Abstract
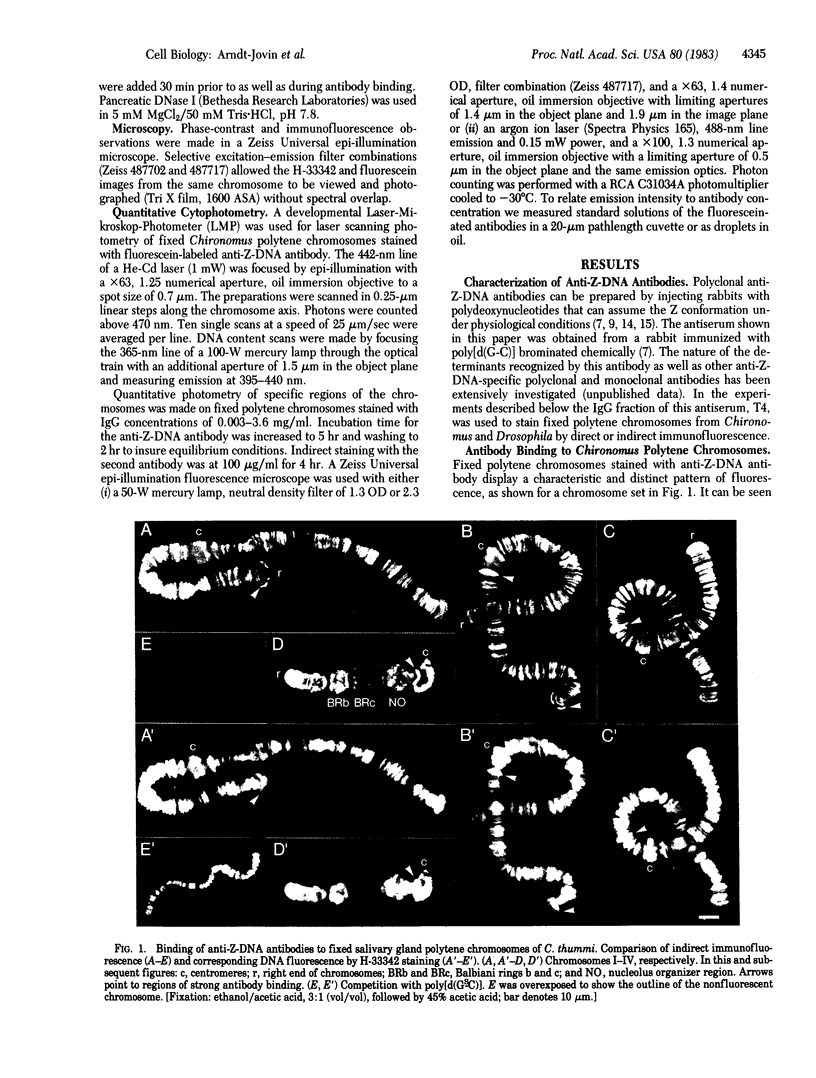
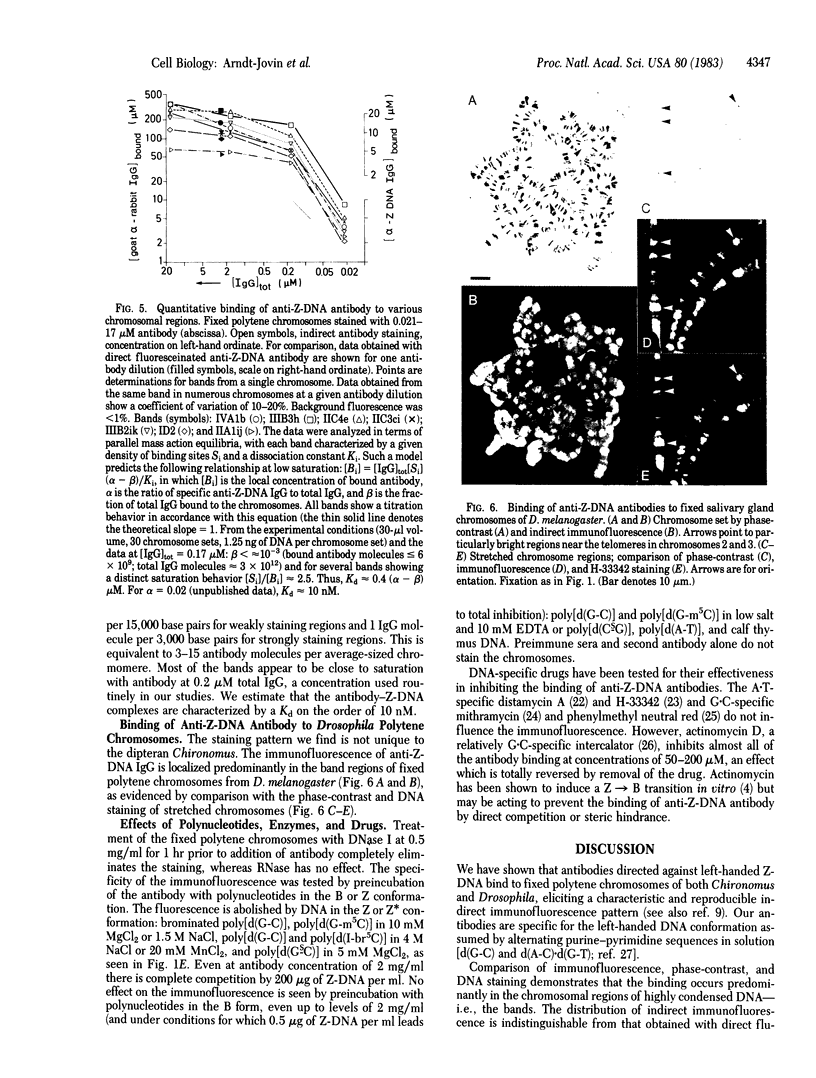
Antibodies to DNA in the left-handed (Z) conformation bind to acid-fixed polytene chromosomes of both Chironomus thummi and Drosophila melanogaster, as shown by direct and indirect immunofluorescence. Comparison of the phase-contrast, immunofluorescence, and DNA staining patterns shows a predominant localization of the antibody to the regions of high contrast and DNA density, the bands. The immunofluorescence is completely abolished by competition with polynucleotides in the Z conformation but not by those in the B form. DNase but not RNase treatment eliminates the antibody staining. Actinomycin D inhibits binding, whereas mithramycin has no effect. The highly reproducible immunofluorescence patterns obtained with the anti-Z-DNA antibodies demonstrate variations in fluorescence intensity between particular bands, which can be quantitated by laser scanning and photon counting techniques. The telomeric regions and DNA strands associated with end-to-end chromosome linkage and ectopic pairing are exceptionally bright. At saturation, average values of 1 IgG molecule per 3,000 base pairs and 1 per 15,000 base pairs are found in the intensely and weakly staining regions, respectively. An alternative statement is that the left-handed Z-DNA conformation is present at a frequency of 0.02-0.1%. The measured differences reflect variations in the local density of Z-DNA sites and not in the affinity for the specific antibody, which appears to be relatively constant throughout the chromosomes (Kd approximately equal to 10 nM). These observations taken together with results of biophysical studies on the properties of Z-DNA in solution suggest that regions of DNA in the left-handed conformation could be involved in higher-order structural organization of chromosomes and possibly in modulation of their functional state.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick C., Johns E. W. The effect of two acetic acid containing fixatives on the histone content of calf thymus deoxyribonucleoprotein and calf thymus tissue. Exp Cell Res. 1968 Aug-Sep;51(2-3):626–632. doi: 10.1016/0014-4827(68)90150-x. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Conformation and dynamics in a Z-DNA tetramer. J Mol Biol. 1981 Nov 15;152(4):723–736. doi: 10.1016/0022-2836(81)90124-8. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Hägele K. DNS-Replikationsmuster der Speicheldrüsen-Chromosomen von Chironomiden. Chromosoma. 1970;31(1):91–138. doi: 10.1007/BF00321159. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., van de Sande J. H., Zarling D. A., Arndt-Jovin D. J., Eckstein F., Füldner H. H., Greider C., Grieger I., Hamori E., Kalisch B. Generation of left-handed Z-DNA in solution and visualization in polytene chromosomes by immunofluorescence. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):143–154. doi: 10.1101/sqb.1983.047.01.019. [DOI] [PubMed] [Google Scholar]
