Abstract
Background
The clonal spread of Acinetobacter baumannii is a global problem, and carbapenems, such as imipenem, remain the first-choice agent against A. baumannii. Using synergy to enhance the antibiotic activity of carbapenems could be useful. Here, amlodipine (AML) was tested alone and with imipenem against A. baumannii isolates.
Methods
Forty-two isolates of A. baumannii were collected. Multilocus sequence typing (MLST) assessed the genetic relationship of the isolates. The resistance phenotypes were determined using disc diffusion. The minimum inhibitory concentrations (MICs) of the drugs were determined by broth microdilution. The combined effects of the drugs were determined by a checkerboard procedure. Metallo-β-lactamase (MBL) was determined using the MBL Etest.
Results
Forty-two A. baumannii isolates were collected from 42 patients who were mostly older than 65 years and had long inpatient stays (≥7 days). A. baumannii was mostly recovered from the respiratory system (N = 35, 83.3%). Most patients (N = 27, 64.3%) received care in intensive care units (ICUs). Disc diffusion testing demonstrated that A. baumannii susceptibility to polymyxin B was 100%, while susceptibility to other antimicrobial agents was less than 30%, classifying the isolates into 10 MDR and 32 XDR strains. MLST grouped the A. baumannii isolates into 4 existing STs and 6 new STs. STn4 carried allele G1, with a T → C mutation at nt3 on the gpi111 locus. STn5 carried allele A1, possessing A → C mutations at nt156 and nt159 on the gltA1 locus. ST195 and ST208 accounted for 68.05% (29/42) of the isolates. Clonal relation analysis showed that ST195 and ST208 belonged to clonal complex (CC) 92. The inhibitory concentration of imipenem ranged from 0.5 to 32 μg/ml, and that of AML ranged from 40 to 320 μg/ml. In combination, the susceptibility rate of A. baumannii isolates increased from 16.7% to 54.8% (P = 0.001). In the checkerboard procedure, half of the isolates (N = 21, 50.0%) demonstrated synergy or partial synergy with the drug combination. The MBL Etest revealed that 1 A. baumannii strain (N = 1, 2.4%) produced MBL.
Conclusions
CC92 was the major clone spreading in our hospital. AML improved the activity of imipenem against A. baumannii isolates in vitro but did not inhibit MBL.
Keywords: Acinetobacter baumannii, MLST, Imipenem, Amlodipine
Background
Acinetobacter baumannii (AB) has emerged as one of the most troublesome pathogens in health care institutions globally. It is characterised by frequent episodes of multi-drug resistance due to multiple mechanisms, and therapeutic options have become increasingly restricted [1-3]. Unfortunately, the present rate of discovery of antibiotics is much slower than that in the past [4]. This problem is exemplified by the fact that it is necessary to identify potential reservoirs of the organism and modes of transmission to control the spread of A. baumannii in hospitals. To distinguish the outbreak strain from epidemiologically unrelated acinetobacters, a comparison of isolates at the subspecies level is required. Multilocus sequence typing (MLST) is a highly discriminative typing system in which typing data are translated into a numerical code that can be obtained in an identical manner at different laboratories using the same protocol. It provides a portable method that may be suitable for global epidemiologic studies and that may allow for the recognition of epidemic, multiresistant, and virulent A. baumannii clones and the monitoring of their national and international spread [1,5,6]. However, data have been limited in Guangzhou.
Recently, an effective approach has been to explore non-antibiotic compounds (helper compounds) that express antibacterial properties, possibly by acting through different mechanisms from those of existing drugs, either by the enhancement of antibiotic activity (synergism) or the reversal of antibiotic resistance, yielding sensitivity comparable to that derived from classical antibiotics in previously drug-resistant microorganisms [7]. Among these non-antibiotics, amlodipine (AML) is the most promising helper compound. It has been shown to exhibit in vitro activity against a wide range of bacteria [8-11], but few studies have reported its potentiating effects on clinical A. baumannii isolates. In this study, AML was tested, alone and in combination with imipenem, against A. baumannii isolates.
Methods
Bacterial isolates and drugs
A total of 42 isolates of A. baumannii were collected from patients treated at our hospitals. The Guangzhou First People’s Hospital Ethics Committee approved the protocol, and all of the patients provided informed consent for inclusion in the study. The total number of beds in the hospitals was 1571. All of the A. baumannii isolates were consecutively collected from April 2011 to February 2012 from clinical specimens (respiratory secretion, blood, cerebrospinal fluid, and urine) from patients who were hospitalised in the general wards and intensive care units. Duplicate isolates from the same patients were excluded. The isolates were identified using the Vitek 2 (bioMerieux, Inc. Durham, North Carolina, USA) automated microbiology system.
