Abstract
The burdens of type 2 diabetes (T2D) and cardiovascular diseases (CVD) are increasingin Africa. T2D and CVD are the result of the complex interaction between inherited characteristics, lifestyle, and environmental factors. The epidemic of obesity is largely behind the exploding global incidence of T2D. However, not all obese individuals develop diabetes and positive family history is a powerful risk factor for diabetes and CVD. Recent implementations of high throughput genotyping and sequencing approaches have advanced our understanding of the genetic basis of diabetes and CVD by identifying several genomic loci that were not previously linked to the pathobiology of these diseases. However, African populations have not been adequately represented in these global genomic efforts. Here, we summarize the state of knowledge of the genetic epidemiology of T2D and CVD in Africa and highlight new genomic initiatives that promise to inform disease etiology, public health and clinical medicine in Africa.
Keywords: Type 2 diabetes, cardiovascular diseases, genetics, epidemiology, Africa
Introduction
Although infectious diseases such as HIV/AIDS, tuberculosis, respiratory infections, diarrhea, and malaria account for the majority of morbidity and mortality in Africa, recent projections indicate that non-communicable diseases (NCD) including type 2 diabetes (T2D) and cardiovascular diseases (CVD) will become the leading causes of morbidity and death across Africa. For example, the prevalence of diabetesin sub-Saharan Africa (SSA)is projected to rise from 7.2 million in the year 2000 to 18.7 million in 2030 representing an alarming 161% increase in the proportion of persons with diabetes.1 The projected increase in the prevalence of diabetes in SSA is much higher than the projected global average of 114% and second only to the projection for the Middle Eastern Crescent.1 In parallel, the incidence and prevalence of CVD are rising in Africa, mainly because of the rise in prevalence of risk factors such as hypertension, diabetes, obesity, physical inactivity, increased tobacco use and dyslipidemia.2 Ischemic heart disease (the principal component of CVD) is now ranked the eighth leading cause of death among men and women in SSA.2 The World Health Organization estimates that by the year 2030, ischemic heart disease will be the leading cause of mortality in low-income countries, and cerebrovascular disease will be the third leading cause of mortality, just following HIV/AIDS.3
Genetics of T2D in Africa
Family History and Heritability
T2D results from complex interactions between genetic and non-genetic factors including lifestyle, sociocultural, ecological and demographic characteristics (see figure 1).4 Positive family history (PFH) is a major risk factor for T2D. For example, the Framingham Offspring Study, a prospective epidemiologic study of over 5,000 young adults in the USA, reported that an individual who has one or both parents with T2D has a lifetime risk of 30–40% and 70% for developing diabetes respectively.5 Moreover, a person who has a sibling with T2D is at 2 to 4 fold increased risk of developing T2D compared to an individual in the general population.6 The effect of PFH on the risk of developing T2D increases dramatically in the presence of obesity. Goldfine et al. (2003) observed that the risk of developing T2D is 16.7 per 1000 person years among obese individuals with PFH compared to 8.8 per 1000 person years among all persons with PFH.7 Interestingly, obesity did not significantly increase T2D risk among persons with negative family history (NFH); the age-adjusted incidence rate for obese persons with NFH was 1.6 per 1000 person years which is comparable to the rate of 1.8 per 1000 person years for all persons with NFH.7
Figure 1.
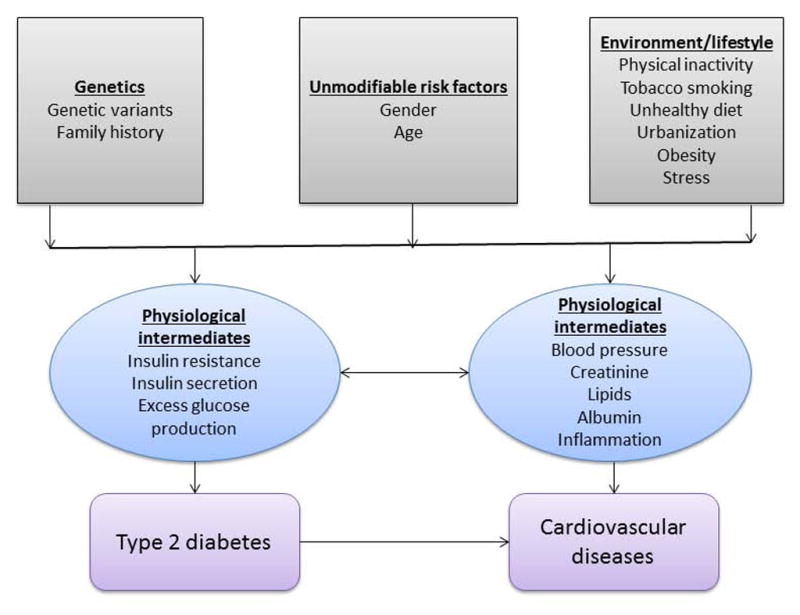
A Schematic Representation of Gene-by-environment Interaction in the Pathophysiology of Type 2 Diabetes and Cardiovascular Diseases
The limited number of studies conducted in African ancestry populations provides evidence for the familial clustering of CVD and predisposing risk factors such as T2D and hypertension. In our study of first degree relatives of 232 AfricanAmerican pedigrees which included 1,420 individuals recruited from the Chicago area, we observed excess disease risk among relatives (parents and offspring) of affected probands compared to relatives of unaffected probands for coronary heart disease (odds ratio [OR] = 5.30; 95% confidence interval [CI] = 2.51–11.23); hypertension (OR = 1.98; CI = 1.41–2.80); stroke (OR = 3.24; CI = 1.08–9.70); and diabetes (OR = 2.95; CI = 1.55–5.62). The results of this study clearly show that coronary heart disease, hypertension, stroke, and T2D aggregate in some AfricanAmerican families and not in others.8
A pedigree study that compared 69 offspring from 26 Cameroonian families with at least one T2D parent with 62 offspring from 25 families with no affected T2D parent provided evidence for increased prevalence of diabetes and impaired glucose tolerance in the offspring of parents with T2D.9 Similarly, the impact of PFH was also demonstrated among Nigerians in a study that assessed glucose and insulin responses to an oral glucose load.10 This study revealed that offspring of parents with T2D had higher levels of fasting plasma glucose, fasting plasma insulin, and 2-hours post glucose load plasma insulin indicating a greater risk for developing diabetes among this group.10 A study involving over one thousand black South African patients with T2D and 687 controls showed that PFH of diabetes is over three-fold more common among diabetic subjects than controls (27.3% vs. 8.4%, p<0.01). Moreover, patients with PFH had an earlier onset of diabetes than those with NFH.11 Together, these findings provide evidence that T2D and CVD in African ancestry populations show familial clustering and have heritable components.
