Abstract
Proper coordination of cholesterol biosynthesis and trafficking is essential to human health. The sterol regulatory element binding proteins (SREBPs) are key transcription regulators of genes involved in cholesterol biosynthesis/uptake. We show here that microRNAs (miR-33a/b) embedded within introns of the SREBP genes target the ATP-binding cassette transporter A1 (ABCA1), an important regulator of high-density lipoprotein (HDL) synthesis and reverse cholesterol transport, for post-transcriptional repression. Antisense inhibition of miR-33 in cell lines causes upregulation of ABCA1 expression and increased cholesterol efflux, and injection of mice on a western-type diet with locked nucleic acid (LNA)-antisense oligonucleotides results in elevated plasma HDL. Collectively, our findings indicate that miR-33 acts in concert with the SREBP host genes to control cholesterol homeostasis, and suggest that miR-33 may represent a therapeutic target for ameliorating cardiometabolic diseases.
Results
Cholesterol and other lipids play key roles in many physiological processes in metazoans, and aberrant cholesterol/lipid homeostasis has been linked to a number of diseases, including atherosclerosis, metabolic syndrome, and type II diabetes, underscoring the importance of understanding fully how cholesterol/lipid homeostasis is regulated (1, 2).
The sterol regulatory element-binding protein (SREBP) transcription factors are critical regulators of cholesterol/lipid homeostasis by controlling the expression of many cholesterogenic and lipogenic genes (e.g. LDL receptor, HMG-CoA reductase, fatty acid synthase) (3-6). While much is known about SREBP-dependent transcriptional mechanisms governing the biosynthesis and uptake of cholesterol and fatty acids, it is unclear how this regulatory circuit is coordinated with opposing pathways mediating cholesterol/lipid efflux or degradation to achieve appropriate cholesterol/lipid levels to satisfy cellular and physiological demands.
In addition to classical transcription regulators, a class of non-coding RNAs termed microRNAs (miRNAs) has emerged as important modulators of numerous cellular processes that impact organism growth, development, homeostasis and disease (7-12). Indeed, recent studies have revealed that a liver-restricted miRNA, miR-122, regulates cholesterol/lipid metabolism in mice and non-human primates, although the mechanism remains unclear (13-16). Data from these studies not only emphasize the important roles played by miRNAs in normal physiology, but also point to the feasibility of antisense-based therapeutic targeting of miRNAs to treat human disease.
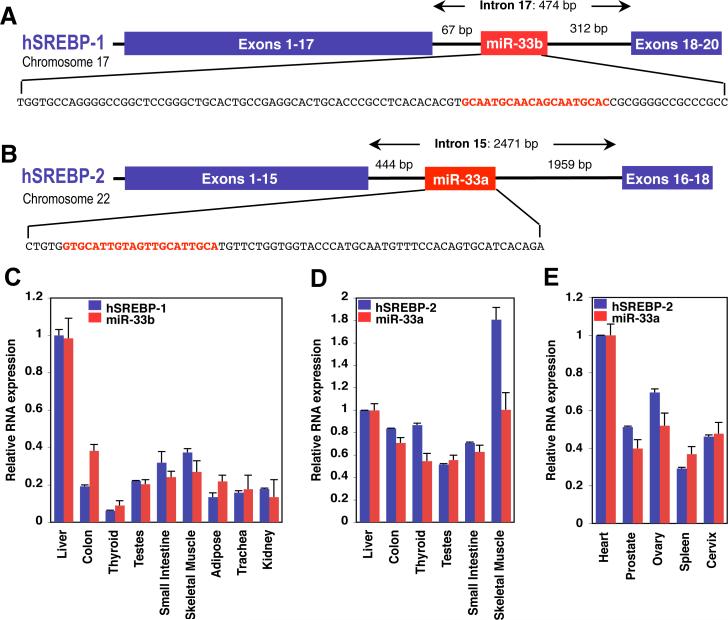
During our investigations of gene regulation by SREBPs (17), we noted the intriguing presence of a highly conserved miRNA family, miR-33, within intronic sequences of the SREBP genes in organisms from Drosophila to humans (Fig. 1A, B, and fig. S1). Two isoforms of miR-33 exist in humans: miR-33b, which is present in intron 17 of the SREBP-1 gene on chromosome 17, and miR-33a, which is located in intron 15 of the SREBP-2 gene on chromosome 22 (Fig. 1A, B). In mice, however, there is only one miR-33 isoform (which is conserved with human miR-33a), located within intron 15 of the mouse SREBP-2 gene, whereas intron 17 in the mouse SREBP-1 gene lacks sequence homology to the human intronic sequences harboring miR-33b (fig. S1).