Antibiotic discs (OXOID) were obtained from Melone Pharmaceutical Co. Ltd. (China). AML was obtained from Melone Pharmaceutical Co. Ltd. (China), and imipenem was obtained from Merck & Co. Inc. Imipenem was dissolved in phosphate-buffered saline (PBS) according to CLSI (M100-S22, 2012) [12]. AML was dissolved in dimethyl sulfoxide (DMSO) [9].
Multilocus sequence typing (MLST)
MLST was performed on A. baumannii, as described by Bartual et al. [5]. Briefly, seven housekeeping genes, i.e., gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD, were amplified, followed by sequencing on the ABI Prism Sequencer 3730 (Applied Biosystems). The sequences of these seven housekeeping genes were analysed using the Pubmlst database [13]. The sequence type (ST) was designated according to the allelic profiles in the database. The eBURST algorithm (version 3) [14] was used to assign STs to CCs and to assess the genetic relationship with the definition of the groups sharing alleles at ≥6 of 7 loci. The CC comprises a founding ST as a common ancestor, as well as several other closely related STs descending from the predicted founding genotype.
Antimicrobial susceptibility testing and metallo-β-lactamase detection
The strains were stored at −80°C in trypticase soy broth supplemented with 15% glycerol and were passaged twice on a blood agar plate before testing. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as the control organisms.
All 42 of the isolates were tested against a panel of antibiotics with the disc diffusion method, as recommended by the Clinical and Laboratory Standards Institute (CLSI; M100-S22, 2012) [12], to determine the resistance phenotype. Multidrug resistance (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, and extensive drug resistance (XDR) was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e., bacterial isolates remaining susceptible to only one or two categories) [3]. The MBL Etest (bioMerieux, Inc. Solna, Sweden) was applied to detect metallo-β-lactamase (MBL) [15].
The MIC of AML was defined as the lowest concentration at which no growth was observed. All of the broth preparations used in the microdilution testing and the Mueller Hinton broth were freshly prepared and used within 24 hours. The checkerboard procedure [10] was used to determine the combined effects of AML and imipenem. Synergy was determined by calculating the fractional inhibitory concentration (FIC) index as follows: FIC index = FICA + FICB = [A]/MICA + [B]/MICB, where [A] is the concentration of drug A; FICA is the FIC of drug A; and [B], MICB, and FICB are defined in the same manner for drug B. The FIC index was interpreted as follows: <0.5, synergy; 0.5 to 0.75, partial synergy; 0.76 to 1.0, additive effect; >1.0 to 4.0, indifference; and >4.0, antagonism.
Statistical analysis
SPSS (version 19.0) was used for all of the calculations. Percentages were compared using the chi-square test. A P value ≤0.05 in a two-tailed test was considered to indicate statistical significance.
Results
Summary of 42 A. baumannii isolates and MLST
A total of 42 A. baumannii isolates were collected from 42 patients; 30 patients were male, and 12 were female. Most of the patients were older than 65 years and had long inpatient stays (≥7 days). The most common site from which A. baumannii was recovered was the respiratory system (N = 35, 83.3%), followed by the blood (N = 3, 7.1%). Most of the patients (N = 27, 64.3%) were receiving care in intensive care units (ICUs), while 15 were in general wards (see Table 1).
Table 1.
Summary of 42 A. baumannii isolates
| Gender (%) | Age (%) | Hospital ward (%) | Type of specimen (%) | Inpatient status (%) |
|---|---|---|---|---|
| Male 30 (71.4%) |
≤17 0 (0%) |
ICU 27 (64.3%) |
Respiratory 35 (83.3%) |
≤2 days 4 (9.5%) |
| Female 12 (28.6%) |
18-65 9 (21.4%) |
Medical 10 (23.8%) |
Blood 3 (7.1%) |
3-6 days 9 (21.4%) |
| |
≥66 33 (78.6%) |
Surgical 4 (9.5%) |
Wound 2 (4.8%) |
≥7 days 29 (69.1%) |
| |
|
Burn 1 (2.4%) |
Urine 1 (2.4%) |
|
| CSF 1 (2.4%) |
ICU, intensive care unit; CSF, cerebrospinal fluid.