Candidate Gene and Genome-wide Approaches to T2D in African Populations
Overall, only a handful of studies have used the genome-wide approach to identify genomic regions linked to or associated with T2D in African populations. The first of such studies was conducted among participants enrolled in the Africa America Diabetes Mellitus (AADM) study, the longest running genetic epidemiology study of T2D in Africa with participants recruited from Ghana and Nigeria.12 Using an affected sibling pair (ASP) approach with 390 short tandem repeat markers typed in 343 families, multipoint linkage analysis identified suggestive evidence of linkage in four regions on chromosomes 12, 19, and 20. The strongest evidence for linkage was observed on chromosome 20q13.3, a locus that had also been reported to be linked to T2D in non-African populations.13 In addition, the AADM 12q24 locus harbors the hepatocyte nuclear factor-1alpha(HNF1A)gene that encodes hepatocyte nuclear factor-1alpha, a transcription factor that has role in tissue-specific regulation of expression of several liver-specific genes. Mutations in the hepatocyte nuclear factor-1alpha(HNF1A)gene have been implicated in maturity onset diabetes of the young type 3 (MODY3), a form of diabetes mellitus found in 2–5% of patients with T2D.14 The AADM study also investigated linkage evidence for several T2D-related quantitative traits including intraocular pressure – a risk factor for retinopathy (5q22 and 14q22),15 renal function – a major consequence of diabetes,16 obesity (e.g. percent body fat on 2p13, fat mass on 2p13 and 5q14),17 serum lipids such as high density lipoprotein cholesterol (HDL-C, 7q31),18 and C-peptide plasma levels (10q23, 4p15).19 The loci found to influence C-peptide plasma levels harbor multiple T2D candidate genes (phosphatase and tensin homolog - PTEN, protein phosphatase 1, regulatory subunit 3C - PPP1R3C, insulin degrading enzyme -IDE, and peroxisome proliferator activated receptor gamma, coactivator 1 alpha- PPARGC1).19 To date, this remains the only published genome-wide linkage study of T2D in Africa.
Not a single genome-wide association study (GWAS) has been conducted on T2D among continental African populations to date. However, West African participants in the AADM study have been included as a replication cohort in several GWAS. For example, a collaboration between deCODE Genetics and AADM investigators successfully fine-mapped and showed the influence of recent selection in the TCF7L2 locus with respect to T2D risk (described in the next section).20, 21 Through similar collaborations with the deCODE Genetics group, the AADM study cohort was used to confirm replication of genome-wide significant associations of variants in Cyclin-dependent kinase 5 regulatory subunit associated protein 1-like 1 (CDKAL1) gene with susceptibility to T2D.22 The AADM cohort also played a key role in the observation that prostate cancer risk variants rs757210 and rs4430796 in the TCF2 gene(also known as HNF1 homeobox B, HNF1β) protect against T2D.23
Several candidate genes for T2D and associated traits have been investigated in African ancestry populations. The AADM study has identified association of the AGRP (agouti-related protein) −38C/T variant with decreased risk for T2D,24 a haplotype incalpain 10 (CAPN10) gene and increased risk for T2D,25 association between FTO(fat mass and obesity associated) gene single nucleotide polymorphisms (SNPs) and obesity,26 and a significant association between the 4a/b polymorphism of the nitric oxide synthase 3 (NOS3) gene (also known as eNOS) and diabetic retinopathy (OR=2.4, 95% CI=1.39–4.09).27 Investigation of variants in the promoter region of the adiponectin (ACDC) gene in a black South African population identified T2D-protective effect of the variant allele G11391A.28
In addition to findings from the AADM study, other published studies on T2D in African populations focused on evaluating the reproducibility of selected variants that have previously been found to confer risk for T2D in non-Africans. A study conducted in South African subjects of Zulu decent comprising 178 individuals with T2D and 200 normoglycemic controls assessed association between T2D and six variants known to be associated with T2D in European populations: rs7903146 (TCF7L2), rs12255372 (TCF7L2), rs1111875 (hematopoietically expressed homeobox- HHEX), rs1801282 (PPARG), rs5215 (potassium inwardly-rectifying channel, subfamily J, member 11 - KCNJ11), and rs9939609 (fat mass and obesity associated gene - FTO). Only rs7903146 (TCF7L2) showed statistically significant association, suggesting that the genetic architecture of T2D in some indigenous African populations may differ from that described in European populations. These findings should, however, be interpreted cautiously given the small sample size. Interestingly, rs1801282 (PPARG) and rs5215 (KCNJ11) had no variant alleles in the Zulu.29 The variant rs1801282 (PPARG Pro12Ala) was also absent in the Beninese,30 and Nigerians, and at low frequency in Kenyan Masai and Ethiopians compared to most non-African global populations (Table 1). Compared to the wild type Pro12 variant of the PPARG gene, the Ala12 variant has been observed to be associated with increased insulin sensitivity and lower body mass index, and a protective effect against T2D in several studies including a meta-analysis involving 32,849 cases and 47,456 controls from 60 studies.31 The PPARG receptor is implicated in regulation of lipid and glucose homeostasis, adipocyte differentiation, and fatty acid storage. The Pro12 allele is suspected to have a role in increased receptor activity resulting in a larger fat storage, making it useful for survival during periods of famine.30 Absence or relatively small frequency of the Ala12 variant, so called “unthrifty allele”, among African populations suggests that it may have been subject to negative selection to enhance survival in previous generations.32 Therefore, investigation of the relationship between this variant and T2D in various African populations is necessary to understand its metabolic consequences as urbanization, changes in diet (towards carbohydrate and fat-rich foods), and reduced physical activity are becoming more common across Africa.
Table 1.