- Kiknadze I. I., Perov N. A., Chentsov Y. S. Electron microscopic studies on the polytene chromosomes of Chironomus thummi salivary glands. I. Ultrastructural mapping. Chromosoma. 1976 Mar 31;55(1):91–103. doi: 10.1007/BF00288332. [DOI] [PubMed] [Google Scholar]
- Lafer E. M., Möller A., Nordheim A., Stollar B. D., Rich A. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3546–3550. doi: 10.1073/pnas.78.6.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latt S. A., Stetten G. Spectral studies on 33258 Hoechst and related bisbenzimidazole dyes useful for fluorescent detection of deoxyribonucleic acid synthesis. J Histochem Cytochem. 1976 Jan;24(1):24–33. doi: 10.1177/24.1.943439. [DOI] [PubMed] [Google Scholar]
- Malfoy B., Leng M. Antiserum to Z-DNA. FEBS Lett. 1981 Sep 14;132(1):45–48. doi: 10.1016/0014-5793(81)80424-3. [DOI] [PubMed] [Google Scholar]
- Malfoy B., Rousseau N., Leng M. Interaction between antibodies to Z-form deoxyribonucleic acid and double-stranded polynucleotides. Biochemistry. 1982 Oct 26;21(22):5463–5467. doi: 10.1021/bi00265a013. [DOI] [PubMed] [Google Scholar]
- Miller F. D., Rattner J. B., van de Sande J. H. Nucleosome-core assembly on B and Z forms of poly[d(G-m5C)]. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):571–575. doi: 10.1101/sqb.1983.047.01.067. [DOI] [PubMed] [Google Scholar]
- Müller W., Bünemann H., Dattagupta N. Interactions of heteroaromatic compounds with nucleic acids. 2. Influence of substituents on the base and sequence specificity of intercalating ligands. Eur J Biochem. 1975 May;54(1):279–291. doi: 10.1111/j.1432-1033.1975.tb04138.x. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Pardue M. L., Lafer E. M., Möller A., Stollar B. D., Rich A. Antibodies to left-handed Z-DNA bind to interband regions of Drosophila polytene chromosomes. Nature. 1981 Dec 3;294(5840):417–422. doi: 10.1038/294417a0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Pohl F. M. Salt-induced transition between two double-helical forms of oligo (dC-dG). Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):113–117. doi: 10.1101/sqb.1983.047.01.014. [DOI] [PubMed] [Google Scholar]
