Gyrodactylus salaris shows lophotrochozoan features. (a) Evolutionary acquisition of miRNA families mapped on a simplified tree of Eumetazoa; (b) detailed table for group-specific conserved miRNAs (edited part of table from Wheeler et al. [2009] and results from Philippe et al. [2011]). Families are designated parenthetically; family names are bold. (c) miRNAs that are expected in G. salaris based on the affiliation to Lophotrochozoans and the actual miRNAs found (black); (d) detailed table for conserved miRNAs found in G. salaris and inferred losses/total number of conserved miRNA families.
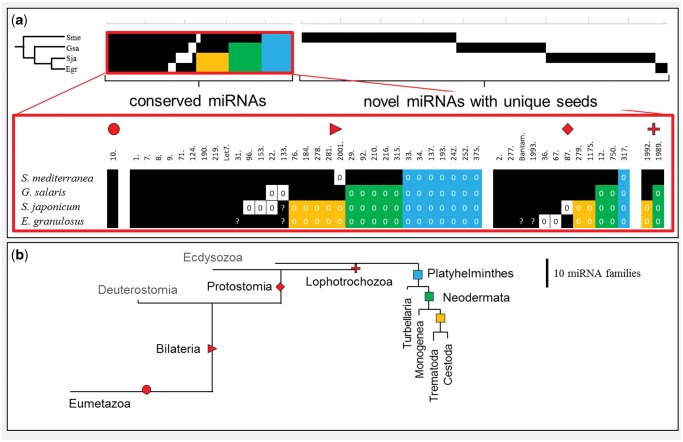
The obtained miRNA complement of G. salaris was compared with the miRNA complements of the free-living flatworm S. mediterranea (Turbellaria), the fluke Schi. japonicum (Trematoda), and the tapeworm E. granulosus (Cestoda). A total of 148 miRNAs are deposited in miRBase for S. mediterranea that represent 37 conserved miRNA families and 38 miRNAs unique to Schmidtea. We included five miRNAs without prior assignments to a conserved family, that is, the bilaterian-specific mir-153 (sme-mir-2163), mir-184 (sme-mir-748), mir-190 (sme-mir-756), and mir-210 (sme-mir-2164) families, as well as the lophotrochozoan specific mir-1989 (sme-mir-2154). For Schi. japonicum, 55 loci that represent 20 conserved miRNA families and 27 novel miRNAs including sja-mir-2162, which is a hitherto unreported member of the protostomian mir-1993 family were retrieved from miRBase. Because of the missing hairpin structures, the previously described mir-92 (sja-mir-310) and mir-279 miRNAs (sja-mir-61) (Wang et al. 2010) were rejected as bona fide miRNAs and thus not included. The miR-133 (sja-mir-133) miRNA (Wang et al. 2010), also without a bona fide hairpin structure, was nevertheless accepted, because of recently published expression data and northern blot evidence (see miRBase entries MI0015298, MI0015290, and MI0015293). Only 23 precursors that consisted of 16 conserved families and just 3 novel miRNA were recovered from miRBase for E. granulosus, including mir-4989 that belongs to the protostomian-specific mir-277 family. A further six miRNA families have been recently proposed for E. granulosus based on a blast-search approach (Jin et al. 2013) out of which we found four representatives (mir-31, mir-133, mir-2162 [1993], and bantam) with properly folding hairpins and reasonable offsets for hypothetical mature and star reads (data available upon request). Eight of the conserved bilaterian, protostomian, and lophotrochozoan miRNA families that were not detected in G. salaris were also not found for the other flatworm miRNA complements (fig. 2); these were mir-33, mir-34, mir-137, mir-193, mir-242, mir-252, mir-317, and mir-375. In addition, the neodermatan Schi. japonicum, E. granulosus, and G. salaris lack the eight conserved miRNA families mir-12, mir-29, mir-92, mir-210, mir-216, mir-315, mir-750, and mir-1989, which sum up to 16 shared losses of miRNA families in this clade. A clade consisting of Cestoda and Trematoda is supported by the shared loss of a further eight miRNA families (mir-76, mir-184, mir-278, mir-279, mir-281, mir-1175, mir-1992, and mir-2001). Thus, 24 of the 46 expected miRNAs, which include all lophotrochozoan-specific miRNAs, were not found in Cestoda and Trematoda. The compiled data set also indicates that each flatworm lineage has lost further miRNA families independently; that is, mir-2001 in S. mediterranea, mir-22, and mir-133 in G. salaris, mir-22, mir-87, mir-96, and mir-153 in Schi. japonicum, and mir-36, and mir-67 in E. granulosus. For each flatworm species, a set of novel miRNAs has been described. Surprisingly, no novel miRNAs were shared between flatworm lineages, that is, they were all species/lineage-specific. The observed specificities of the novel miRNA include the hairpin and particularly the family-and-function determining seed sequences. Although nothing can be said about their function, they must have different target sequences.