Frequency of the PPARG Ala12 (rs1801282 G) Allele in African Populations Rotmi-Table 1
| Population (population group sampled) | Sample size | Allele frequency | Data Source [reference number] |
|---|---|---|---|
| Africans | |||
|
| |||
| South Africans (Zulu) | 378 | 0 | Pirie et al, 2010 [29] |
| Beninese (Berba and Bariba) | 97 | 0 | Scacchi et al, 2007 [30] |
| Nigerians (Yoruba) | 120 | 0 | HapMap-YRI (http://hapmap.ncbi.nlm.nih.gov/) |
| Kenyans (Masai) | 176 | 0.017 | HapMap-MKK (http://hapmap.ncbi.nlm.nih.gov/) |
| Ethiopians (Oromo and Amhara) | 166 | 0.036 | Scacchi et al, 2007 [30] |
|
| |||
| Non-Africans | |||
|
| |||
| Japanese (Japanese from Tokyo) | 172 | 0.035 | HapMap-JPT (http://hapmap.ncbi.nlm.nih.gov/) |
| Chinese (Han Chinese) | 82 | 0.073 | HapMap-CHB (http://hapmap.ncbi.nlm.nih.gov/) |
| Europeans (Utah residents of Northern and Western European ancestry) | 226 | 0.097 | HapMap-CEU (http://hapmap.ncbi.nlm.nih.gov/) |
Transcription Factor 7-like 2 (TCF7L2) Variants and T2D in Africa
The gene transcription factor 7-like 2 (TCF7L2), particularly the intronic SNP rs7903146, has been one of the strongest and most widely reproduced associations with T2D in several ethnic groups.33 This association was first reported by the deCODE group of investigators.20 Initially, it was unclear if it was present in other populations. A collaboration between deCODE and the AADM investigators conducted a replication and fine-mapping study of the TCF7L2T2D risk variantsin West African populations (621 cases and 448 controls) enrolled in the AADM study. This study confirmed and fine-mapped the association between rs7903146 T allele and increased risk for T2D (RR=1.45, 95% CI =1.19–1.77, p=0.00021), taking advantage of the weaker linkage disequilibrium (LD, a measure of degree of correlation among loci on the same chromosome) in the West African population. This study was one of the first empirical demonstrations of trans-ethnic fine-mapping of a disease locus, in this case exploiting the known weaker LD structure of African populations to refine association signals detected in other populations.21
The AADM study was also instrumental in identifying selective pressure episodes, environmental changes that are more favorable to the survival and reproduction of individuals that carry advantageous alleles, on TCF7L2, focusing on a haplotype that contains the TCF7L2 gene SNPs rs7903146 and rs10885406 (defined as HapA). The estimated time of the selective event coincided with the historical time of onset of agriculture in several ethnic groups including those of West Africa. Moreover, HapAhas shown associations with body mass index and levels of hunger-satiety hormones ghrelin and leptin in males implying the selective advantage of HapA may have been mediated through effects on energy metabolism.21 Such studies that combine genetic-epidemiology, anthropology, and history of a specific population are useful to detect genetic variants associated with a disease and understand the interactions among dietary, ecological and cultural history of a population, prevalence of risk genetic variants and disease burden. The mechanism through which TCF7L2 influences susceptibility to T2D has not been clearly understood. Present evidence has shown that the bipartite transcription factor cat/TCF7L2 regulates the gene proglucagon, which encodes the insulinotropic hormone glucagon-like peptide 1 that coordinates with insulin and plays a role in blood glucose homeostasis. Based on these relationships, TCF7L2 gene polymorphisms are speculated to influence T2D susceptibility by alteringglucagon-like peptide 1 levels.34 In human islets TCF7L2 mRNA levels are higher in T2D patients than controls, and the levels increase with the number of T2D risk alleles. As such, over-expression of TCF7L2 is speculated to influence susceptibility to T2D by inhibiting insulin secretion.35
Four SNPs in TCF7L2 gene have been well studied with regards to susceptibility to T2D: a C-to-T substitution at rs7903146 of intron 3 (IVS3C>T), a G-to-T substitution at rs12255372 of intron 4 (IVS4G>T), a T-to-C substitution at rs7901695 of intron 3 (IVS3T>C), and a G-to-C substitution at rs11196205 of intron 4 (IVS4G>C).36 A recent study characterized the global distribution of the four T2D associated TCF7L2 SNPs in individuals from more than 50 populations and global sub-regions. In the global comparison, African regions had high frequencies of rs7903146 T, rs12255372 T, and rs7901695 C alleles, and lower frequency of rs11196205 C. Within Africa, rs7903146 T allele distribution showed high variability, with southern, central, and North African populations showing high frequencies and eastern and western African populations exhibiting lower frequencies.37 Analysis of the HapMap dataset revealed that the frequency of rs7901695 C allele ranges from 0.01 to 0.49, with African-ancestry populations having up to 40-fold higher frequency.38 This finding of extensive genetic variations along with vast environmental contrast at the global level and within different regions in Africa may shed light on the role of genetics in the observed differences in the distribution of T2D across populations.
Recent Positive Selection and T2D Risk Loci in Africa
Selection pressures on the genome can alter allele frequencies at disease risk loci. A study that plotted the genetic risk map of T2D using the allelic frequency of 16 variants known to be associated with T2D in 51 populations suggested that Africans face the highest known genetic risk for T2D (http://geneworld.stanford.edu/hgdp.html). In parallel, a recent analysis performed in 938 individuals in 53 populations from the HGDP-CEPH human diversity panel (Human Genome Diversity Panel, Center for the Study of Human Polymorphisms, www.cephb.fr/en/cephdb/) using 30 variants associated with T2D in European-ancestry populations reported high degree of differentiation for T2D loci in sub-Saharan Africans and East Asians. Unlike East Asians for whom one locus (hematopoietically expressed homeobox, HHEX) is the most strongly differentiated in comparison with other groups, in sub-Saharan Africans no single locus stands out to explain the overall T2D trend. These findings provide evidence supporting the thinking that sub-Saharan Africans experienced recent positive natural selection at loci associated with T2D, and that many moderately selected loci or loci that have not been identified by genome-wide scans in non-African populations may contribute to the genetic risk for T2D in Africa.39
Genetics of CVD in Africa
Currently, little is known about the genetic basis of the major types of CVD in sub-Saharan Africa. Few candidate-gene studies have been conducted on coronary artery disease (CAD), a disorder that results from the complex interaction between multiple environmental and genetic factors (figure 1),40 in Egypt and Tunisia. A recent study that investigated the association between polymorphisms of the renin-angiotensin system (RAS) and CAD in Egyptians observed that the deletion (allele DD) at the 278-bp variant in the angiotensin I converting enzyme (ACE)gene, the TT genotype at rs699 (M235T) located on exon 2 of the angiotensinogen (AGT) gene, and the CC genotype of rs5186 (A1166C) in the ADP-ribosyltransferase 1 (ATR1) gene are associated with increased risks for both premature CAD and late-onset CAD.41 Other independent studies have also reported that the AGT TT genotype was significantly elevated in Egyptian CAD patients compared to controls.42, 43 In Tunisians, rs699 (M235T), and rs4762 (T174M) were also associated with CAD.44 The Glu298Asp polymorphism in exon 7 of the nitric oxide synthase 3 (eNOS) gene was more frequently observed in Egyptians with CAD than controls.43