According to the MLST, the 42 A. baumannii isolates could be grouped into four existing STs [Additional file 1] (ST195, 1-3-3-2-2-96-3; ST208, 1-3-3-2-2-97-3; ST254, 21-15-3-2-35-111-4; and ST457, 1-15-3-2-2-153-3) and six new STs (STn1, 1-3-3-2-2-96-5; STn2, 1-15-3-2-2-96-3; STn3, 1-81-3-2-2-96-4; STn4, 21-15-3-2-35-G1-3; STn5, A1-15-3-2-2-153-4; and STn6, 11-65-3-20-37-96-15). STn4 carried a new allele, G1, which had a T → C mutation at nt3 of the gpi111 locus. STn5 had a new allele, A1, that had two mutations at the gltA1 locus, which were A → C mutations at nt156 and nt159. All of the isolates belonging to ST195 and ST208 accounted for 68.05% (29/42) of the isolates. Clonal relation analysis showed that both ST195 and ST208 belonged to the globally disseminated CC92 for A. baumannii (see Table 2 and Figure 1).
Table 2.
Multilocus sequence typing of 42 isolates
| Number of isolates | Specimens | Wards | gltA | gyrB | gdhB | recA | cpn60 | gpi | rpoD | ST |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 2 |
Blood |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
5 |
STn1 |
| 3 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 4 |
Blood |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 5 |
BALF |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 6 |
Sputum |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 7 |
Sputum |
Respiratory |
1 |
15 |
3 |
2 |
2 |
153 |
3 |
ST457 |
| 8 |
Sputum |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 9 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 10 |
Sputum |
Respiratory |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 11 |
Sputum |
Neurosurgery |
1 |
15 |
3 |
2 |
2 |
96 |
3 |
STn2 |
| 12 |
Sputum |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 13 |
Sputum |
Respiratory |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 14 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 15 |
BALF |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 16 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 17 |
Wound |
Gastroenterology |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 18 |
Sputum |
Respiratory |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 19 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 20 |
Urine |
Geriatrics ICU |
21 |
15 |
3 |
2 |
35 |
111 |
4 |
ST254 |
| 21 |
BALF |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 22 |
BALF |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 23 |
Sputum |
Respiratory |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 24 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 25 |
Sputum |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 26 |
Sputum |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 27 |
Sputum |
Neurosurgery |
1 |
81 |
3 |
2 |
2 |
96 |
4 |
STn3 |
| 28 |
Sputum |
Respiratory |
21 |
15 |
3 |
2 |
35 |
111 |
4 |
ST254 |
| 29 |
Sputum |
RICU |
21 |
15 |
3 |
2 |
35 |
111 |
4 |
ST254 |
| 30 |
Sputum |
RICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 31 |
Sputum |
Respiratory |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 32 |
Sputum |
Neurosurgery |
1 |
81 |
3 |
2 |
2 |
96 |
4 |
STn3 |
| 33 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 34 |
BALF |
RICU |
1 |
15 |
3 |
2 |
2 |
153 |
3 |
ST457 |
| 35 |
CSF |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 36 |
Sputum |
Respiratory |
21 |
15 |
3 |
2 |
35 |
G1 |
3 |
STn4 |
| 37 |
Sputum |
ICU |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 38 |
Wound |
Burn |
A1 |
15 |
3 |
2 |
2 |
153 |
4 |
STn5 |
| 39 |
Sputum |
Geriatrics ICU |
21 |
15 |
3 |
2 |
35 |
G1 |
3 |
STn4 |
| 40 |
Blood |
Urinary surgery |
1 |
3 |
3 |
2 |
2 |
96 |
3 |
ST195 |
| 41 |
Sputum |
Nephrology |
1 |
3 |
3 |
2 |
2 |
97 |
3 |
ST208 |
| 42 | BALF | RICU | 11 | 65 | 3 | 20 | 37 | 96 | 15 | STn6 |
BALF, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; ICU, intensive care unit; RICU, respiratory intensive care unit; G1, a new allele that has a T → C mutation at nt3 in the gpi111 locus; A1, a new allele possessing two mutations at the gltA1 locus (A → C mutations at nt156 and nt159).
Figure 1.
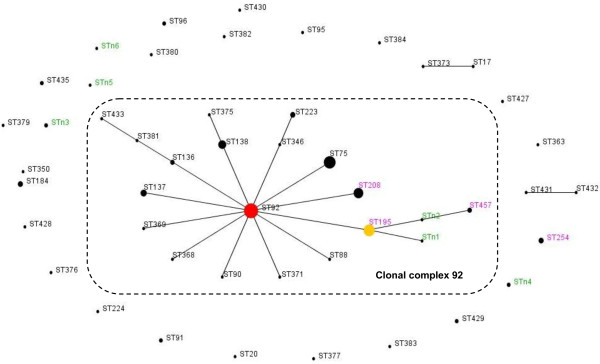
Population snapshot of A. baumannii in this study and existing isolates in China. Population snapshot of A. baumannii in this study and existing isolates in China, based on the data contained in the Pubmlst database [13] as of 27 April 2013, represented by an eBURST algorithm. Circles represent STs, and their sizes correspond to the number of isolates. The red circle represents the founder ST (ST92). The broken line indicates clonal complex (CC) 92. The ST labels are coloured as follows: black, STs found only in the Pubmlst database; green, STs found only in this study; and purple, STs found in both the Pubmlst database and this study. STn3, STn4, STn5, STn6, and ST254 were the singletons in this study.