Fig. 2.
Loss and gain of miRNAs in flatworms. (a) miRNA distribution in flatworms shows pattern of taxa-specific losses of conserved miRNA families and sets of novel miRNAs with unique seeds for each group. Red blow-up: presence/absence pattern of taxa-specific groups of conserved miRNAs (Eumetazoa, Bilateria, Protostomia, and Lophotrochozoa); black squares represent the presence of a miRNA family (note: predicted miRNAs without sufficient experimental evidence for expression, or expressed miRNAs without genomic confirmation are depicted by “?”). Losses are depicted as white fields or highlighted in colors (blue: shared losses in flatworms, green: shared losses in Neodermata, yellow: shared losses in Trematoda + Cestoda); (b) gain and loss of miRNA families in Eumetazoa and Platyhelminthes, branch lengths corresponds to number of gains/losses.
Multicopy miRNAs
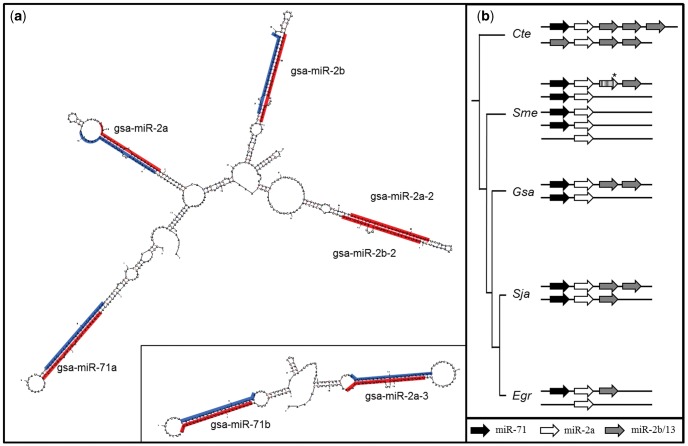
The miRNA complement of G. salaris includes several multi-copy miRNA families. Four hairpins consisted of mature sequences with seeds from the miR-2 family, three miRNAs representing the miR-10 family, two copies for each of the miR-96, miR-124, miR-184, and miR-277 families. Two miRNAs with high similarity to the miR-71 family were found in clusters that include other miRNAs with seed sequences known for members of the extended miR-2 family (fig. 3). All members of the extended mir-2 family from G. salaris were arranged in clusters, that is, gsa-mir-71a clustered together with gsa-mir-2a;-2b;-2a-2/b-2, and gsa-mir-71b with gsa-mir-2a-3 (fig. 3a). Clustered miR-2 family members have been described previously from several organisms, including flatworms (Marco et al. 2012). However, when comparing the mir-2 data of G. salaris with those of S. mediterranea, E. granulosus, and Schi. japonicum, it becomes likely that there are at least two extended mir-2 miRNA clusters that contain a mir-71 each in flatworms (fig. 3b).
Fig. 3.
Secondary structure of extended mir-2 family in Gyrodactylus salaris and their genomic organization in flatworms. (a) Secondary structure of the two extended mir-2 clusters found in G. salaris (mfold). Red marked sequences represent mature miRNAs, blue marked are star sequences; (b) comparison of mir-2 gene-arrangements in flatworms and as outgroup representative Capitella teleta (arrows: black mir71 seed: GAAAGA, white mir-2a seed: CACAGC, gray mir-2b/-13 seed AUCACA). *sme-mir-752 evolved by arm-switching from a mir-2a precursor.