Among known risk factors for CVD in Africa, hypertension is the most studied. There are a number of genetic studies on hypertension in Africa.45–49 A meta-analysis of genome-wide linkage scans for blood pressure (BP) in Nigerian and African American individuals reported suggestive associations with two loci: 2p14-p13.1 and 7p21.3-p15.3. Specifically, the second locus had not been previously observed, and the meta-analysis signal (Meta LOD 3.5) was primarily attributed to the Nigerian population sample(LOD 3.3), suggesting a novel quantitative trait locus for BP variation in individuals of African ancestry.50 Analysis of six SNPs in the genes angiotensinogen -AGT, cytochrome P450, family 4, subfamily F, polypeptide 12 (CYP4F12), solute carrier family 4 (sodium bicarbonate cotransporter) member 5 (SLC4A5), and Matrix metallopeptidase 3 (MMP3) in West African women from the Dogan region of Mali showed that rs8179526 (SLC4A5) was significantly associated with systolic BP (SBP). The encoded proteinregulates intracellular pH, and transports sodium and bicarbonate across cell membranes.51 The gain-of-function variant CLCNKB-T481S (chloride channel voltage-sensitive Kb with a variant threonine change to serine at position 481) reported to increase the entry of sodium chloride into the circulation thereby increasing extracellular fluid volume has been associated with essential hypertension. The prevalence of the mutant allele was found to be greater in Ghanaians (22%) compared to Europeans (12%).52 Follow-up studies performed in different ethnic populations failed to replicate the association of this gain-of-function variant with hypertension; however, it was replicated in Ghanaian males (OR=1.62, 95% CI 1.08–2.42, p=0.02 in the additive model; OR=3.29, 95% CI 1.17–9.20, p=0.024 in a recessive model). Absence of association in Ghanaian women suggests that the effect of the variant may be sex specific.53 Two SNPs that have shown suggestive associations with SBP (rs8039294, in synaptic vesicle glycoprotein 2B (SV2B) gene) and diastolic BP (rs1867226, in protein regulator of cytokinesis 1 (PRC1) gene) in African Americans from the Howard University Family Study (HUFS) population cohort were replicated in West Africans.54
In addition, the HUFS has contributed to our understanding of the genomic basis of BP regulation in African ancestry populations. Using a dense panel of over 800,000 SNPs on the Affymetrix SNP 6.0 chip in a sample of 1,017 African Americans, we identified multiple SNPs reaching genome-wide significance for SBP in or near the genes:postmeiotic segregation increased 1 - PMS1, solute carrier family 24 (sodium/potassium/calcium exchanger) member 4 - SLC24A4, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta polypeptide -YWHAZ, importin 7 - IPO7, and calcium channel, voltage-dependent, T type, alpha 1H subunit- CACNA1H. Two of these genes, SLC24A4 (a sodium/potassium/calcium exchanger) and CACNA1H (a voltage-dependent calcium channel), are potential candidate genes for BP regulation and the latter is a drug target for a class of calcium channel blockers. Some of these findings were replicated in a sample of West Africans.54 Also, the HUFS cohort has contributed to several consortia that have shed light on the genetic basis of BP control and risk of developing hypertension in African and non-African ancestry populations; these include the International Consortium for BP Genome-wide Association Studies conducted on over 200,000 persons,55 the Candidate Gene Association Resource African American cohorts for BP,56 and Candidate Gene Association Resource for Renal Function.57 Finally, the HUFS cohort participates in the Minority Health Genomics and Translational Research Bio-respositoryDatabase (MH-GRID) Network Infrastructure project funded by the NIH. The MH-GRID focuses on a sub-phenotype of primary hypertension that is common among African Americans and enriched for genetic determinants based on its early-onset, severity and associated kidney damage. A major deliverable of the MH-GRID project is to conduct whole exome sequencing on over 2,000 African Americans with the ultimate goal of shedding light on the patho-biology of BP regulation and the risk of developing hypertension in African ancestry populations.
A study that characterized the worldwide patterns of risk allele frequencies (RAF) of 158 SNPs associated with CVD and related quantitative traits using genotype data from the Human Genome Diversity Projectpanel revealed some interesting findings. First, in comparison with the rest of the world, the largest differences in RAF were observed among Africans. Second, the risk allele of SNP rs174570 in the fatty acid desaturase 2 (FADS2)gene that regulates unsaturation of fatty acids is fixed/nearly fixed in Africans (RAF=1) but absent in Pima (RAF=0).58 The degree of correlation of this glaring contrast in variant allele frequency with LDL-cholesterol among Africans is not clearly understood, but the finding is congruent with observations of low LDL cholesterol in the Pima Indians living in central and southern Arizona and Mexico.59 Third, these findings demonstrate possible roles of positive selection in explaining observed large differences in RAF among global populations; importantly, the variation in the prevalence of CVDs could partly be due to differential RAFs among different populations.58 Silander et al. evaluated the Human Genome Diversity Project populations from seven global sub-regions with respect to allele frequencies of SNPs on chromosome 9p21.3 that were previously reported to be associated with CAD and myocardial infarction.60, 61 They found that two of the investigated variants (rs10116277 and rs2383207 located in the intron of cyclin-dependent kinase inhibitor 2B antisense RNA 1 (CDKN2B-AS1/ANRIL) gene) were by far most common in populations from sub-Saharan Africa.62
Recently, analysis of the AADM cohort along with other population datasets revealed that the relationship between HDL-C and kidney function varies by ancestry with observed inverse relationship in African ancestry populations (West Africans and African Americans), and positive relationship in East Asians (Han Chinese) and European Americans.63 Interestingly, the inverse relationship between HDL and kidney function in African ancestry populations was strongest in the subset of individuals with the nephropathy risk genotype of SNP rs73885319 in the apolipoprotein L, 1 (APOL1)gene. Notably, higher HDL-C was associated with worse kidney function in those with the risk genotype, while no association was observed among those without this genotype. It is believed that the APOL1 risk variant rose to high frequency in West African populations presumably to provide protection against a deadly form of African sleeping sickness.63–65 In sum, the increasing incidence of CVD in Africa along with evidence of genetic variants that increase susceptibility to CVD signal the need for large scale genetic epidemiology studies on CVD in Africa.
