Abstract
Background
Plant calcium (Ca2+) signals are involved in a wide array of intracellular signalling pathways following pathogen invasion. Ca2+-binding sensory proteins such as Ca2+-dependent protein kinases (CPKs) have been predicted to mediate signalling following Ca2+ influx after pathogen infection. However, to date this prediction has remained elusive.
Results
We conducted a genome-wide identification of the Malus x domestica CPK (MdCPK) gene family and identified 30 CPK genes. Comparative phylogenetic analysis of Malus CPKs with CPKs of Arabidopsis thaliana (AtCPKs), Oryza sativa (OsCPKs), Populous trichocarpa (PtCPKs) and Zea mays (ZmCPKs) revealed four different groups. From the phylogenetic tree, we found that MdCPKs are closely related to AtCPKs and PtCPKs rather than OsCPKs and ZmCPKs, indicating their dicot-specific origin. Furthermore, comparative quantitative real time PCR and intracellular cytosolic calcium ([Ca2+]cyt) analysis were carried out on fire blight resistant and susceptible M. x domestica apple cultivars following infection with a pathogen (Erwinia amylovora) and/or mechanical damage. Calcium analysis showed an increased [Ca2+]cyt over time in resistant cultivars as compared to susceptible cultivars. Gene expression studies showed that 11 out of the 30 MdCPKs were differentially expressed following pathogen infection.
Conclusions
We studied the genome-wide analysis of MdCPK gene family in Malus x domestica and analyzed their differential gene expression along with cytosolic calcium variation upon pathogen infection. There was a striking difference in MdCPKs gene expressions and [Ca2+]cyt variations between resistant and susceptible M. x domestica cultivars in response to E. amylovora and mechanical wounding. Our genomic and bioinformatic analysis provided an important insight about the role of MdCPKs in modulating defence responses in susceptible and resistant apple cultivars. It also provided further information on early signalling and downstream signalling cascades in response to pathogenic and mechanical stress.
Keywords: Malus x domestica, Calcium dependent Protein Kinases, Erwinia amylovora, Phylogenetic analysis, Gene expression, Cytosolic calcium variations
Background
Calcium ions (Ca2+) plays a central role as a second messenger in nearly every aspect of cellular signalling. In plants, regulation of cytosolic Ca2+-concentration ([Ca2+]cyt) occurs in response to various endogenous and external signals, including changes in hormone status, abiotic stress stimuli such as drought, high and low temperature or light, biotic stress stimuli such as pathogen infection, microbial elicitors and symbiotic nodulation factors, as well as mechanical wounding [1-4]. Ca2+ influx is also a prerequisite for programmed cell death in plants [5,6]. These Ca2+ signatures are perceived by different Ca2+ sensor molecules which subsequently transduce the signal to downstream signalling cascades such as phosphorylation of target proteins [3,7,8].
Plants have four different classes of Ca2+ sensors: clamodulins (CaM), clamodulin-like proteins (CaML), calcineurin B-like proteins (CBL) and calcium-dependent protein kinases (CPKs) [9]. CaM, CaML and CBL lack an effector domain and contain only a Ca2+ binding domain; hence, they can sense and transmit Ca2+ signals by interacting with target proteins [10]. In Arabidopsis, the CaM-like protein (CML24) is required for nitric oxide (NO) production and AvrRpt2-mediated programmed cell death (PCD) [5], whereas CML42-mediated Ca2+ signalling coordinates responses to herbivory and abiotic stress [11].