Phylogenetic Analysis of miRNA Data
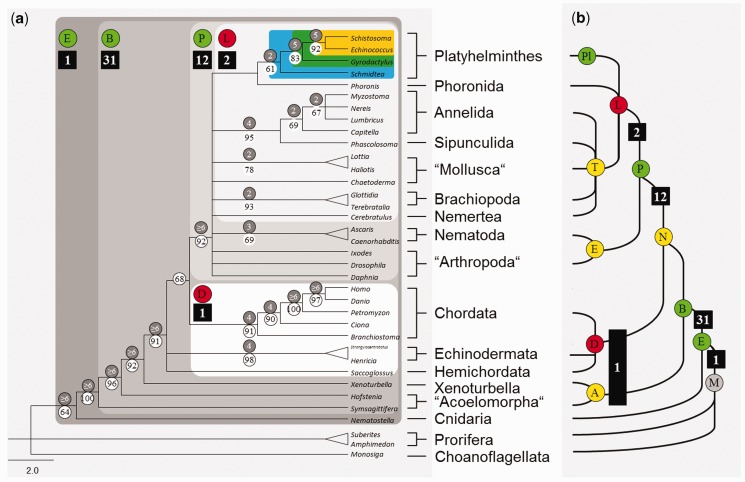
A presence/absence matrix was compiled for 153 conserved miRNA families from 36 metazoan taxa (supplementary matrix, Supplementary Material online). Only miRNAs were included that were unambiguously identified on the basis of well-characterized seed regions. The subsequent maximum parsimony analysis with 151 parsimony-informative loci yielded 72 equally parsimonious trees each with 306 steps. The strict consensus tree (fig. 4) confirmed the monophyly of Platyhelminthes with moderate bootstrap and Bremer support and their affiliation to Lophotrochozoa. The free-living S. mediterranea was basal to the Neodermata. Within the Neodermata, the Monogenea (G. salaris) were basal to a clade comprising of Trematoda (Schi. japonicum) and Cestoda (E. granulosus). Within Neodermata, there was also moderate statistical support for the respective nodes.
Fig. 4.
(a) Strict consensus of 24 equally parsimonious trees of a maximum parsimony analysis of 153 miRNA families in 36 metazoan taxa. The tree was rooted with Monosiga, Amphimedon, and Suberites. Numbers in black squares show evolutionary acquisitions of miRNA families; numbers in white circles show bootstrap values for 1,000 replicates, gray are Bremer support values. Squares colored in shades of gray depict Eumetazoa (E), Bilateria (B), Protostomia (P), Lophotrochozoa (L); white: polyphyletic Deuterostomia (D); blue: flatworms; green: Neodermata; yellow: Trematoda + Cestoda. (b) Simplified tree taken from Edgecombe et al. (2011) for the comparison with our results. Letters in circles represent abbreviation for major animal groups and are colored corresponding to their recovery in our data set: gray, not tested; red, miRNA data, but not recovered; yellow, no miRNA data, not recovered; green, recovered.
Discussion
The miRNA complement of the monogenean flatworm G. salaris was comprehensively characterized using the newly developed bioinformatics pipeline miRCandRef. A major advantage of miRCandRef is that it does not require the availability of a reference genome. Instead, it utilizes a local assembly approach based on small RNA and genomic DNA read-pools. Considering only genomic reads that contain a miRNA-candidate sequence, that is, a small RNA read, the computational requirements (in particular CPU time) for the sequence assembly are significantly reduced. MiRCandRef also takes rather small contigs with a length of less than 200 bp into account; these are usually not considered in de novo genome assemblies. Small modifications to miRDeep2 have been implemented that allow detecting additional miRNA-hairpins with unusual length (supplementary information, Supplementary Material online). It is likely they had been overlooked in a standard approach. Therefore, miRCandRef recovers more miRNAs candidate loci from NGS read-pools than other approaches that rely on de novo genome assemblies (e.g., with the default miRDeep2).