Future of Genomics Research in T2D and CVD in Africa
The successful application of high-throughput genotyping, sequencing and computational capabilities in the identification of common genetic variants associated with T2D and CVD in non-African populations needs to be extended to Africans through collaboration and targeted grant funding. Given the well-documented genetic diversity of Africans and the key evolutionary contribution of Africa to the genomic landscape of all human populations, it is critically important to apply new genomic tools in African populations to shed light on the genetic architecture of human traits including T2D and CVD. The ongoing GWAS and sequencing activities in the AADM study and the newly funded Wellcome Trust Human Heredity and Health in Africa Initiative (H3Africa,http://h3africa.org/) T2D project in twelve sub-Saharan countries with the goal of enrolling 12,000 T2D cases and 12,000 population-based controls promise to contribute to our understanding of the genetic and non-genetic bases of T2D in African populations.
The genetic structure of African people presents both opportunities and challenges. On average, African populations display the highest genetic diversity and the lowest levels of linkage disequilibrium across human populations.66 While these characteristics are hugely important in the more efficient localization (fine mapping) of disease loci identified in non-African populations with longer LD, they pose considerable challenges at the discovery stages for several reasons including the need for larger sample sizes and better designed genotyping platforms. For example, it is well recognized that several of the current genotyping platforms display limited efficiency when used to identify risk variants for diseases in African populations.66 This is so because, the type and density of the genetic markers (SNPs) on these genotyping platforms are, for the most, not as informative in African compared to Europeans and South East Asians.67 In this regard, there is an urgent need for the development of more efficient genotyping tools that will facilitate genome-wide search for disease susceptibility loci across African populations. An important step to achieving this goal is a more robust understanding of the genetic diversity across the African continent through comprehensive sampling of multiple ethnic/geographic groups. Whole-genome sequencing and genotyping efforts of the 1000 Genomes Project and the African Genome Variation Project (http://www.sanger.ac.uk/research/initiatives/globalhealth/research/africangenome.html) are making significant contributions to this effort.
An open question – one that has not yet been adequately addressed – is whether and to what extent GWAS signals identified in non-African populations would transfer across diverse African populations. Initial evaluation of this question has produced mixed results. While some variants transfer consistently across African ethnic groups (e.g., rs7903146(TCF7L2) for T2D),20, 21 others did not at least in some African populations (e.g., rs1801282 (PPARG) for T2D in the Zulu).29 The observed mixed results are likely due to several factors including differences in underlying genetic architecture and inadequate sample size. Overall, systematic approaches that take into consideration these limitations are needed to adequately answer the question of how well loci identified in non-Africa populations transfer across African populations. For example, it may be necessary to conduct targeted re-sequencing of GWAS loci identified in Europeans and Asians in sufficiently powered samples of Africans. This strategy will create opportunities to identify both globally cosmopolitan and African specific alleles in disease etiology.
In addition to evaluating the transferability of known loci, genomic studies that aim to identify novel common and rare variants in African populations are needed. Sequence-based variant discovery is becoming the standard approach for identifying rare and low-frequency variants that may have large effect size. Currently, whole-exome and whole-genome sequencing studies are underway in a small number of African populations (Nigerians, Ghanaians, and Ugandans) with the goal of identifying both common and rare variants that may have implications for disease susceptibility. These sequencing projects are being conducted in population-base unrelated individuals as well as in families with multiply affected members and in persons displaying extreme phenotypic characteristics. These types of projects need to be implemented in more African populations to enable the establishment of much larger cohorts that are not feasible with the resources available to one or two investigators.
An important lesson from GWAS performed in non-African populations on common complex diseases is that the majority of the reported variants lie in introns or intergenic regions; hence, their primary function may be regulation of gene expression or splicing.68 In addition, SNPs associated with common traits have been found to be enriched for expression quantitative trait loci (eQTLs) – genetic variations affecting gene expression.69 Therefore, additional insights on the genetic architecture of diabetes and CVD in Africa can be leveraged through transcriptomics, quantification and expression profiling of all genes in specific tissues using microarrays and high throughput RNA sequencing technologies (RNAseq).70 Therefore, ongoing and future genomic studies in T2D and CVD in Africa could take advantage of these advances to discover novel loci and interpret previously identified GWAS signals.
How much do the genetic and epigenetic factors, already known or yet to be identified, interact? This is an important question that has not been adequately explored in most human populations. The vast contrast in the distribution of non-genetic factors (e.g., diet) and the huge genetic diversity of African people present researchers with wonderful opportunities to investigate epigenetic questions. Illustratively, the effect of PPARG gene on insulin sensitivity and T2D development has been reported to be modulated by dietary lipid intake. Increased rs1801282 G allele frequency of the PPARG gene was associated with less prevalence of T2D in populations where energy from lipids is higher than the recommended 30% of the total daily energy intake, but not in populations such as sub-Saharan Africans where energy from dietary fat is lower than 30%.30 Therefore, better characterization of non-genetic risk factors and investigation of their synergistic effects with putative genetic risk variants will be needed for the proper tailoring of clinical and public health intervention strategies.
Public Health and Clinical Relevance of Genomics Research in T2D and CVD in Africa
Genomic science promises to shed light on the knowledge gaps that exist in current understanding of the pathophysiology of and treatment approaches to diabetes and CVD in Africans and other human populations. For example, genomic research has provided novel insights into the molecular understanding of several types of diabetes especially the monogenic forms.71 Although most of the new genetic discoveries for T2D lie outside of the coding regions of genes, the hope is that these findings will provide information on the transcript regulation aspects of the maintenance of glucose homeostasis.71 It is also anticipated that genomic discoveries will provide insights into the identification of novel preventive targets as illustrated by the observation of a robust association between insulin secretion and variants in a gene encoding a zinc transporter protein (SLC30A8) illuminating the role of zinc in islet function and inspired public health interest in using dietary zinc to prevent T2D.72, 73 Genetic factors have also been found to contribute to individual variations in response to diabetes and CVD medications. Some success stories from pharmacogenomics studies in T2D include associations between KCNJ11 variants and glibenclamide treatment failure,74 TCF7L2 variants and sulfonylurea treatment response,75 and variants near the ataxia telangiectasia mutated (ATM) gene and glycemic response to metformin.76 One striking example of pharmacogenomics applications for CVD therapy is the testing for selected genetic variants to determine dosage of warfarin, the most widely prescribed oral anticoagulant. Among the variants routinely tested, rs1799853 in cytochrome P450, family 2, subfamily C, polypeptide 9(CYP2C9) gene, rs17708472 and rs9934438 in vitamin K epoxide reductase complex, subunit 1 (VKORC1) gene are either not polymorphic or have low variant allele frequencies in some African ancestry populations; this observation implies that the clinical utility of these variants in those African populations are likely limited and points to the need to have population-specific studies to better inform clinical decisions.77, 78 In the future, along with comprehensive data from socio-economical, demographic, and other biological sources, genome-based information promises to facilitate more precise clinical classification of the subtypes of T2D thus aiding better treatment decision and perhaps facilitating individual and population-level risk stratification and development of targeted prevention measures to those at varying risk of developing diabetes.