Antimicrobial susceptibility testing and metallo-β-lactamase detection
The data from the disc diffusion testing (see Table 3) demonstrated that the susceptibility of A. baumannii to polymyxin B was 100%, while its susceptibility to other antimicrobial agents was lower than 30%, thus classifying the 42 A. baumannii isolates into 10 MDR and 32 XDR strains. In particular, no A. baumannii isolates were susceptible to ceftazidime, ceftriaxone, cefepime, aztreonam, levofloxacin, ciprofloxacin, or doxycycline, which have been routinely used in clinical practice in China. Only one A. baumannii strain (N = 1, 2.4%) demonstrated MBL production, according to the MBL Etest.
Table 3.
Isolates were tested against antibiotics by disc diffusion
| Isolate no. | SCF | TZP | CAZ | CRO | FEP | ATM | IPM | MEM | LEV | CIP | AK | DO | TGC | PB | TOB | Phenotype/MBL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
R |
XDR/Neg |
| 2 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
R |
XDR/Neg |
| 3 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
R |
XDR/Neg |
| 4 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 5 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 6 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
I |
S |
S |
MDR/Neg |
| 7 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 8 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 9 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 10 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 11 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 12 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
I |
S |
S |
MDR/Neg |
| 13 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 14 |
S |
R |
R |
R |
R |
R |
I |
I |
R |
R |
R |
R |
S |
S |
R |
MDR/Neg |
| 15 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 16 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
XDR/Neg |
| 17 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 18 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 19 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 20 |
I |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
S |
R |
XDR/Neg |
| 21 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
XDR/Neg |
| 22 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
R |
R |
S |
S |
MDR/Neg |
| 23 |
I |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
R |
XDR/Neg |
| 24 |
S |
R |
R |
R |
R |
R |
S |
S |
R |
R |
S |
R |
I |
S |
S |
MDR/Neg |
| 25 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
R |
XDR/Neg |
| 26 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
R |
XDR/Neg |
| 27 |
R |
R |
R |
R |
R |
R |
S |
I |
R |
R |
R |
R |
I |
S |
R |
MDR/Neg |
| 28 |
I |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
S |
S |
R |
XDR/Neg |
| 29 |
I |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
S |
R |
XDR/Neg |
| 30 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 31 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 32 |
I |
R |
R |
R |
I |
R |
S |
S |
R |
R |
R |
I |
I |
S |
R |
MDR/Neg |
| 33 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 34 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 35 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 36 |
I |
R |
R |
R |
R |
I |
R |
R |
R |
R |
R |
S |
S |
S |
R |
MDR/Neg |
| 37 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 38 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 39 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
S |
R |
XDR/Pos |
| 40 |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
I |
S |
R |
XDR/Neg |
| 41 |
I |
R |
R |
R |
R |
R |
S |
I |
R |
R |
R |
R |
R |
S |
R |
MDR/Neg |
| 42 |
I |
S |
R |
R |
R |
R |
S |
S |
I |
R |
R |
I |
R |
S |
R |
MDR/Neg |
| Susceptible (%) | 2.4 | 2.4 | 0.0 | 0.0 | 0.0 | 0.0 | 11.9 | 7.1 | 0.0 | 0.0 | 9.5 | 0.0 | 28.6 | 100.0 | 9.5 | - |
Metallo-β-lactamase was detected by MBL Etest.
R, resistance; S, sensitivity; I, intermediate; XDR, extensively drug-resistant; MDR, multidrug-resistant; MBL, metallo-β-lactamase; Pos, positive; Neg, negative; PB, polymyxin B (300 units); TGC, tigecycline (15 μg); IPM, imipenem (10 μg); AK, amikacin (30 μg); TOB, tobramycin (10 μg); MEM, meropenem (10 μg); TZP, piperacillin/tazobactam (100/10 μg); SCF, cefoperazone/sulbactam (70/35 μg); CAZ, ceftazidime (30 μg); CRO, ceftriaxone (30 μg); FEP, cefepime (30 μg); ATM, aztreonam (30 μg); LEV, levofloxacin (5 μg); CIP, ciprofloxacin (5 μg); DO, doxycycline (30 μg).