With respect to 46 conserved miRNAs earlier described for Metazoa, Bilateria, Protostomia, and Lophotrochozoa (Wheeler et al. 2009; Erwin et al. 2011), a significant loss of conserved miRNAs was detected in Monogenea. Altogether 18 of the expected conserved miRNA families were not detected in G. salaris. When including miRNA complements of S. mediterranea (Turbellaria), Schi. japonicum (Trematoda), and E. granulosus (Cestoda) into the analyses, it turned out that the loss of conserved miRNA loci is common in flatworms. In the free-living S. mediterranea, approximately 20% of the conserved miRNA families could not be detected, whereas in the more derived Schi. japonicum and E. granulosus approximately 50% had been lost. Conserved miRNA families are not lost randomly, but there is a pattern that is congruent with the phylogenetic relationship Turbellaria+Neodermata ([Monogenea {Trematoda + Cestoda}]). This means that losses of conserved miRNA families in platyhelminths are phylogenetically informative. It is important to note that the miRNA complement of E. granulosus representing Cestoda is derived from cloning data and is therefore most likely incomplete. Blast approaches have revealed further miRNAs (Jin et al. 2013) that were at least considered partly based on the available information about their structure. These hairpins were considered provisional for this study although conclusive proof is still pending. However, the extent of the inferred losses of conserved miRNAs in the closely related trematode Schi. japonicum is very similar, which was taken as support for assuming that the majority of miRNAs are included in the published complement of E. granulosus. Losses of more than 50% of conserved miRNA families as seen in endoparasitic platyhelminths have not yet been reported from any major lineage within other bilaterian phyla. Given the particular importance of miRNAs for the functionality of gene expression (Bartel and Chen 2004), it seems rather unlikely that about half of the otherwise conserved miRNAs just got lost in platyhelminths. Indeed, it has been suggested that miRNA losses might relate to loss of targets (Sperling and Peterson 2009) or reduced morphological complexity (Erwin et al. 2011). For Neodermata, there are some morphological reductions, such as, for example, the loss of the gut in tapeworms. Recently, Tsai et al. (2013) also reported genomic reductions such as, for example, the loss of several homeobox gene families, and the absence of most genes essential for peroxisomes and fatty acid biosynthesis in flukes and tapeworms. It seems therefore likely that not only morphological changes but also metabolic changes relate to the loss of miRNAs. However, several novel miRNAs were identified for each platyhelminth lineage but function could not be proposed for these. Some recent studies demonstrated differentially expressed conserved miRNAs from various developmental stages and sexes of platyhelminth worms, pointing to functional differences at different life stages (Friedlaender et al. 2009; Cai et al. 2011; Cucher et al. 2011). However, there is only very little detailed information relating to how miRNAs actually target mRNAs in animals. Rather, it is likely that miRNAs themselves are also frequently targets of other messenger RNAs (Sumazin et al. 2011). The few published studies on miRNA targets in flatworms were all performed in trematodes using either a bioinformatics approach for miRNA prediction from EST data that lack crucial small RNA read evidence (Schi. mansoni) (de Souza Gomes et al. 2011) or predicted miRNAs without a genomic reference specific to the respective species prior to target prediction (Fasciola [Xu et al. 2012] and Orientobilharzia [Wang et al. 2012]). Accordingly, these studies are of only limited value for understanding the functions of miRNA targets. For the monogenean G. salaris, there is currently no fully assembled and annotated genome or EST data set available, which would be a prerequisite for a similar bioinformatics target analysis.
A phylogenetic analyses of the presence/absence of 153 miRNAs from 36 metazoan taxa recovered platyhelminths as monophyletic (fig. 4). In the current analysis, the number of loci included in the data matrix was more than doubled as compared with the previously published study of Helm et al. (2012). The increased matrix may largely explain the improved resolution of the tree, and the higher support for many nodes within (supplementary matrix, Supplementary Material online).