Summary
For several decades and understandably so, national and international attention was focused on decreasing the devastating health consequences of infectious diseases and under-nutrition in African communities. These efforts have met with varying degree of successes with the eradication/elimination of small pox, dracunculiasis (guinea worm disease), and poliomyelitis, and decreasing morbidity and mortality from malaria and HIV/AIDS. However, during those years of almost a singular focus on infectious diseases, the health burden of NCD including CVD, diabetes, and obesity grew to such an extent that most African communities now deal with the simultaneous health burden of communicable and NCD – the so called incomplete epidemiologic transition. Notably, the ongoing demographic changes, including urbanization, changes in physical activity, diet, political and economic instability continue to fuel the burden of NCD such as diabetes and CVD in Africa.
To begin to address the health burden of NCD in Africa communities, there is an urgent need for the design of sufficiently large-scale population-based studies that will systematically document the prevalence, incidence and risk factors of NCD in Africa. A comprehensive integration of genomics into these epidemiological studies will provide researchers with opportunities to understand the interactions between genetic and non-genetic factors in the pathophysiology of NCD. Examples of these types of studies include recently funded large-scale multi-center and multinational genetic epidemiology projects of T2D, cardiometabolic diseases, rheumatic heart disease and chronic kidney disease under the umbrella of the H3Africa that areco-funded by the U.S. National Institutes of Health and the U.K. Wellcome Trust. The H3Africa T2D project is designed to study the burden, etiology and spectrums of diabetes in Africa. The study is projected to enroll and examine 12,000 cases of T2D and 12,000 ethnically matched control from 12 centers across nine sub-Saharan African countries. The Wits-INDEPTH (International Network for the Demographic Evaluation of populations and their Health in low and middle income countries) H3Africa Collaborative Centre, with field sites in four sub-Saharan African countries, has been funded to study genomic and environmental risk factors for cardiometabolic diseases in Africa. These studies will provide opportunities to investigate the co-evolution of infectious and non-infectious diseases within the unique demographic, historic and cultural experiences of African people with the ultimate goal of identifying factors that influence disease development and progression, and response to therapy. These genomic projects are also expected to build research capacity that will facilitate future pan-African biomedical research activities.
Acknowledgments
This work was supported by the Intramural Research Program of the Center for Research on Genomics and Global Health. The Center for Research on Genomics and Global Health is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology, and the Office of the Director at the National Institutes of Health (1ZIAHG200362-02).
Abbreviations and Acronyms
- BP
Blood pressure
- CAD
Coronary artery disease
- CVD
Cardiovascular diseases
- GWAS
Genome-wide association study/studies
- H3Africa
Human Heredity and Health in Africa Initiative
- HapMap
International project that aims to catalogue genetic variations in human beings
- HDL-c
High density lipoprotein cholesterol
- HUFS
The Howard University Family Study
- LD
Linkage disequilibrium - a measure of the degree of correlation between loci on the same chromosome
- LOD
Logarithm of the odds (LOD) is a statistical test commonly used in genetic linkage analysis. It compares the likelihood of obtaining the test data if two loci are indeed linked, to the likelihood of observing the same data purely by chance
- MH-GRID
Minority Health Genomics and Translational Research Bio-respositoryDatabase
- NCD
Non-communicable diseases
- NFH
Negative family history
- OR
Odds ratio
- PFH
Positive family history
- PPARG
Peroxisome proliferator-activated receptor gamma (gene)
- RAF
Risk allele frequency
- SBP
Systolic blood pressure
- SNP
Single nucleotide polymorphism
- SSA
Sub-Saharan Africa
- T2D
Type 2 diabetes
- TCF7L2
Transcription factor 7-like 2 (gene)
Footnotes
Statement of Conflict of Interest
All authors declare that there are no conflicts of interest. The contents of this review are the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Fasil Tekola-Ayele, Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, 20892.
Adebowale A. Adeyemo, Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, 20892.
Charles N. Rotimi, Center for Research on Genomics and Global Health, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, 20892.
References
- 1.Wild S, Roglic G, Green A, et al. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(5):1047–53. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Mensah GA. Ischaemic heart disease in Africa. Heart. 2008;94(7):836–43. doi: 10.1136/hrt.2007.136523. [DOI] [PubMed] [Google Scholar]
- 3.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson FL. Ethnogenetic layering (EL): an alternative to the traditional race model in human variation and health disparity studies. Ann Hum Biol. 2008;35(2):121–44. doi: 10.1080/03014460801941752. [DOI] [PubMed] [Google Scholar]
- 5.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49(12):2201–7. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 6.Hemminki K, Li X, Sundquist K, et al. Familial risks for type 2 diabetes in Sweden. Diabetes Care. 2010;33(2):293–7. doi: 10.2337/dc09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfine AB, Bouche C, Parker RA, et al. Insulin resistance is a poor predictor of type 2 diabetes in individuals with no family history of disease. Proc Natl Acad Sci U S A. 2003;100(5):2724–9. doi: 10.1073/pnas.0438009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotimi C, Cooper R, Cao G, et al. Familial aggregation of cardiovascular diseases in African-American pedigrees. Genet Epidemiol. 1994;11(5):397–407. doi: 10.1002/gepi.1370110502. [DOI] [PubMed] [Google Scholar]
- 9.Mbanya JC, Pani LN, Mbanya DN, et al. Reduced insulin secretion in offspring of African type 2 diabetic parents. Diabetes Care. 2000;23(12):1761–5. doi: 10.2337/diacare.23.12.1761. [DOI] [PubMed] [Google Scholar]
- 10.Adeleye JO, Abbiyesuku FM. Glucose and insulin responses in offspring of Nigerian Type 2 diabetics. Afr J Med Med Sci. 2002;31(3):253–7. [PubMed] [Google Scholar]
