Abstract
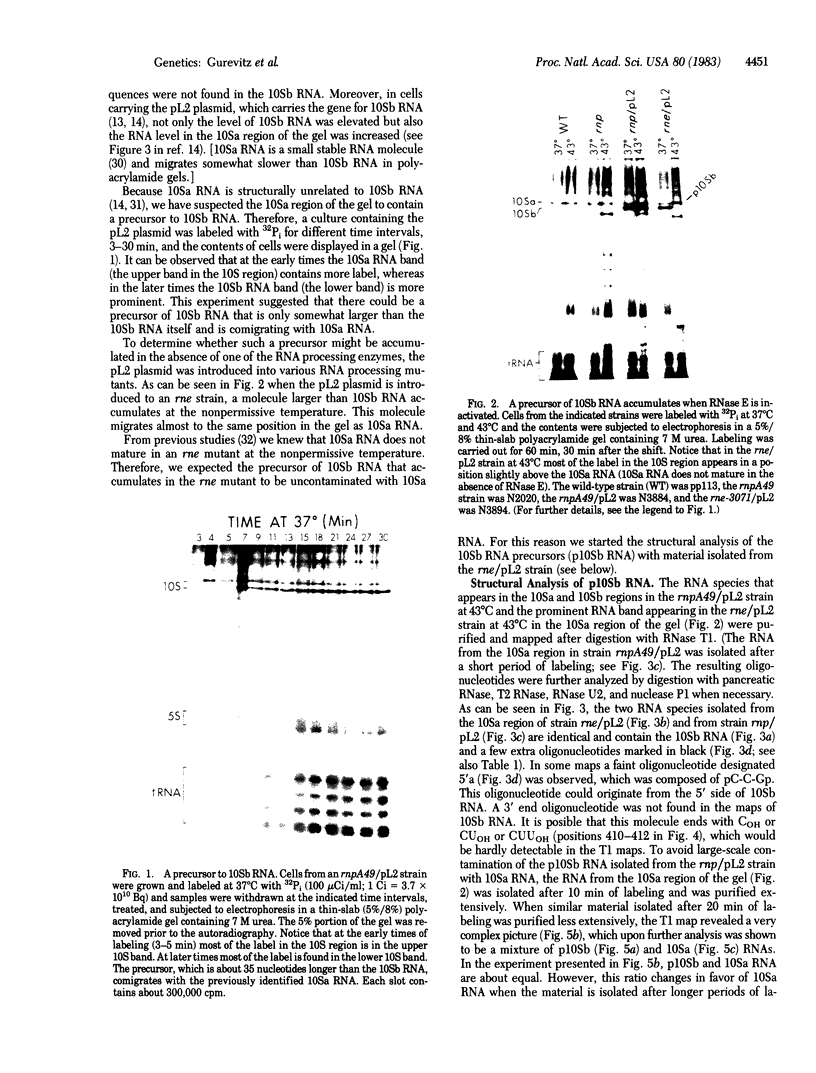
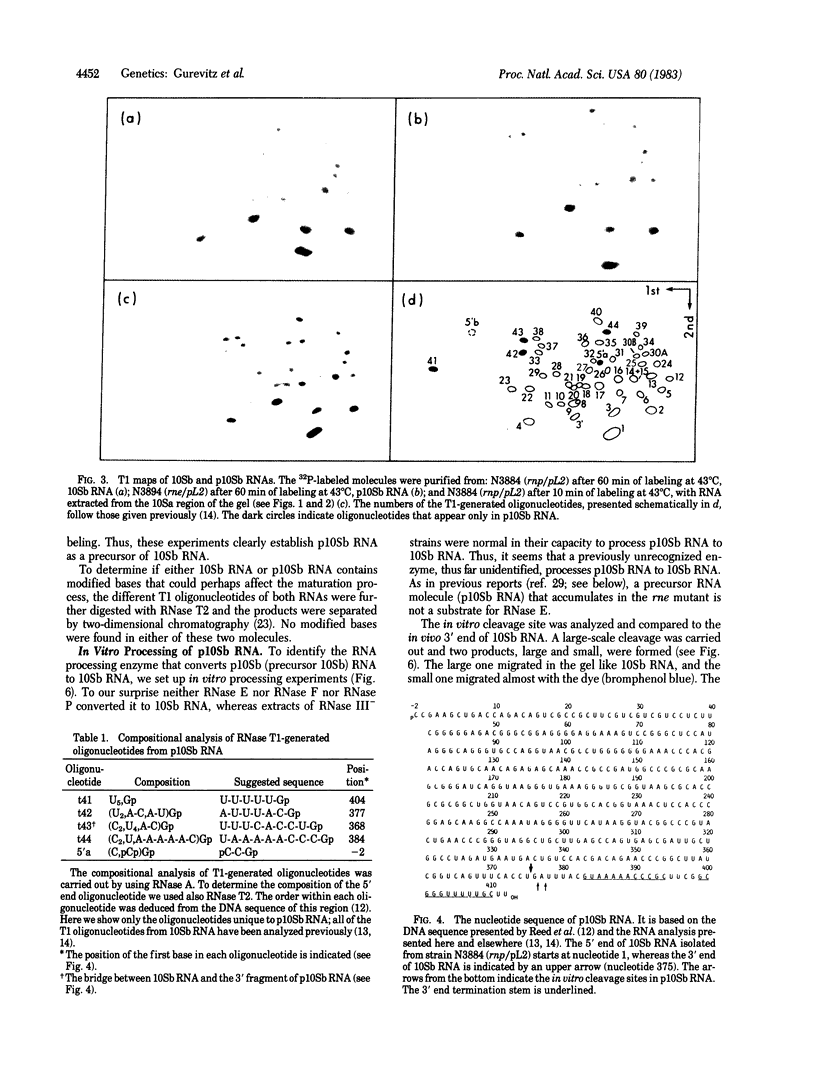
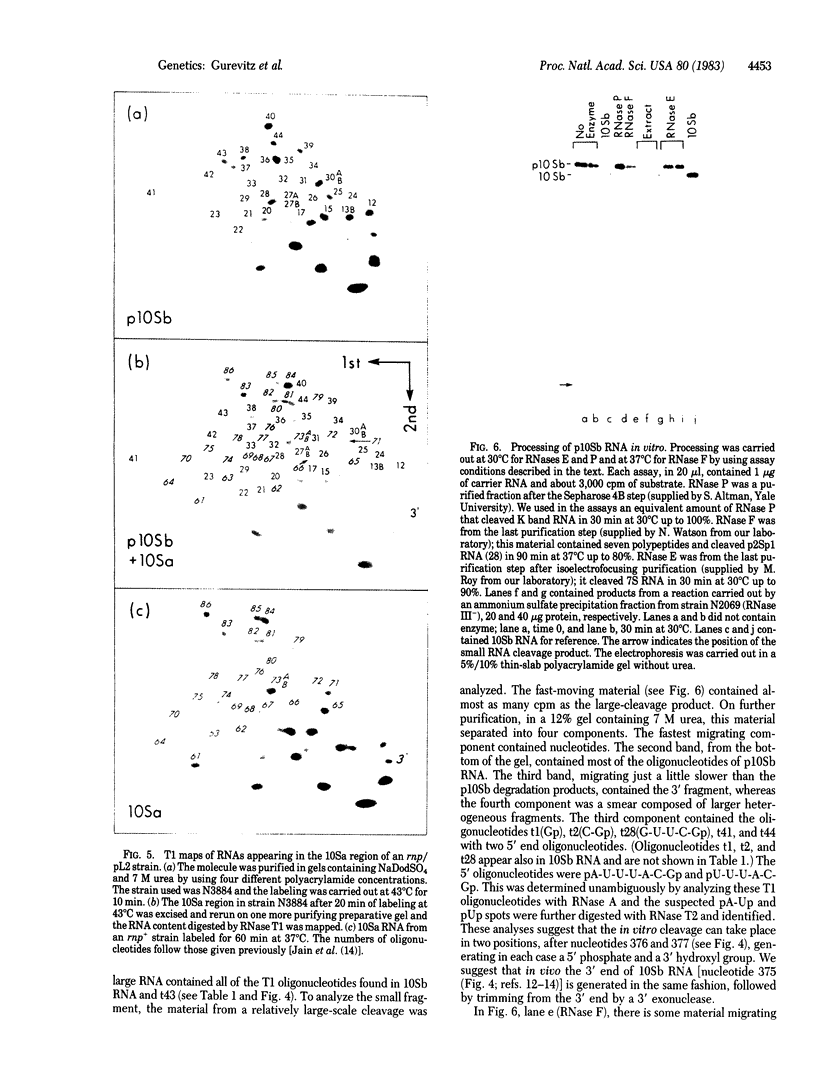
A precursor molecule for 10Sb (M1) RNA, the RNA moiety of the RNA processing enzyme ribonuclease P (EC 3.1.26.5), is accumulated transiently in an Escherichia coli strain containing a plasmid that carries the 10Sb RNA gene. The same RNA precursor molecule is accumulated, in relatively large quantities, in a temperature-sensitive RNase E- mutant at the nonpermissive temperature. The RNA precursor includes 10Sb RNA and an extra 3' fragment that contains a termination stem and loop. It can be processed in vitro to a molecule the size of 10Sb RNA. None of the four endoribonucleases of E. coli--RNase III, RNase E, RNase F, or RNase P--takes part in this cleavage reaction. Therefore, we suggest that the processing of the precursor-10Sb RNA to 10Sb RNA is carried out by a thus-far unidentified endoribonuclease. The accumulation of a RNA molecule in a RNase E- mutant that does not contain a cleavage site for RNase E has been encountered previously and can be explained by assuming the existence of a RNA processing complex in the E. coli cell.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Altman S. Biosynthesis of transfer RNA in Escherichia coli. Cell. 1975 Jan;4(1):21–29. doi: 10.1016/0092-8674(75)90129-4. [DOI] [PubMed] [Google Scholar]
- Apirion D., Gegenheimer P. Processing of bacterial RNA. FEBS Lett. 1981 Mar 9;125(1):1–9. doi: 10.1016/0014-5793(81)80984-2. [DOI] [PubMed] [Google Scholar]
- Apirion D. Genetic mapping and some characterization of the rnpA49 mutation of Escherichia coli that affects the RNA-processing enzyme ribonuclease P. Genetics. 1980 Feb;94(2):291–299. doi: 10.1093/genetics/94.