The obtained phylogenetic tree was largely in agreement with the current view on metazoan phylogeny, which is largely based on phylogenomic data sets as a gold standard (Edgecombe et al. 2011). Bilateria and Chordata are both monophyletic and well supported. However, there was no support for the monophyly of Lophotrochozoa/Spiralia, Deuterostomia, and Acoela. This can likely be attributed to the lack (Nephrozoa, Acoela, Ecdysozoa, and Trochozoa) or the low number (Deuterostomes [1], Lophotrochozoa [2]) of apomorphic miRNAs. Surprisingly, no novel miRNA was found apomorphic for Platyhelminthes, Neodermata, or the Trematoda + Cestoda clade. However, the shared losses of miRNAs were phylogentically informative, and reasonable statistical support was found for Platyhelminthes, and for the Monogenea being basal in the Neodermata with a sister group consisting of the endoparasitic Trematoda and Cestoda. Although novel miRNAs provided no information for the phylogenetic relationships of flatworms, the duplication of the mir-2 cluster, each including a further mir-71 hairpin, is likely a synapomorphy of Platyhelminthes. Two copies were described previously also for Schi. mansoni (two miR-71) (de Souza Gomes et al. 2011), C. sinensis (two miR-71) (Xu et al. 2010), and O. turkestanicum (two mir-71) (Wang et al. 2012). In E. granulosus and E. multilocularis, a second mir-71 that is associated with a mir-2 is missing, even though a second mir-2 locus has been described (Cucher et al. 2011). This implies a secondary loss of a second mir-71 copy in cestodes. In contrast, S. mediterranea showed altogether 5 extended mir-2 loci, of which 4 included also an additional mir-71. This indicates a secondary acquisition of further mir-71/2 clusters in Turbellaria. Despite all limitations of the miRNA data currently available for platyhelminth species, that is, limited taxonomic coverage, incorrectly annotated miRNA loci in miRBase and/or incomplete data, there is no question about the significant loss of otherwise conserved miRNAs. Interestingly, the observed numbers of lost miRNA families in flatworms is lowest in the free-living S. mediterranea, somewhat higher in the ectoparasitic G. salaris and highest in the endoparasitic Schi. japonicum and E. granulosus. Currently, there is however no convincing evidence that would tie loss of miRNAs to parasitic lifestyle. Flatworms, and in particular the parasitic Neodermata, therefore offer an excellent model for addressing functional aspects of miRNA evolution. The pipeline miRCandRef offers a straightforward analysis of miRNAs from nonmodel organisms by exploiting genomic information on the level of NGS reads and user-optimized usability at the hyperbrowser (http://hyperbrowser.uio.no, last accessed September 26, 2013).
Materials and Methods
Next-Generation Sequencing of G. salaris Short RNA and Total Genomic DNA
The G. salaris (Lier strain) parasites were cultured and harvested as described before (Fromm et al. 2011). A total RNA sample enriched for small RNA was extracted from 100 pooled individuals using the ZR RNA MicroPrep kit (Zymo Research). A small RNA library was prepared using the ScriptMiner Small RNA-Seq Multiplex Library Preparation Kit (Epicentre) and subsequently sequenced on an Illumina GA2 in a 55 bp, single-end run (one lane). A total genomic DNA extraction of approximately 20,000 pooled individuals was performed using the E.Z.N.A-Tissue Kit (Omega Bio-tek). The genomic DNA was subsequently used for library preparation and subjected to a 76 bp, paired-end run on an Illumina GA2 instrument (one lane).
Bioinformatics
The algorithm miRCandRef was developed for assembling high-quality filtered NGS data into relatively short genomic contigs (crystal-contigs) that match predicted miRNA candidate loci. The pipeline includes a modified version of miRDeep2 (Friedländer et al. 2012) and delivers a multi-fasta file of assembled and clustered miRNA crystal-contigs that can be used as a reference for miRNA prediction software like miRDeep2 rather than a properly assembled genome (for details see supplementary information, Supplementary Material online). To distinguish between missing and not expressed miRNAs, the crystal-contigs were additionally screened for absent miRNA families. Known miRNAs in G. salaris were identified using a reference file that contained all miRNAs from S. mediterranea, Schi. japonicum and E. granulosus retrievable from miRBase. All miRNAs were compared with a database of 46 miRNAs that represent proposed ancestral miRNA complements of Eumetazoa, Bilateria, Protostomia, and Lophotrochozoa. Sequence similarity in the seed sequence of the mature miRNA and other regions of the hairpin were the key criteria for miRNA family assignment. miRNAs from the flatworms S. mediterranea, Schi. japonicum, and E. granulosus were retrieved from miRBase and analyzed accordingly.
Phylogenetic Analysis
The character matrix for 153 miRNAs from 36 taxa was analyzed using PAUP* v.4.0b10 (Swofford 2003). All characters were encoded for presence or absence and assigned equal weight. The analysis was performed with unordered characters and a heuristic search implemented under a Dollo model. Hereby, each character is allowed to evolve only once and all homoplasy is explained by reversals from the ancestral state. Node support in the strict consensus tree of equally parsimonious trees was estimated by running 1,000 bootstrap replicates. Bremer support values were generated including all trees with up to six more steps (Bremer 1994).