CPKs constitute a large family of serine/threonine protein kinases that are widely distributed in the plant kingdom. For example, the Arabidopsis genome is predicted to have 34 different CPKs, Zea mays has 34, Populus 30, Oryza 31 and Triticum 24 CPKs[9,12-14], which can be classified into four groups (I-IV) based on sequence similarity [15]. CPKs are of special interest, since they represent a new class of Ca2+ sensors, having both a protein kinase domain and a calmodulin-like domain (including an EF-hand calcium-binding site) in a single polypeptide [9,15]. CPKs are activated by the binding of Ca2+ at the EF-motifs, resulting in protein conformational changes that in turn drive the auto inhibitory domain to become detached from the protein kinase domain [16]. Activated CPKs can mediate Ca2+ signalling by phosphorylating their substrate proteins [3]. The N- and C-terminal domains are variable, differing in their length and amino acid composition in various CPK proteins and it has been suggested that these variable domains determine the specific functions of individual CPKs[17,18]. Arabidopsis CPK1 was the first CPK to be characterised, and is known to be activated by phospholipids and 14-3-3 proteins [19]. AtCPKs 3, 4, 6, 11 and 32 act as abscisic acid (ABA) signalling components, and are involved in ABA-responsive gene expression, seed germination, seedling growth, and stomatal movement [20-22]. In plant immunity, four Arabidopsis CPKs (CPKs 4/5/6/11) have been shown to play important roles, together with mitogen activated protein kinase (MAPK) cascades, in relaying primary microbe associated molecular pattern (MAMP) immune signalling [23]. Recently, six Arabidopsis CPKs have been identified in sensing and transducing Ca2+, indicating the specificity and redundancy of individual CPKs in nucleotide-binding domain leucine-rich repeat (NLR) signalling events: CPK4/5/6/11 modulate immune response expression, CPK1/2/4/11 ROS production, and CPK1/2/5/6 are involved in programmed cell death (PCD), as revealed by integrative molecular analyses [6,24]. Apparently, specific CPKs are engaged in diverse immune responses via phosphorylation and activation of WRKY transcription factors. For example, activation of CPK4/5/6/11 phosphorylates WRKY8/28/48 for transcription reprogramming of immune genes; CPK1/2/4/11 phosphorylates NADPH oxidases for ROS production and contributes to PCD [6]. These results indicate that CPKs are involved in the bifurcation of NLR signalling mechanisms.
The most economically important fruit and ornamental trees and bushes, such as apple (Malus × domestica), pear (Pyrus communis), peach (Prunus persica), cherry (Prunus avium), strawberry (Fragaria spp.), apricot (Prunus armeniaca), almond (Prunus amygdalus) and rose (Rosa hybrida) all belong to the Rosaceae family [25]. M. × domestica is one of the most economically important woody plants cultivated worldwide as a fruit crop, however the function of apple CPKs in the immune response to pathogens has never been reported.
The enterobacterial phytopathogen Erwinia amylovora causes fire blight, an invasive disease that threatens a wide range of commercial and ornamental Rosaceae host plants [26]. It has been difficult to eradicate or reduce the incidence of fire blight epidemics. Management practices include the use of a few size-controlling rootstocks that are resistant to fire blight and chemical treatments to enhance host resistance [26]. Molecular investigations of the E. amylovora-Malus interaction have been limited to a restricted number of plant defences previously characterised in other plant-pathogen interactions [27], or via untargeted analysis [28-31]. These different molecular approaches have provided a long list of up or down regulated genes in susceptible or resistant plants, but have not elucidated the mechanism of apple susceptibility or resistance to fire blight.
Here we undertook a genome wide study to identify and to classify the CPKs involved in the defence response of M. × domestica against the pathogen E. amylovora. A gene encoding CPK was shown to be up-regulated in the blossom of susceptible apple cultivars after E. amylovora infection, suggesting that Ca2+ may be one of the key signals that initiates stress resistance reactions in blossom [31]. In order to identify genes implicated in the control of fire blight resistance, we evaluated [Ca2+]cyt, the role of CPKs in early signalling cascades in the cultivars Golden delicious 'GD' (susceptible) and 'M.7' (resistant) [28] following challenge with a virulent strain of E. amylovora (Ea273) or mechanical damage.
The purpose of this study was to understand the mechanisms of interaction between M. × domestica and E. amylovora in resistant and susceptible apple cultivars. The results will help to design new strategies to improve apple resistance to E. amylovora and facilitate development of resistant transgenic lines for economically important susceptible cultivars.