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D., Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975 Oct;124(1):317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner K., Pace N. R. RNase P of Bacillus subtilis has a RNA component. J Biol Chem. 1980 Aug 25;255(16):7507–7509. [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of procaryotic ribonucleic acid. Microbiol Rev. 1981 Dec;45(4):502–541. doi: 10.1128/mr.45.4.502-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of rRNA by RNAase P: spacer tRNAs are linked to 16S rRNA in an RNAase P RNAase III mutant strain of E. coli. Cell. 1978 Oct;15(2):527–539. doi: 10.1016/0092-8674(78)90021-1. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Structural characterization and in vitro processing of Escherichia coli ribosomal RNA transcripts containing 5- triphosphates, leader sequences, 16 S rRNA, and spacer tRNAs. J Mol Biol. 1980 Nov 5;143(3):227–257. doi: 10.1016/0022-2836(80)90188-6. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P., Watson N., Apirion D. Multiple pathways for primary processing of ribosomal RNA in Escherichia coli. J Biol Chem. 1977 May 10;252(9):3064–3073. [PubMed] [Google Scholar]
- Goldblum K., Apririon D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J Bacteriol. 1981 Apr;146(1):128–132. doi: 10.1128/jb.146.1.128-132.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitz M., Watson N., Apirion D. A cleavage site of ribonuclease F. A putative processing endoribonuclease from Escherichia coli. Eur J Biochem. 1982 Jun;124(3):553–559. [PubMed] [Google Scholar]
- Guthrie C. The nucleotide sequence of the dimeric precursor to glutamine and leucine transfer RNAs coded by bacteriophage T4. J Mol Biol. 1975 Jul 15;95(4):529–547. doi: 10.1016/0022-2836(75)90315-0. [DOI] [PubMed] [Google Scholar]
- Jain S. K., Gurevitz M., Apirion D. A small RNA that complements mutants in the RNA processing enzyme ribonuclease P. J Mol Biol. 1982 Dec 15;162(3):515–533. doi: 10.1016/0022-2836(82)90386-2. [DOI] [PubMed] [Google Scholar]
- Jain S. K., Pragai B., Apirion D. A possible complex containing RNA processing enzymes. Biochem Biophys Res Commun. 1982 Jun 15;106(3):768–778. doi: 10.1016/0006-291x(82)91777-6. [DOI] [PubMed] [Google Scholar]