- 11.Erasmus RT, Blanco Blanco E, Okesina AB, et al. Importance of family history in type 2 black South African diabetic patients. Postgrad Med J. 2001;77(907):323–5. doi: 10.1136/pmj.77.907.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rotimi CN, Dunston GM, Berg K, et al. In search of susceptibility genes for type 2 diabetes in West Africa: the design and results of the first phase of the AADM study. Ann Epidemiol. 2001;11(1):51–8. doi: 10.1016/s1047-2797(00)00180-0. [DOI] [PubMed] [Google Scholar]
- 13.Rotimi CN, Chen G, Adeyemo AA, et al. A genome-wide search for type 2 diabetes susceptibility genes in West Africans: the Africa America Diabetes Mellitus (AADM) Study. Diabetes. 2004;53(3):838–41. doi: 10.2337/diabetes.53.3.838. [DOI] [PubMed] [Google Scholar]
- 14.Yamagata K, Oda N, Kaisaki PJ, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384(6608):455–8. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 15.Rotimi CN, Chen G, Adeyemo AA, et al. Genomewide scan and fine mapping of quantitative trait loci for intraocular pressure on 5q and 14q in West Africans. Invest Ophthalmol Vis Sci. 2006;47(8):3262–7. doi: 10.1167/iovs.05-1537. [DOI] [PubMed] [Google Scholar]
- 16.Chen G, Adeyemo AA, Zhou J, et al. A genome-wide search for linkage to renal function phenotypes in West Africans with type 2 diabetes. Am J Kidney Dis. 2007;49(3):394–400. doi: 10.1053/j.ajkd.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Chen G, Adeyemo AA, Johnson T, et al. A genome-wide scan for quantitative trait loci linked to obesity phenotypes among West Africans. Int J Obes (Lond) 2005;29(3):255–9. doi: 10.1038/sj.ijo.0802873. [DOI] [PubMed] [Google Scholar]
- 18.Adeyemo AA, Johnson T, Acheampong J, et al. A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis. 2005;181(2):389–97. doi: 10.1016/j.atherosclerosis.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Adeyemo A, Zhou J, et al. Genome-wide search for susceptibility genes to type 2 diabetes in West Africans: potential role of C-peptide. Diabetes Res Clin Pract. 2007;78(3):e1–6. doi: 10.1016/j.diabres.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–3. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 21.Helgason A, Palsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39(2):218–25. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 22.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39(6):770–5. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 23.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39(8):977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 24.Bonilla C, Panguluri RK, Taliaferro-Smith L, et al. Agouti-related protein promoter variant associated with leanness and decreased risk for diabetes in West Africans. Int J Obes (Lond) 2006;30(4):715–21. doi: 10.1038/sj.ijo.0803047. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Kittles R, Zhou J, et al. Calpain-10 gene polymorphisms and type 2 diabetes in West Africans: the Africa America Diabetes Mellitus (AADM) Study. Ann Epidemiol. 2005;15(2):153–9. doi: 10.1016/j.annepidem.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Adeyemo A, Chen G, Zhou J, et al. FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes. 2010;59(6):1549–54. doi: 10.2337/db09-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Huang H, Zhou J, et al. Polymorphism of the endothelial nitric oxide synthase gene is associated with diabetic retinopathy in a cohort of West Africans. Mol Vis. 2007;13:2142–7. [PubMed] [Google Scholar]
- 28.Olckers A, Towers GW, van der Merwe A, et al. Protective effect against type 2 diabetes mellitus identified within the ACDC gene in a black South African diabetic cohort. Metabolism. 2007;56(5):587–92. doi: 10.1016/j.metabol.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Pirie FJ, Motala AA, Pegoraro RJ, et al. Variants in PPARG, KCNJ11, TCF7L2, FTO, and HHEX genes in South African subjects of Zulu descent with type 2 diabetes. African J Diabetes Med. 2010;18:12–16. [Google Scholar]
- 30.Scacchi R, Pinto A, Rickards O, et al. An analysis of peroxisome proliferator-activated receptor gamma (PPAR-gamma 2) Pro12Ala polymorphism distribution and prevalence of type 2 diabetes mellitus (T2DM) in world populations in relation to dietary habits. Nutr Metab Cardiovasc Dis. 2007;17(9):632–41. doi: 10.1016/j.numecd.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Gouda HN, Sagoo GS, Harding AH, et al. The association between the peroxisome proliferator-activated receptor-gamma2 (PPARG2) Pro12Ala gene variant and type 2 diabetes mellitus: a HuGE review and meta-analysis. Am J Epidemiol. 2010;171(6):645–55. doi: 10.1093/aje/kwp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Narvaez E. Is the Ala12 variant of the PPARG gene an “unthrifty allele”? J Med Genet. 2005;42(7):547–50. doi: 10.1136/jmg.2004.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cauchi S, El Achhab Y, Choquet H, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med (Berl) 2007;85(7):777–82. doi: 10.1007/s00109-007-0203-4. [DOI] [PubMed] [Google Scholar]
- 34.Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem. 2005;280(2):1457–64. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- 35.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117(8):2155–63. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong Y, Lin Y, Zhang Y, et al. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet. 2009;10:15. doi: 10.1186/1471-2350-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guinan KJ. Worldwide distribution of type II diabetes-associated TCF7L2 SNPs: evidence for stratification in Europe. Biochem Genet. 2012;50(3–4):159–79. doi: 10.1007/s10528-011-9456-2. [DOI] [PubMed] [Google Scholar]
- 38.Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics. 2010;13(2):72–9. doi: 10.1159/000218711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klimentidis YC, Abrams M, Wang J, et al. Natural selection at genomic regions associated with obesity and type-2 diabetes: East Asians and sub-Saharan Africans exhibit high levels of differentiation at type-2 diabetes regions. Hum Genet. 2011;129(4):407–18. doi: 10.1007/s00439-010-0935-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection E and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143–421. [PubMed] [Google Scholar]
- 41.Abd El-Aziz TA, Hussein YM, Mohamed RH, et al. Renin-angiotensin system genes polymorphism in Egyptians with premature coronary artery disease. Gene. 2012;498(2):270–5. doi: 10.1016/j.gene.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 42.Shaker OG, Eldemellawy HH, Kassem HH. Angiotensinogen gene (M235T) polymorphism and coronary artery disease in the Egyptian population. A genetic association study. Heart Mirror J. 2009;3(2):86–91. [Google Scholar]
- 43.Motawi T, Shaker O, Taha M, et al. Endothelial nitric oxide synthase and angiotensinogen gene polymorphism in coronary artery diseases in Egypt. Angiology. 2011;62(2):191–7. doi: 10.1177/0003319710373094. [DOI] [PubMed] [Google Scholar]