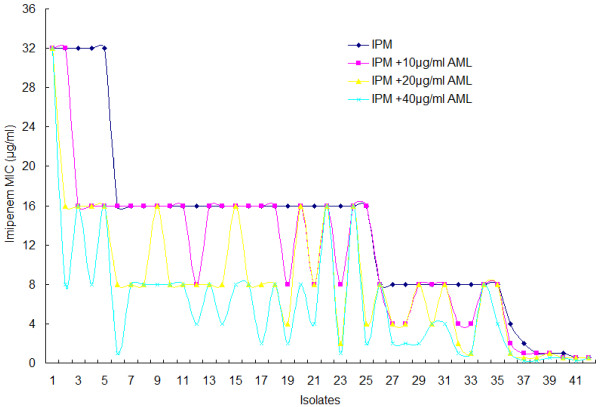
Table 4 shows the resistance reversal effects in A. baumannii isolates tested with imipenem. The inhibitory concentrations of imipenem alone against all of the strains tested ranged from 0.5 to 32 μg/ml, and the AML inhibitory concentrations ranged from 40 to 320 μg/ml. In combination with AML, the susceptibility rate of A. baumannii isolates to imipenem increased from 16.7% to 54.8% (P = 0.001). Figure 2 clearly shows the effects on imipenem MIC changes when different concentrations of AML were added. The imipenem MICs decreased significantly with the addition of 40 μg/ml of AML. Note that the resistance reversal by AML in combination with imipenem occurred at concentrations that were much lower than the MIC90 required for the growth inhibition of 90% of A. baumannii isolates by the compound.
Table 4.
Imipenem potency in the absence and presence of AML in 42 A. baumannii isolates
| Compound/combination of compounds |
MIC (μg/ml) |
No. susceptible (%) | No. nonsusceptible (%)a |
Statistical analysisb |
|||
|---|---|---|---|---|---|---|---|
| MICRange | MIC50 | MIC90 | χ2 -values | P values | |||
| AML |
40-320 |
80 |
160 |
- |
- |
- |
- |
| IPM |
0.5-32 |
16 |
16 |
7 (16.7%) |
35 (83.3%) |
- |
- |
| IPM +10 μg/ml AML |
0.5-32 |
8 |
16 |
13 (31.0%) |
29 (69.0%) |
2.363 |
0.200 |
| IPM +20 μg/ml AML |
0.5-32 |
8 |
16 |
15 (35.7%) |
27 (64.3%) |
3.941 |
0.081 |
| IPM +40 μg/ml AML | <0.5-32 | 4 | 16 | 23 (54.8%) | 19 (45.2%) | 13.274 | 0.001 |
MIC, minimum inhibitory concentration; MIC50 and MIC90, MICs for 50% and 90% of strains, respectively; anonsusceptible, including intermediate resistance; bstatistical analysis compared with the IPM group.
Figure 2.
The change in the imipenem MIC by adding AML. Figure 2 shows the distribution of imipenem MICs. The change in the imipenem MIC with the addition of 10, 20, and 40 μg/ml of AML compared with the MIC resulting from no AML addition. The imipenem MICs decreased significantly with the addition of 40 μg/ml of AML.
In synergy testing with the checkerboard procedure (see Table 5), half of the isolates (N = 21, 50.0%) demonstrated synergy or partial synergy with the imipenem and AML combination. The FIC index values ranged from 0.375 to 0.75. Ten isolates (N = 10, 23.8%) demonstrated additive effects; 11 (26.2%) isolates demonstrated indifference; and no cases of antagonism were observed.
Table 5.
Fractional inhibitory concentration (FIC) index of the checkerboard procedure in 42 A. baumannii isolates
| Combination of compounds |
FIC index (N,%) |
||||
|---|---|---|---|---|---|
| <0.5 Synergy | 0.5 to 0.75 partial synergy | 0.76 to 1.0 additive effect | >1.0 to 4.0 indifference | >4.0 antagonism | |
| Imipenem + AML | 6 (14.3%) | 15 (35.7%) | 10 (23.8%) | 11 (26.2%) | 0 (0%) |
The concentrations of imipenem and AML used in the checkerboard procedure depended on the MICs of each A. baumannii isolate tested.