- Kole R., Altman S. Properties of purified ribonuclease P from Escherichia coli. Biochemistry. 1981 Mar 31;20(7):1902–1906. doi: 10.1021/bi00510a028. [DOI] [PubMed] [Google Scholar]
- Kole R., Altman S. Reconstitution of RNase P activity from inactive RNA and protein. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3795–3799. doi: 10.1073/pnas.76.8.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R., Baer M. F., Stark B. C., Altman S. E. coli RNAase P has a required RNA component. Cell. 1980 Apr;19(4):881–887. doi: 10.1016/0092-8674(80)90079-3. [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Bailey S. C., Apirion D. Small stable RNAs from Escherichia coli: evidence for the existence of new molecules and for a new ribonucleoprotein particle containing 6S RNA. J Bacteriol. 1978 Feb;133(2):1015–1023. doi: 10.1128/jb.133.2.1015-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem. 1979 Nov 10;254(21):11154–11159. [PubMed] [Google Scholar]
- Motamedi H., Lee K., Nichols L., Schmidt F. J. An RNA species involved in Escherichia coli ribonuclease P activity. Gene cloning and effect on transfer RnA synthesis in vivo. J Mol Biol. 1982 Dec 15;162(3):535–550. doi: 10.1016/0022-2836(82)90387-4. [DOI] [PubMed] [Google Scholar]
- Plautz G., Apirion D. Processing of RNA in Escherichia coli is limited in the absence of ribonuclease III, ribonuclease E and ribonuclease P. J Mol Biol. 1981 Jul 15;149(4):813–819. doi: 10.1016/0022-2836(81)90360-0. [DOI] [PubMed] [Google Scholar]
- Pragai B., Apirion D. Processing of bacteriophage T4 transfer RNAs. Structural analysis and in vitro processing of precursors that accumulate in RNase E-strains. J Mol Biol. 1982 Jan 25;154(3):465–484. doi: 10.1016/s0022-2836(82)80007-7. [DOI] [PubMed] [Google Scholar]