- Robert M. Einfluss von Ionenstärke und pH auf die differentielle Dekondensation der Nukleoproteide isolierter speicheldrüsen-Zellkerne und -Chromosomen von Chironomus thummi. Chromosoma. 1971;36(1):1–33. doi: 10.1007/BF00326419. [DOI] [PubMed] [Google Scholar]
- Robert M. Isolation and manipulation of salivary gland nuclei and chromosomes. Methods Cell Biol. 1975;9(0):377–390. doi: 10.1016/s0091-679x(08)60083-7. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Isolation of a telomeric DNA sequence from Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1041–1046. doi: 10.1101/sqb.1978.042.01.104. [DOI] [PubMed] [Google Scholar]
- Serfling E., Majorov V. I., Mikichur N. I., Popova T. G., Sandakchiev L. S. DNA and RNA content of Chironomus thummi polytene chromosomes determined by micro column gel filtration. Cell Differ. 1975 Mar;3(6):361–370. doi: 10.1016/0045-6039(75)90004-4. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. The stereochemistry of actinomycin binding to DNA and its implications in molecular biology. Prog Nucleic Acid Res Mol Biol. 1973;13:153–190. doi: 10.1016/s0079-6603(08)60103-8. [DOI] [PubMed] [Google Scholar]
- Sorsa V. Volume of chromatin fibers in interbands and bands of polytene chromosomes. Hereditas. 1982;97(1):103–113. doi: 10.1111/j.1601-5223.1982.tb00718.x. [DOI] [PubMed] [Google Scholar]
- Wang A. J., Quigley G. J., Kolpak F. J., van der Marel G., van Boom J. H., Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981 Jan 9;211(4478):171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Klysik J., Stirdivant S. M., Wells R. D. Conditions which cause the right-handed to left-handed DNA conformational transitions. Evidence for several types of left-handed DNA structures in solution. J Biol Chem. 1982 Mar 25;257(6):2775–2782. [PubMed] [Google Scholar]
- Zimmer C. Effects of the antibiotics netropsin and distamycin A on the structure and function of nucleic acids. Prog Nucleic Acid Res Mol Biol. 1975;15(0):285–318. doi: 10.1016/s0079-6603(08)60122-1. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B. The three-dimensional structure of DNA. Annu Rev Biochem. 1982;51:395–427. doi: 10.1146/annurev.bi.51.070182.002143. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Jovin T. M. Z* DNA, the left-handed helical form of poly[d(G-C)] in MgCl2-ethanol, is biologically active. EMBO J. 1982;1(1):115–120. doi: 10.1002/j.1460-2075.1982.tb01133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sande J. H., McIntosh L. P., Jovin T. M. Mn2+ and other transition metals at low concentration induce the right-to-left helical transformation of poly[d(G-C)]. EMBO J. 1982;1(7):777–782. doi: 10.1002/j.1460-2075.1982.tb01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]