Discussion
A. baumannii infections have become more difficult to treat due to the emergence of isolates that are resistant to multiple antimicrobial drugs. Data from the Chinese Network for Bacterial Resistance Surveillance demonstrated that almost 17% of these isolates were resistant to all of the antimicrobial agents that are routinely used in clinical practice and that the susceptibility of A. baumannii to carbapenems has decreased to less than 50% [16]. As a consequence, infections caused by A. baumannii strains have been associated with high mortality and treatment failure [17]. According to our data, A. baumannii has been widely distributed in our hospital and has been isolated from the respiratory system, blood, wounds, urine, and cerebrospinal fluid, especially from ICU patients who are older and have longer inpatient stays, similar to the results reported in many other studies [1,18-20]. The susceptibility of these isolates to imipenem was 16.1% (according to the MIC method). Only 1 (2.4%) MBL-producing A. baumannii strain was found in our study, which was similar to the findings of Hui Wang [21], in which only 1 isolate among 221 nonrepetitive imipenem-resistant clinical isolates of Acinetobacter spp. carried the bla(IMP-8) MBL gene. The present study demonstrated that MBL was not the primary cause of A. baumannii resistance to imipenem in our hospital. Class D β-lactamases, the modification of penicillin-binding proteins and porins, or the upregulation of the AdeABC efflux system may have played more important roles in their potential resistance mechanisms [22]; thus, further investigation is needed.
MLST is an unambiguous typing method that has achieved notable success in global epidemiological investigations of A. baumannii[5,6]. Recently, some studies [6,23-26] have revealed that CC92 (previously known as CC22) has worldwide dissemination. The ST92 A. baumannii isolate is the most common clone, and it has been predicted to be the founder of this clonal complex. Yiqi Fu et al. [26] confirmed that CC92 represented the most epidemic STs in China. In addition to ST92, the other STs belong to CC92 and vary by area. A study by He et al. [25] showed that more ST138 isolates are associated with ST92 in western China. However, results from Qiao Zhong et al. [23] indicated that ST75, accompanied by ST92, may be the most common epidemical sequence types in eastern China. In our study, we assumed that ST195 and ST208 may be more common in southern China. These differences may be caused by different antibiotic usage habits, which could have influenced the evolutionary direction of ST92. Why has this clonal complex been so successful? Studies have shown that antibiotic susceptibility of CC92 is variable [27]. These findings may suggest that adaptation to the hospital environment, as well as antibiotic resistance, has been more important for the success of A. baumannii as a nosocomial pathogen.
Because imipenem remains the first-choice agent in China against A. baumannii strains [28], finding a way to enhance the antibiotic activity (synergism) of carbapenems against A. baumannii strains could be encouraging. Calcium channel blockers (CCBs) are a heterogeneous class of drugs used for the control of hypertension, angina, and cardiac arrhythmias [29]. However, many studies have reported a wide variety of broad-spectrum antibacterial activities of CCBs [30], the most powerful of which was AML. Kumar [9] reported in vitro antimicrobial activity of AML against several Gram-positive and Gram-negative standard and clinical strains, including Bacillus pumilus NCTC 8241, Staphylococcus aureus NCTC 6571, Escherichia coli K12Row, Salmonella typhimurium NCTC74, Shigella dysenteriae 7 NCTC 519/66, Shigella sonnei 1NCTC 5/59, Shigella flexneri 4a24, Shigella boydii 8 NCTC 254/66, Klebsiella pneumoniae 14, Vibrio cholerae ATCC 14033, and P. aeruginosa APC. The inhibitory concentrations of AML against most of the strains tested ranged from 25 to 200 μg/ml, but its effects on P. aeruginosa, E. coli, and K. pneumoniae were limited, and the inhibitory concentrations were >800, >800, and 400 μg/ml, respectively. A further study demonstrated that AML in combination with streptomycin had a synergistic effect [10]. In addition to its in vitro and in vivo activity against Gram-positive and Gram-negative bacteria, AML showed a two- to eightfold reduction in its MIC when combined with streptomycin, and vice versa [10]. Similarly, when paired with levofloxacin, AML showed potential synergistic effects in the eradication of P. aeruginosa biofilms [11]. Our research demonstrated the antimicrobial activities of AML against clinical A. baumannii strains, and the MICs ranged from 40 to 320 μg/ml. In combination with imipenem, half of the isolates (N = 21, 50.0%) demonstrated synergy or partial synergy. Further investigations, such as the development of a time-killing curve, which may demonstrate killing synergistic effects against A. baumannii strains, are needed.