Figure 1.
SREBPs are host genes to conserved intronic miRNAs, miR-33a/b, which are co-expressed with SREBPs. Human SREBP-1 (A) and SREBP-2 (B) genes harbor related intronic miRNAs (miR-33b and miR-33a, respectively). The sequences encoding the pri-miRNAs are shown, with the mature miRNA sequences highlighted in red. C-E Expression profile of miR-33a/b and SREBP host genes in selected human tissues. Error bars represent experimental S.D.
Similar to miR-33, many mammalian miRNAs are located within introns of protein-coding genes rather than in their own unique transcription units (18). Intronic miRNAs are typically coordinately expressed and processed with the pre-mRNA in which they reside (19-21). Accordingly, the mature forms of miR-33a/b appear to be co-expressed with the SREBP host genes in a number of human and mouse tissues examined (Fig. 1C-D and figs. S2-4).
We wished to determine the potential function(s) of miR-33a/b and whether they exhibit functional association with the SREBP host genes. MiRNAs have been shown to target mRNAs for post-transcriptional repression by base-pairing with mRNA sequences typically located in the 3’ untranslated regions (3’UTRs) and causing translational inhibition or mRNA cleavage (22). We initially employed commonly used bioinformatics tools that predict miRNA targets largely based on the ability of the miRNA sequence to undergo specific base-pairing with its putative 3’UTR target, known as “seed pairing” (22). The most significant predicted conserved target for miR-33a/b among vertebrates is the ATP-binding cassette A1 (ABCA1) cholesterol transporter (table S1 and fig. S5A). ABCA1 is a key mediator of intracellular cholesterol efflux from liver to apoA-I for generation of high-density lipoprotein (HDL) (1, 23, 24). It is also important for HDL-cholesterol trafficking from peripheral tissues (e.g. macrophages) by the reverse cholesterol transport (RCT) pathway back to the liver for processing and excretion into bile and feces (25).
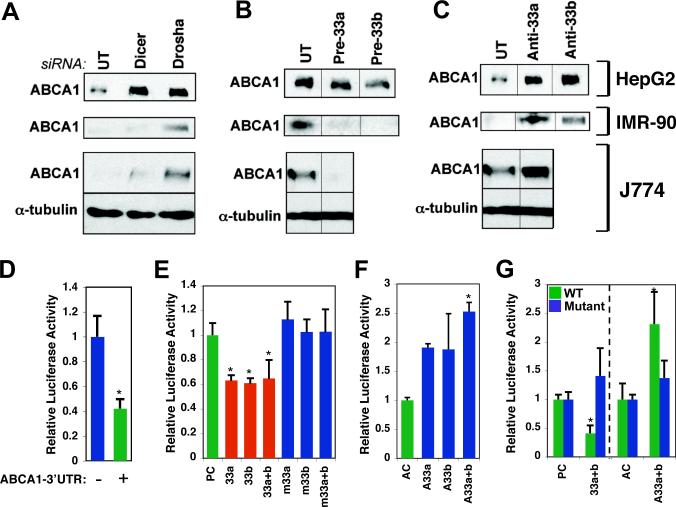
As SREBPs promote cholesterol uptake and synthesis through transactivation of the LDL receptor and cholesterol biosynthesis genes, we hypothesized that miR-33-mediated inhibition of ABCA1 and cholesterol efflux could potentially act in cooperation with SREBPs to boost intracellular cholesterol levels. To begin to test this hypothesis, we first carried out RNAi-mediated knockdown of components of the miRNA biogenesis pathway to determine whether ABCA1 protein expression is regulated by miRNAs. Indeed, transfection with siRNAs directed against the Drosha and Dicer miRNA processing enzymes resulted in a marked increase in ABCA1 protein expression in several human and mouse cell lines, including human HepG2 liver carcinoma cells, IMR-90 normal human fibroblasts, and the mouse macrophage cell line J774 (Fig. 2A). These results are thus consistent with regulation of ABCA1 by miRNAs. To evaluate the specific effect of miR-33 on ABCA1 expression, we transfected cells with synthetic miR-33 precursor oligonucleotides (Pre-miR-33a/b) to increase the intracellular levels of miR-33a and b, respectively. These studies clearly show that ABCA1 expression is repressed by excess miR-33, especially in mouse J774 macrophages and the human IMR-90 fibroblasts (Fig. 2B). To further determine whether miR-33a/b are specifically involved in regulating ABCA1 expression, we transfected the three cell lines with antisense oligonucleotides directed against miR-33a and/or b (Anti-miR-33a/b). Introduction of Anti-miR-33a/b indeed resulted in strongly increased levels of ABCA1 in the three cell lines (Fig. 2C). Taken together, these findings demonstrate that ABCA1 protein expression is regulated by miR-33a/b.
Figure 2.
MiR-33 regulates the cholesterol transporter ABCA1. A. Transfection of siRNA against Drosha and Dicer results in increased protein levels of ABCA1 in human HepG2 hepatoma cells (top row), human IMR-90 fibroblasts (middle row), and the mouse macrophage cell line J774 (bottom row). B. Introduction of miR-33a or miR-33b precursors into the three cell lines represses ABCA1 protein levels. C. MiR-33a/b antisense oligonucleotides (A33a/b) increase ABCA1 levels. E. Insertion of the ABCA1 3’UTR fragment into a Luciferase reporter results in decreased Luciferase expression in HEK293 cells. F. Co-transfection with wild-type, but not mutated, miR-33a/b precursors causes further repression of the Luciferase-ABCA1 3’UTR reporter. A scrambled sequence was used as precursor control (PC). G. MiR-33a/b antisense oligonucleotides result in de-repression of the Luciferase-ABCA1 3’UTR reporter. A scrambled sequence was used as antisense control (AC). Error bars represent S.D. * denotes p<0.05.
To determine whether miR-33a/b specifically target ABCA1 post-transcriptionally through its 3’UTR, we fused a fragment of the human ABCA1 3’UTR harboring the predicted miR-33 target sequences to a Luciferase reporter plasmid (fig. S5B). As shown in Fig. 2D, the Luciferase-ABCA1 3’UTR reporter was expressed at markedly lower levels in HEK293 cells as compared with the Luciferase vector without insert. Introduction of excess wild-type human miR-33a/b precursor oligonucleotides resulted in further repression of the wild-type ABCA1 3’UTR reporter (Fig. 2E). By contrast, miR-33a/b precursors mutated in the seed base-pairing sequence had no effect, as compared with precursor control (Fig. 2E and fig. S5C). Together, these data show that miR-33a/b are capable of repressing expression from the ABCA1 3’UTR. Importantly, co-transfection with miR-33a/b antisense oligonucleotides caused complete de-repression of the Luciferase-ABCA1 3’UTR reporter, consistent with regulation of ABCA1 3’UTR by endogenous miR-33a/b (Fig. 2F). Finally, mutations in the seed base-pairing sequences of the predicted miR-33 target sites in the ABCA1 3’UTR reporter abolished regulation by miR-33a/b precursors as well as Anti-miR-33a/b (Fig. 2G). Together, these results suggest that miR-33a/b inhibit expression of human ABCA1 by targeting the ABCA1 3’UTR for translation repression or mRNA degradation.