- 44.Abboud N, Ghazouani L, Kaabi B, et al. Evaluation of the contribution of renin angiotensin system polymorphisms to the risk of coronary artery disease among Tunisians. Genet Test Mol Biomarkers. 2010;14(5):661–6. doi: 10.1089/gtmb.2010.0070. [DOI] [PubMed] [Google Scholar]
- 45.Cooper RS, Luke A, Zhu X, et al. Genome scan among Nigerians linking blood pressure to chromosomes 2, 3, and 19. Hypertension. 2002;40(5):629–33. doi: 10.1161/01.hyp.0000035708.02789.39. [DOI] [PubMed] [Google Scholar]
- 46.Adeyemo A, Luke A, Wu X, et al. Genetic effects on blood pressure localized to chromosomes 6 and 7. J Hypertens. 2005;23(7):1367–73. doi: 10.1097/01.hjh.0000173519.06353.8b. [DOI] [PubMed] [Google Scholar]
- 47.Tayo BO, Luke A, Zhu X, et al. Association of regions on chromosomes 6 and 7 with blood pressure in Nigerian families. Circ Cardiovasc Genet. 2009;2(1):38–45. doi: 10.1161/CIRCGENETICS.108.817064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reder NP, Tayo BO, Salako B, et al. Adrenergic alpha-1 pathway is associated with hypertension among Nigerians in a pathway-focused analysis. PLoS One. 2012;7(5):e37145. doi: 10.1371/journal.pone.0037145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouzekri N, Zhu X, Jiang Y, et al. Angiotensin I-converting enzyme polymorphisms, ACE level and blood pressure among Nigerians, Jamaicans and African-Americans. Eur J Hum Genet. 2004;12(6):460–8. doi: 10.1038/sj.ejhg.5201166. [DOI] [PubMed] [Google Scholar]
- 50.Rice T, Cooper RS, Wu X, et al. Meta-analysis of genome-wide scans for blood pressure in African American and Nigerian samples. The National Heart, Lung, and Blood Institute GeneLink Project. Am J Hypertens. 2006;19(3):270–4. doi: 10.1016/j.amjhyper.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Taylor JY, Sampson D, Taylor AD, et al. Genetic and BMI risks for predicting blood pressure in three generations of West African Dogon women. Biol Res Nurs. 2013;15(1):105–11. doi: 10.1177/1099800411419026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeck N, Waldegger S, Lampert A, et al. Activating mutation of the renal epithelial chloride channel ClC-Kb predisposing to hypertension. Hypertension. 2004;43(6):1175–81. doi: 10.1161/01.HYP.0000129824.12959.f0. [DOI] [PubMed] [Google Scholar]
- 53.Sile S, Velez DR, Gillani NB, et al. CLCNKB-T481S and essential hypertension in a Ghanaian population. J Hypertens. 2009;27(2):298–304. doi: 10.1097/hjh.0b013e3283140c9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adeyemo A, Gerry N, Chen G, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5(7):e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fox ER, Young JH, Li Y, et al. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Human molecular genetics. 2011;20(11):2273–84. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu CT, Garnaas MK, Tin A, et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet. 2011;7(9):e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding K, Kullo IJ. Geographic differences in allele frequencies of susceptibility SNPs for cardiovascular disease. BMC Med Genet. 2011;12:55. doi: 10.1186/1471-2350-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howard BV, Davis MP, Pettitt DJ, et al. Plasma and lipoprotein cholesterol and triglyceride concentrations in the Pima Indians: distributions differing from those of Caucasians. Circulation. 1983;68(4):714–24. doi: 10.1161/01.cir.68.4.714. [DOI] [PubMed] [Google Scholar]
- 60.Shen GQ, Rao S, Martinelli N, et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008;53(2):144–50. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 61.Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Human molecular genetics. 2008;17(6):806–14. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 62.Silander K, Tang H, Myles S, et al. Worldwide patterns of haplotype diversity at 9p21.3, a locus associated with type 2 diabetes and coronary heart disease. Genome Med. 2009;1(5):51. doi: 10.1186/gm51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bentley AR, Doumatey AP, Chen G, et al. Variation in APOL1 Contributes to Ancestry-Level Differences in HDLc-Kidney Function Association. Int J Nephrol. 2012;2012:748984. doi: 10.1155/2012/748984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–5. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bentley AR, Rotimi C. Interethnic variation in lipid profiles: implications for underidentification of African – Americans at risk for metabolic disorders. Expert Rev Endocrinol Metab. 2012;7(6):659–67. doi: 10.1586/eem.12.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–33. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363(2):166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 69.Nicolae DL, Gamazon E, Zhang W, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pedrotty DM, Morley MP, Cappola TP. Transcriptomic biomarkers of cardiovascular disease. Prog Cardiovasc Dis. 2012;55(1):64–9. doi: 10.1016/j.pcad.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCarthy MI. Genomics, type 2 diabetes, and obesity. The New England journal of medicine. 2010;363(24):2339–50. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 72.Sun Q, van Dam RM, Willett WC, et al. Prospective study of zinc intake and risk of type 2 diabetes in women. Diabetes Care. 2009;32(4):629–34. doi: 10.2337/dc08-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Travers ME, McCarthy MI. Type 2 diabetes and obesity: genomics and the clinic. Hum Genet. 2011;130(1):41–58. doi: 10.1007/s00439-011-1023-8. [DOI] [PubMed] [Google Scholar]
- 74.Sesti G, Laratta E, Cardellini M, et al. The E23K variant of KCNJ11 encoding the pancreatic beta-cell adenosine 5′-triphosphate-sensitive potassium channel subunit Kir6.2 is associated with an increased risk of secondary failure to sulfonylurea in patients with type 2 diabetes. J Clin Endocrinol Metab. 2006;91(6):2334–9. doi: 10.1210/jc.2005-2323. [DOI] [PubMed] [Google Scholar]
- 75.Pearson ER, Donnelly LA, Kimber C, et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56(8):2178–82. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 76.Zhou K, Bellenguez C, Spencer CC, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet. 2011;43(2):117–20. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dandara C, Lombard Z, Du Plooy I, et al. Genetic variants in CYP (-1A2, -2C9, -2C19, -3A4 and -3A5), VKORC1 and ABCB1 genes in a black South African population: a window into diversity. Pharmacogenomics. 2011;12(12):1663–70. doi: 10.2217/pgs.11.106. [DOI] [PubMed] [Google Scholar]
- 78.Ramos E, Doumatey A, Elkahloun AG, et al. Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenomics J. 2013 doi: 10.1038/tpj.2013.24. [DOI] [PubMed] [Google Scholar]