To date, research on the antimicrobial efficacy of AML has been conducted only in vitro and in animal models. Chronic toxicity tests with healthy mice showed that the therapeutic dose of AML (10 mg/kg) was within a safe and acceptable margin. Low doses of AML at 1.5 mg/kg/day continue to show good efficacy in mice [7]. According to our study results, AML must be used at doses of up to 40 μg/ml to improve the activity of imipenem against A. baumannii isolates. However, in humans, the recommended dose of AML is much lower, at only 10 mg/day (Cmax 6.14 ng/ml) as an antihypertensive agent, thus currently limiting the clinical application of AML [31]. However, AML has been able to cure highly virulent bacterial infections in mice. This ability suggests that AML could be employed as a ‘lead compound’ to synthesise more active novel agents that may be free of side effects, such as hypotension. Because both drugs have been satisfactorily used for a long time in clinical medicine for different purposes, their combination in humans is feasible.
Conclusions
CC92 was the major clone that spread in our hospital. AML improved the activity of imipenem against A. baumannii isolates in vitro, but it did not work by inhibiting MBL.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YjL supervised the study, performed the susceptibility testing and metallo-β-lactamase detecting, and wrote the manuscript. ZxZ and WbL contributed to the susceptibility testing and metallo-β-lactamase detection. HlC provided advice regarding the susceptibility testing technology. CzP discussed the data and helped to finalise the manuscript. ZwZ planned and supervised the experiments. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Summary of 125 existing A. baumannii isolates in China. The file’s format is excel spreadsheet, the file contains data of 125 existing A. baumannii isolates in China, which were downloaded from the Pubmlst database (http://pubmlst.org/abaumannii/), up to 27 April 2013.
Contributor Information
Yu jun Li, Email: liyujun_0110@163.com.
Chu zhi Pan, Email: fanshusi@163.com.
Zi wen Zhao, Email: zhaozw@yeah.net.
Zhu xiang Zhao, Email: zhaozhuxiang@126.com.
Hui ling Chen, Email: huiling1515@163.com.
Wei bo Lu, Email: luweibo-520@163.com.
Acknowledgements
This work was funded by the Nature and Science Project of Guangdong Province (no. 10151006001000015) and the Guangzhou Medical and Health Science and Technology Key Project (no. 201102A212019). We thank Hui fen YE, of the Department of Clinic Laboratory, Guangzhou First People’s Hospital, for technical support in culturing and reisolating the strains used in this study and for contributions to the interpretation of the data.
References
- Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo SC, Chang SC, Wang HY, Lai JF, Chen PC, Shiau YR, Huang IW, Lauderdale TL. Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: nationwide data from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program. BMC Infect Dis. 2012;12:200. doi: 10.1186/1471-2334-12-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria expert proposal for standard definitions for acquired resistance. Clin Microbiol Infect. 2011;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. J Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karah N, Sundsfjord A, Towner K, Samuelsen O. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat. 2012;15:237–247. doi: 10.1016/j.drup.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Mazumdar K, Asok Kumar K, Dutta NK. Potential role of the cardiovascular non-antibiotic (helper compound) amlodipine in the treatment of microbial infections: scope and hope for the future. Int J Antimicrob Agents. 2010;36:295–302. doi: 10.1016/j.ijantimicag.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Kristiansen JE, Thomsen VF, Martins A, Viveiros M, Amaral L. Non-antibiotics reverse resistance of bacteria to antibiotics. In Vivo. 2010;24:751–754. [PubMed] [Google Scholar]
- Kumar KA, Ganguly K, Mazumdar K, Dutta NK, Dastidar SG, Chakrabarty AN. Amlodipine: a cardiovascular drug with powerful antimicrobial property. Acta Microbiol Pol. 2003;52:285–292. [PubMed] [Google Scholar]
- Asok Kumar K, Mazumdar K, Dutta NK, Karak P, Dastidar SG, Ray R. Evaluation of synergism between the aminoglycoside antibiotic streptomycin and the cardiovascular agent amlodipine. Biol Pharm Bull. 2004;27:1116–1120. doi: 10.1248/bpb.27.1116. [DOI] [PubMed] [Google Scholar]
- Elkhatib WF, Haynes VL, Noreddin AM. Microbiological appraisal of levofloxacin activity against Pseudomonas aeruginosa biofilm in combination with different calcium chanel blockers in vitro. J Chemother. 2009;21:135–143. doi: 10.1179/joc.2009.21.2.135. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standard Institute. Performance standards for antimicrobial susceptibility testing. Nineteenth informational supplement. CLSI document M100 S22. PA, USA: Wayne; 2012. [Google Scholar]