Supplementary Material
Supplementary file S1, figs. S1 and S2, information, and matrix are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank T.D. Otto for discussions and inspiration on the design of miRCandRef, and M. Friedländer and S. Mackowiak for the help and assistance adapting miRDeep2 to miRCandRef. They are grateful to K. Trengereid for implementing miRCandRef into the hyperbrowser and thank A. Marco for useful insights and discussions on the extended mir-2 cluster. The authors also thank T.A. Bakke and P.D. Harris for animal collection and keeping animal cultures.
References
- Ai L, Xu MJ, Chen MX, Zhang YN, Chen SH, Guo J, Cai YC, Zhou XN, Zhu XQ, Chen JX. Characterization of microRNAs in Taenia saginata of zoonotic significance by Solexa deep sequencing and bioinformatics analysis. Parasitol Res. 2012;110:2373–2378. doi: 10.1007/s00436-011-2773-x. [DOI] [PubMed] [Google Scholar]
- Baehrecke EH. miRNAs: micro managers of programmed cell death. Curr Biol. 2003;13:R473–R475. doi: 10.1016/s0960-9822(03)00405-6. [DOI] [PubMed] [Google Scholar]
- Baguna J, Riutort M. Molecular phylogeny of the Platyhelminthes. Can J Zool. 2004;82:168–193. [Google Scholar]
- Bakke TA, Cable J, Harris PD. The biology of gyrodactylid monogeneans: the “Russian-doll killers”. Adv Parasitol. 2007;64:161–376. doi: 10.1016/S0065-308X(06)64003-7. [DOI] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304. [Google Scholar]
- Brennecke J, Cohen SM. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 2003;4:228. doi: 10.1186/gb-2003-4-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P, Hou N, Piao X, Liu S, Liu H, Yang F, Wang J, Jin Q, Wang H, Chen Q. Profiles of small non-coding RNAs in Schistosoma japonicum during development. PLoS Negl Trop Dis. 2011;5:e1256. doi: 10.1371/journal.pntd.0001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LI, Rota-Stabelli O, Edgecombe GD, Marchioro T, Longhorn SJ, Telford MJ, Philippe H, Rebecchi L, Peterson KJ, Pisani D. MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proc Natl Acad Sci U S A. 2011;108:15920–15924. doi: 10.1073/pnas.1105499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li WX, Xie D, Peng JR, Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell. 2004;16:1302–1313. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou F, Raible F, Tomer R, Simakov O, Trachana K, Klaus S, Snyman H, Hannon GJ, Bork P, Arendt D. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucher M, Prada L, Mourglia-Ettlin G, Dematteis S, Camicia F, Asurmendi S, Rosenzvit M. Identification of Echinococcus granulosus microRNAs and their expression in different life cycle stages and parasite genotypes. Int J Parasitol. 2011;41:439–448. doi: 10.1016/j.ijpara.2010.11.010. [DOI] [PubMed] [Google Scholar]
- de Souza Gomes M, Muniyappa MK, Carvalho SG, Guerra-Sa R, Spillane C. Genome-wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni. Genomics. 2011;98:96–111. doi: 10.1016/j.ygeno.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Edgecombe G, Giribet G, Dunn C, Hejnol A, Kristensen R, Neves R, Rouse G, Worsaae K, Sørensen M. Higher-level metazoan relationships: recent progress and remaining questions. Organ Diversity Evol. 2011;11:151–172. [Google Scholar]
- Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- Friedlaender MR, Adamidi C, Ting H, et al. (13 co-authors) High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci U S A. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Harris PD, Bachmann L. MicroRNA preparations from individual monogenean Gyrodactylus salaris—a comparison of six commercially available totalRNA extraction kits. BMC Res Notes. 2011;4:217. doi: 10.1186/1756-0500-4-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Nabholz B, Glemin S, Hurst GDD. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 2009;18:4541–4550. doi: 10.1111/j.1365-294X.2009.04380.x. [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, et al. (13 co-authors) Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao LL, Cai PF, Jiang N, Wang H, Chen QJ. Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum. BMC Genomics. 