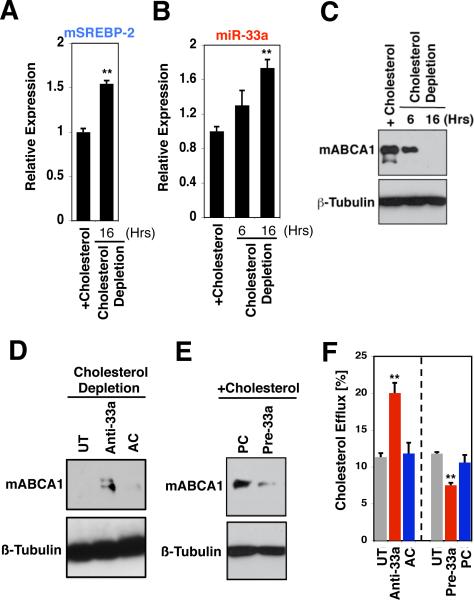
Reverse cholesterol transport from atherogenic macrophages/foam cells is of clinical importance because of the prominent role played by foam cells in cardiovascular disease, and a number of studies have highlighted the biomedical implications of ABCA1-dependent cholesterol efflux and HDL biogenesis for atherosclerosis (23, 26-28). We have investigated the potential biomedical and physiological relevance of the miR-33/SREBP/ABCA1 cholesterol regulatory circuit using the mouse J774 macrophage model (29). SREBPs are regulated in a classic negative feedback manner by cholesterol (3, 5), and we find that depletion of cholesterol by lovastatin/β-cyclodextrin treatment results in increased expression of both miR-33a and the mSREBP-2 host gene in J774 macrophages, with a concomitant decrease in ABCA1 protein expression (Fig. 3A-C). The strong suppression of ABCA1 protein levels in response to cholesterol depletion is at least partially reversed by miR-33a antisense oligonucleotides (Fig. 3D), consistent with the notion that miR-33a mediates cholesterol-regulated post-transcriptional control of ABCA1 levels in mouse macrophages. We also found that the high protein levels of ABCA1 observed in J774 macrophages cultured in the presence of serum/cholesterol are substantially suppressed by additional miR-33a precursor (Fig. 3E), in keeping with miR-33 regulation of ABCA1. By contrast with the apparent regulation of ABCA1 levels in J774 cells by miR-33a, expression of the intracellular cholesterol transporter NPC1 is unaffected by miR-33a precursor or antisense treatment (fig. S6), demonstrating specific regulation of ABCA1 by miR-33a. Our data together show that mouse miR-33a and the SREBP-2 host gene are co-regulated by cholesterol, and suggest the existence of a reciprocal regulatory network of SREBP, miR-33, ABCA1, and cholesterol in macrophages.
Figure 3.
Reciprocal regulation by cholesterol and SREBP/miR-33. Lovastatin/β-cyclodextrin-mediated depletion of cholesterol in J774 mouse macrophages results in increased expression of (A) SREBP-2 and (B) miR-33a, and (C) in decreased ABCA1 protein levels. D. Cholesterol-depleted J774 mouse macrophages exhibit upregulation of ABCA1 protein in response to miR-33a antisense inhibition. E. Transfection of J774 macrophages cultured in the presence of serum/cholesterol with miR-33a precursor results in decreased levels of ABCA1. F. Introduction of miR-33a antisense oligonucleotides into J774 macrophages loaded with radio-labeled cholesterol results in increased cholesterol efflux, whereas miR-33a precursors inhibit cholesterol efflux. Error bars represent S.D. ** denotes p<0.01.