- Acinetobacter baumannii MLST Databases. http://pubmlst.org/abaumannii/
- eBURSTv3 algorithm. http://eburst.mlst.net/
- Lee K, Yong D, Yum JH, Lim YS, Bolmstrom A, Qwarnstrom A, Karlsson A, Chong Y. Evaluation of Etest MBL for detection of blaIMP-1 and blaVIM-2 allele-positive clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2005;43:942–944. doi: 10.1128/JCM.43.2.942-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu DM, Hu FP, Ruan FY, Ni YX, Song JY. CHINET 2009 surveillance of bacterial resistance in China. Chin J Infect Chemother. 2010;10:325–334. [Google Scholar]
- Sunenshine RH, Wright MO, Maragakis LL, Harris AD, Song X, Hebden J, Cosgrove SE, Anderson A, Carnell J, Jernigan DB, Kleinbaum DG, Perl TM, Standiford HC, Srinivasan A. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerg Infect Dis. 2007;13:97–103. doi: 10.3201/eid1301.060716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JY, Park MS, Kim SE, Park BH, Son JY, Kim EY, Lim JE, Lee SK, Lee SH, Lee KJ, Kang YA, Kim SK, Chang J, Kim YS. Risk factors for multi-drug resistant Acinetobacter baumannii bacteremia in patients with colonization in the intensive care unit. BMC Infect Dis. 2010;10:228. doi: 10.1186/1471-2334-10-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect Dis. 2010;10:196. doi: 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souli M, Galani I, Giamarellou H. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro Surveill. 2008;13:19045. [PubMed] [Google Scholar]
- Wang H, Guo P, Sun H, Wang H, Yang Q, Chen M, Xu Y, Zhu Y. Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from chinese hospitals. Antimicrob Agents Chemother. 2007;51:4022–8402. doi: 10.1128/AAC.01259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrilli R, Giannouli M, Tomasone F, Triassi M, Tsakris A. Carbapenem resistance in Acinetobacter baumannii: the molecular epidemic features of an emerging problem in health care facilities. J Infect Dev Ctries. 2009;3:335–341. doi: 10.3855/jidc.240. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Xu W, Wu Y, Xu H. Clonal spread of carbapenem non-susceptible Acinetobacter baumannii in an intensive care unit in a teaching hospital in China. Ann Lab Med. 2012;32:413–419. doi: 10.3343/alm.2012.32.6.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Xie Y, Fan H, Kang M, Tao C, Zhang R, Hu Y, Chen Z, Wang L. Spread of imipenem-resistant Acinetobacter baumannii of European clone II in western China. Int J Antimicrob Agents. 2011;38:257–260. doi: 10.1016/j.ijantimicag.2011.04.015. [DOI] [PubMed] [Google Scholar]
- He C, Xie Y, Zhang L, Kang M, Tao C, Chen Z, Lu X, Guo L, Xiao Y, Duo L, Fan H. Increasing imipenem resistance and dissemination of the ISAba1-associated blaOXA-23 gene among Acinetobacter baumannii isolates in an intensive care unit. J Med Microbiol. 2011;60:337–341. doi: 10.1099/jmm.0.022681-0. [DOI] [PubMed] [Google Scholar]
- Fu Y, Zhou J, Zhou H, Yang Q, Wei Z, Yu Y, Li L. Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J Antimicrob Chemother. 2010;65:644–650. doi: 10.1093/jac/dkq027. [DOI] [PubMed] [Google Scholar]
- Runnegar N, Sidjabat H, Goh HM, Nimmo GR, Schembri MA, Paterson DL. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a single institution over a 10-year period. J Clin Microbiol. 2010;48:4051–4056. doi: 10.1128/JCM.01208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Liu XF, Huang J, Zhu DM, Li J, Zhang J. Activities of colistin- and minocycline-based combinations against extensive drug resistant Acinetobacter baumannii isolates from intensive care unit patients. BMC Infect Dis. 2011;11:109. doi: 10.1186/1471-2334-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott WJ, Ram CV. Calcium channel blockers. J Clin Hypertens (Greenwich) 2011;13:687–689. doi: 10.1111/j.1751-7176.2011.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen JE, Amaral L. The potential management of resistant infections with non-antibiotics. J Antimicrob Chemother. 1997;40:319–327. doi: 10.1093/jac/40.3.319. [DOI] [PubMed] [Google Scholar]
- Chung M, Calcagni A, Glue P, Bramson C. Effect of food on the bioavailability of amlodipine besylate/atorvastatin calcium combination tablet. J Clin Pharmacol. 2006;46:1212–1216. doi: 10.1177/0091270006291097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of 125 existing A. baumannii isolates in China. The file’s format is excel spreadsheet, the file contains data of 125 existing A. baumannii isolates in China, which were downloaded from the Pubmlst database (http://pubmlst.org/abaumannii/), up to 27 April 2013.