2010;11:55. doi: 10.1186/1471-2164-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg AM, Cowper-Sallari R, Semon M, Donoghue PCJ, Peterson KJ. microRNAs reveal the interrelationships of hagfish, lampreys, and gnathostomes and the nature of the ancestral vertebrate. Proc Natl Acad Sci U S A. 2010;107:19379–19383. doi: 10.1073/pnas.1010350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, et al. (17 co-authors) Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm C, Bernhart SH, Siederdissen CHZ, Nickel B, Bleidorn C. Deep sequencing of small RNAs confirms an annelid affinity of Myzostomida. Mol Phylogenet Evol. 2012;64:198–203. doi: 10.1016/j.ympev.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Hurst GDD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc Biol Sci. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Lu L, Su H, Lou Z, Wang F, Zheng Y, Xu GT. Comparative analysis of known miRNAs across platyhelminths. FEBS J. 2013;280:3944–3951. doi: 10.1111/febs.12395. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Jondelius U, Wallberg A, Hooge M, Raikova OI. How the worm got its pharynx: phylogeny, classification and Bayesian assessment of character evolution in Acoela. System Biol. 2011;60:845–871. doi: 10.1093/sysbio/syr073. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(1 Suppl):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood DTJ. The evolution of parasitism in flatworms. In: Maule AG, Marks NJ, editors. Parasitic flatworms: molecular biology, biochemistry, immunology and physiology. Wallingford (UK): CABI Publishing; 2006. pp. 1–36. [Google Scholar]
- Littlewood DTJ, Rohde K, Bray RA, Herniou EA. Phylogeny of the Platyhelminthes and the evolution of parasitism. Biol J Linnean Soc. 1999;68:257–287. [Google Scholar]
- Lockyer AE, Olson PD, Littlewood DTJ. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): implications and a review of the cercomer theory. Biol J Linnean Soc. 2003;78:155–171. [Google Scholar]
- Lu YC, Smielewska M, Palakodeti D, Lovci MT, Aigner S, Yeo GW, Graveley BR. Deep sequencing identifies new and regulated microRNAs in Schmidtea mediterranea. RNA. 2009;15:1483–1491. doi: 10.1261/rna.1702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Hooks K, Griffiths-Jones S. Evolution and function of the extended miR-2 microRNA family. RNA Biol. 2012;9:242–248. doi: 10.4161/rna.19160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the Planarian Schmidtea mediterranea: a model system for stem cell biology. RNA. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Kim KH, Kang S, Kim W, Eom KS, Littlewood DTJ. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes) BMC Evol Biol. 2007;7:11. doi: 10.1186/1471-2148-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Perkins E. Family ties molecular phylogenetics, evolution and radiation of flatworm parasites (Monogenea: capsalidae) [PhD thesis] Adelaide (Australia): The University of Adelaide; 2010. [Google Scholar]
- Perkins EM, Donnellan SC, Bertozzi T, Whittington ID. Closing the mitochondrial circle on paraphyly of the Monogenea (Platyhelminthes) infers evolution in the diet of parasitic flatworms. Int J Parasitol. 2010;40:1237–1245. doi: 10.1016/j.ijpara.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Butterfield NJ. Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc Natl Acad Sci U S A. 2005;102:9547–9552. doi: 10.1073/pnas.0503660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, Wallberg A, Peterson KJ, Telford MJ. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470:255–258. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinar G. A rhabdocoel turbellarian (Platyhelminthes, Typhloplanoida) in Baltic amber with a review of fossil and sub-fossil platyhelminths. Invertebrate Biol. 2003;122:308–312. [Google Scholar]
- Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz H, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rohde K. The origins of parasitism in the platyhelminthes. Int J Parasitol. 1994;24:1099–1115. doi: 10.1016/0020-7519(94)90185-6. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, Campbell L, Brinkmann H, Edgecombe GD, Longhorn SJ, Peterson KJ, Pisani D, Philippe H, Telford MJ. A congruent solution to arthropod phylogeny: phylogenomics, microRNAs and morphology support monophyletic Mandibulata. Proc Biol Sci. 2011;278:298–306. doi: 10.1098/rspb.2010.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve GK, Gundersen S, Johansen M, et al. (21 co-authors) The Genomic HyperBrowser: an analysis web server for genome-scale data. Nucleic Acids Res. 2013;14:W133–W141. doi: 10.1093/nar/gkt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. 2006;4:651–659. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- Sperling EA, Peterson KJ. microRNAs and metazoan phylogeny: big trees from little genes. In: Telford MJ, Littlewood DTJ, editors. Animal evolution—genomes, trees and fossils. Oxford: Oxford University Press; 2009. [Google Scholar]
- Sperling EA, Pisani D, Peterson KJ. Molecular paleobiological insights into the origin of the Brachiopoda. Evol Dev. 2011;13:290–303. doi: 10.1111/j.1525-142X.2011.00480.x. [DOI] [PubMed] [Google Scholar]
- Sperling EA, Vinther J, Moy VN, Wheeler BM, Semon M, Briggs DE, Peterson KJ. MicroRNAs resolve an apparent conflict between annelid systematics and their fossil record. Proc Biol Sci. 2009;276:4315–4322. doi: 10.1098/rspb.2009.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Yang X, Chiu HS, et al. (11 co-authors) An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) 2003. Version 4. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Tarver JE, Donoghue PC, Peterson KJ. Do miRNAs have a deep evolutionary history? BioEssays. 2012;34:857–866. doi: 10.1002/bies.201200055. [DOI] [PubMed] [Google Scholar]
- Tarver JE, Sperling EA, Nailor A, Heimberg AM, Robinson JM, King BL, Pisani D, Donoghue PCJ, Peterson KJ. miRNAs: small genes with big potential in metazoan phylogenetics. Mol Biol Evol. 2013 doi: 10.1093/molbev/mst133. Advance Access published August 28, 2013, doi:10.1093/molbev/mst133. [DOI] [PubMed] [Google Scholar]
- Tsai IJ, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013;496:57–63. doi: 10.1038/nature12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A, Curini-Galletti M, Ahmadzadeh A, Jondelius U. Dismissal of Acoelomorpha: Acoela and Nemertodermatida are separate early bilaterian clades. Zool Scripta. 2007;36:509–523. [Google Scholar]
- Wang C-R, Xu M-J, Fu J-H, Nisbet AJ, Chang Q-C, Zhou D-H, Huang S-Y, Zou F-C, Zhu X-Q. Characterization of microRNAs from Orientobilharzia turkestanicum, a neglected blood fluke of human and animal health significance. PLoS One. 2012;7:e47001. doi: 10.1371/journal.pone.0047001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX, Xue XY, Sun J, Luo R, Xu XD, Jiang YY, Zhang QF, Pan WQ. An “in-depth” description of the small non-coding RNA population of Schistosoma japonicum Schistosomulum. PLoS Negl Trop Dis. 2010;4:e596. doi: 10.1371/journal.pntd.0000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler B, Heimberg A, Moy V, Sperling E, Holstein T, Heber S, Peterson K. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11:50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Trautwein MD, Winkler IS, et al. (27 co-authors) Episodic radiations in the fly tree of life. Proc Natl Acad Sci U S A. 2011;108:5690–5695. doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M-J, Ai L, Fu J-H, Nisbet AJ, Liu Q-Y, Chen M-X, Zhou D-H, Zhu X-Q. Comparative characterization of microRNAs from the liver flukes Fasciola gigantica and F. hepatica. PLoS One. 2012;7:e53387. doi: 10.1371/journal.pone.0053387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Liu QA, Nisbet AJ, Cai XQ, Yan C, Lin RQ, Yuan ZG, Song HQ, He XH, Zhu XQ. Identification and characterization of microRNAs in Clonorchis sinensis of human health significance. BMC Genomics. 2010;11:521. doi: 10.1186/1471-2164-11-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Chen M, Ren Z, Zhang N, Xu H, Liu X, Tian G, Song L, Yang H. Deep sequencing identifies regulated small RNAs in Dugesia japonica. Mol Biol Rep. 2013;40:4075–4081. doi: 10.1007/s11033-012-2485-z. [DOI] [PubMed] [Google Scholar]
- Xue XY, Sun J, Zhang QF, Wang ZX, Huang YF, Pan WQ. Identification and characterization of novel microRNAs from Schistosoma japonicum. PLoS One. 2008;3:e4034. doi: 10.1371/journal.pone.0004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.