Next, we examined the effects of miR-33 manipulations on cholesterol efflux from J774 macrophages. Cells were first labeled with [3H]-cholesterol and then transfected with miR-33a precursor, miR-33a antisense, or control oligonucleotides, followed by measurements of the level of [3H]-cholesterol efflux to serum/apoA-I. As shown in Fig. 3F, introduction of miR-33a antisense oligonucleotides into mouse macrophages caused a marked (two-fold) increase in [3H]-cholesterol efflux, whereas miR-33a precursor treatment led to a significant decrease in [3H]-cholesterol efflux, as compared with control oligonucleotides (AC/PC) or mock transfected cells (UT). These findings demonstrate that miR-33 post-transcriptional repression of ABCA1 results in retention of intracellular cholesterol in macrophages, and are in keeping with our hypothesis that miR-33 regulates cholesterol trafficking by controlling ABCA1 levels.
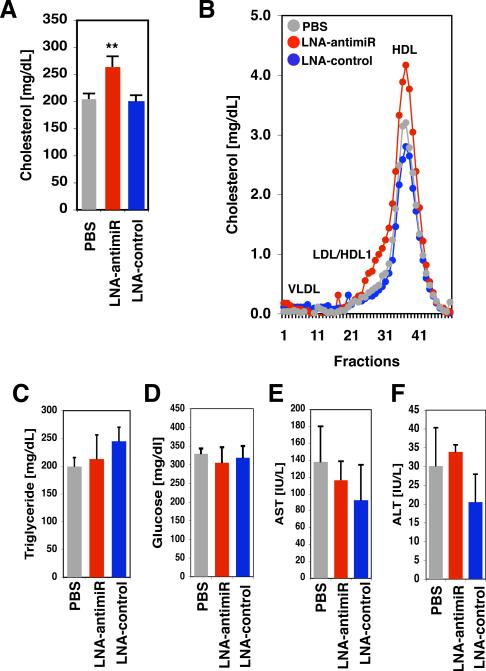
Based on our in vitro findings we hypothesized that manipulation of ABCA1 levels by miR-33 antisense approaches might lead to augmented HDL-cholesterol levels in a mammalian in vivo model. Previous studies in mice and non-human primates have supported the feasibility of locked nucleic acid (LNA) antisense approaches to modulate miRNA pathways with important physiological consequences in vivo (13, 14). We therefore performed tail vein injections of mice on a western-type diet with PBS (vehicle), LNA-miR-33a antisense, or LNA-control oligonucleotides. Following three injections over 5 days, mice were sacrificed and serum was collected. We find that plasma HDL-cholesterol concentrations were significantly increased in LNA-miR-33a antisense-treated animals as compared to LNA-control-treated mice (Fig. 4A, B and table S2). By contrast, there were no significant effects on plasma concentrations of LDL-cholesterol, triglycerides, or glucose levels (Fig. 4C-F and table S2). In these experiments, there was also no apparent hepatotoxicity (plasma AST/ALT) (Fig. 4E, F and table S2). The observed elevation in plasma HDL-cholesterol in response to LNA-miR-33a antisense treatment is consistent with regulation of ABCA1-dependent cholesterol efflux by miR-33a in vivo. Hepatic ABCA1 is a major contributor to HDL production in normal mice (23, 24), however we find only modest effects of miR-33a LNA-antimiR on hepatic ABCA1 mRNA/protein as compared with LNA-control (fig. S7). Based on these findings, we speculate that liver might not be the sole target tissue in mediating the effect of miR-33a LNA-antimiR on HDL production, with possible additional contribution of miR-33a regulation of reverse cholesterol transport from extrahepatic tissues/cells.
Figure 4.
Regulation of HDL by miR-33a in vivo. A. Tail vein injection of mice fed a western-type diet with Locked Nucleic Acid (LNA)-antisense oligonucleotides directed against mouse miR-33a results in elevated total serum cholesterol. B. FPLC analysis of pooled serum from 5 mice on a western-type diet treated revealed an increase in the magnitude of the HDL-cholesterol peak in the LNA-antimiR-treated animals. Elution of lipoprotein standards is indicated by the labels. Plasma triglycerides (C), glucose (D), AST (E), and ALT (F) were unaffected by the treatments. Error bars represent s.e.m. ** denotes p<0.01.