- Ray B. K., Apirion D. Characterization of 10S RNA: a new stable rna molecule from Escherichia coli. Mol Gen Genet. 1979 Jul 2;174(1):25–32. doi: 10.1007/BF00433301. [DOI] [PubMed] [Google Scholar]
- Ray B. K., Apirion D. Transfer RNA precursors are accumulated in Escherichia coli in the absence of RNase E. Eur J Biochem. 1981 Mar;114(3):517–524. doi: 10.1111/j.1432-1033.1981.tb05175.x. [DOI] [PubMed] [Google Scholar]
- Reed R. E., Baer M. F., Guerrier-Takada C., Donis-Keller H., Altman S. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell. 1982 Sep;30(2):627–636. doi: 10.1016/0092-8674(82)90259-8. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Roy M. K., Singh B., Ray B. K., Apirion D. Maturation of 5-S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur J Biochem. 1983 Mar 1;131(1):119–127. doi: 10.1111/j.1432-1033.1983.tb07238.x. [DOI] [PubMed] [Google Scholar]
- Saneyoshi M., Oashi Z., Harada F., Nishimura S. Isolation and characterization of 2-methyladenosine from Escherichia coli tRNA Glu 2 , tRNA Asp 1 , tRNA His 1 and tRNA Arg . Biochim Biophys Acta. 1972 Feb 23;262(1):1–10. [PubMed] [Google Scholar]
- Schedl P., Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. D. Transcription and processing of transfer RNA precursors. Prog Nucleic Acid Res Mol Biol. 1976;16:25–73. doi: 10.1016/s0079-6603(08)60755-2. [DOI] [PubMed] [Google Scholar]
- Uchida T., Arima T., Egami F. Specificity of RNase U2. J Biochem. 1970 Jan;67(1):91–102. doi: 10.1093/oxfordjournals.jbchem.a129239. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Fiers W. Micro thin-layer techniques for rapid sequence analysis of 32P-labeled RNA: double digestion and pancreatic ribonuclease analyses. Anal Biochem. 1977 Nov;83(1):228–239. doi: 10.1016/0003-2697(77)90531-0. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Jou W. M., Fiers W. Analysis of 32P-labeled bacteriophage MS2 RNA by a mini-fingerprinting procedure. Anal Biochem. 1976 May 7;72:433–446. doi: 10.1016/0003-2697(76)90551-0. [DOI] [PubMed] [Google Scholar]