Taken together, we have shown that members of the mammalian SREBP family of transcription factors, key regulators of cholesterogenic and lipogenic genes, are hosts to conserved miRNAs (miR-33a/b) that may function in concert with the SREBP host gene products to govern intracellular cholesterol levels and cholesterol homeostasis in vertebrates (fig. S8). Indeed, our data reveal that miR-33 exerts post-transcriptional control of the ABCA1 cholesterol transporter, with important consequences for cholesterol trafficking in vitro and HDL synthesis in vivo. We believe that this represents an initial example of miRNA-host gene cooperation in regulating a metabolic process. Our findings also point to the possibility of antisense therapeutic targeting of miR-33a/b as a novel strategy to increase HDL in individuals suffering from cardiometabolic diseases.
Supplementary Material
Acknowledgments
We thank Shobha Vasudevan and Amy Walker for critical reading of the manuscript, and Kathryn Coser for technical assistance. This work was supported by the following funding sources: NIH R01GM071449 and R21DK084459 (A.M.N.), R01DK56626 and R01DK48873 (D.E.C.), American Heart Association Established Investigator Award (R.E.G.), the Fondation Leducq (R.E.G.), and MGH Tosteson Award (H.N.-S.).
References and Notes
- 1.Maxfield FR, Tabas I. Nature. 2005;438:612. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 2.Moller DE, Kaufman KD. Ann. Rev. Med. 2005;56:45. doi: 10.1146/annurev.med.56.082103.104751. [DOI] [PubMed] [Google Scholar]
- 3.Brown MS, Goldstein JL. J. Lipid Res. 2009;50(Suppl):S15. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborne TF, Espenshade PJ. Genes Dev. 2009;23:2578. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown MS, Goldstein JL. Cell. 1997;89:331. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 6.Horton JD, Goldstein JL, Brown MS. J. Clin. Invest. 2002;109:1125. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams AH, Liu N, van Rooij E, Olson EN. Curr. Opin. Cell Biol. 2009;21:461. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Croce CM. J. Clin. Oncol. 2009;27:5848. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. Cell. 2004;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. Nat. Rev. Genet. 2004;5:522. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 11.Krutzfeldt J, Stoffel M. Cell Metab. 2006;4:9. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Stefani G, Slack FJ. Nat. Rev. Mol. Cell. Biol. 2008;9:219. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 13.Elmen J, et al. Nature. 2008;452:896. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 14.Elmen J, et al. Nucleic Acids Res. 2008;36:1153. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esau C, et al. Cell Metab. 2006;3:87. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Krutzfeldt J, et al. Nature. 2005;438:685. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, et al. Nature. 2006;442:700. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Genome Res. 2004;14:1902. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, et al. PLoS One. 2009;4:e4421. doi: 10.1371/journal.pone.0004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baskerville S, Bartel DP. RNA. 2005;11:241. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YK, Kim VN. EMBO J. 2007;26:775. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. Cell Metab. 2008;7:365. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Rader DJ. Curr. Opin. Cardiol. 2007;22:368. doi: 10.1097/HCO.0b013e3281ec5113. [DOI] [PubMed] [Google Scholar]
- 25.Rader DJ, Alexander ET, Weibel GL, Billheimer J, Rothblat GH. J. Lipid Res. 2009;50(Suppl):S189. doi: 10.1194/jlr.R800088-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singaraja RR, et al. J. Clin. Invest. 2002;110:35. doi: 10.1172/JCI15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, et al. J. Clin. Invest. 2007;117:2216. doi: 10.1172/JCI32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Eck M, et al. Arterioscler. Thromb. Vasc. Biol. 2006;26:929. doi: 10.1161/01.ATV.0000208364.22732.16. [DOI] [PubMed] [Google Scholar]
- 29.de la Llera-Moya M, et al. Arterioscler. Thromb. Vasc. Biol. 2010;30:796. doi: 10.1161/ATVBAHA.109.199158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.