Results
MdCPK gene family is distributed in 14 out of 17 chromosomes
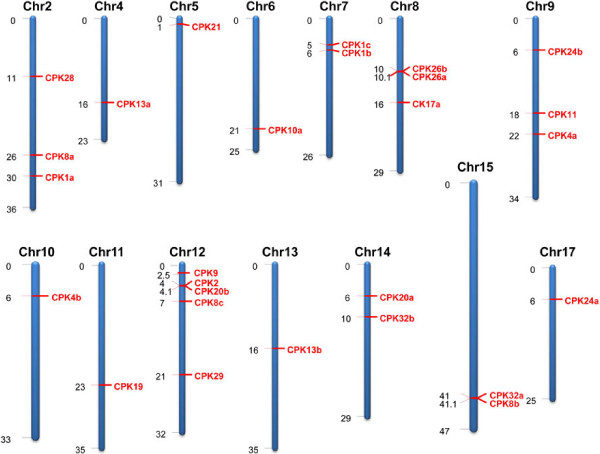
M. × domestica has a diploid genome that underwent a whole genome duplication for 50 million years ago. It has x = 17 chromosomes containing 26,374 loci and 63,541 transcripts, organised in a 881.3 Mb genome [32,33]. A genome-wide search for memebrs of the MdCPK gene family led to identification of 30 CPK genes. Most of the CPK genes have alternative transcript variants. MdCPK11 has 15 possible transcript variants (Table 1). Transcript organisation of MdCPKs shows an average of 8 exons per gene, with the exception of MdCPK11, which has no introns (Table 1, Additional file 1: Figure S1). CPK genes are distributed in 14 of the 17 apple chromosomes (Figure 1). Most CPK genes are present in clusters rather than displaying tandem distribution. Chromosome 12 contains five MdCPKs (MdCPK2, MdCPK8c, MdCPK9, MdCPK20b, and MdCPK29) whereas, chromosome 8 and 9 contain three CPKs (MdCPK17a, MdCPK26a, MdCPK26b and MdCPK4a, MdCPK11, MdCPK24b respectively).
Table 1.
Phytozome locus ID and transcript details of Malus CPKs
| Sl.No | Phytozome locus ID | Location in chromosome | Gene name | ORF | No.of a.a | No.of Introns | No.of alternative splicing variants | 5′-3′Coordinates | Phylogenetic group |
|---|---|---|---|---|---|---|---|---|---|
| 1 |
MDP0000153100 |
2 |
MdCPK1a |
1694 |
566 |
6 |
5 |
MDC017159.84: 8453 - 14350 |
I |
| 2 |
MDP0000142687 |
7 |
MdCPK1b |
1763 |
618 |
8 |
5 |
MDC021045.283: 1756 - 8958 |
I |
| 3 |
MDP0000128057 |
7 |
MdCPK1c |
1943 |
660 |
8 |
5 |
MDC013839.354: 42 - 7131 |
I |
| 4 |
MDP0000232344 |
12 |
MdCPK2 |
2296 |
775 |
8 |
4 |
MDC012227.366: 34198 - 38360 |
I |
| 5 |
MDP0000260834 |
9 |
MdCPK4a |
1553 |
517 |
6 |
4 |
MDC020449.143: 14625 - 18251 |
I |
| 6 |
MDP0000232885 |
10 |
MdCPK4b |
1544 |
518 |
6 |
2 |
MDC010220.255: 18291 - 21903 |
I |
| 7 |
MDP0000269423 |
2 |
MdCPK8a |
1612 |
553 |
8 |
3 |
MDC001073.515: 2333 - 8854 |
IV |
| 8 |
MDP0000119457 |
15 |
MdCPK8b |
1417 |
476 |
6 |
3 |
MDC001073.498: 3281 - 6157 |
IV |
| 9 |
MDP0000260857 |
12 |
MdCPK8c |
1881 |
665 |
9 |
5 |
MDC021346.204: 29191 - 34837 |
IV |
| 10 |
MDP0000169895 |
12 |
MdCPK9 |
1451 |
491 |
2 |
4 |
MDC003603.228: 1126 - 2789 |
I |
| 11 |
MDP0000218522 |
6 |
MdCPK10a |
1692 |
570 |
7 |
5 |
MDC020438.169: 10660 - 14149 |
IV |
| 12 |
MDP0000301254 |
Unanchored |
MdCPK10b |
1618 |
548 |
7 |
8 |
MDC016267.124: 15630 - 19053 |
IV |
| 13 |
MDP0000308706 |
Unanchored |
MdCPK10c |
1613 |
548 |
7 |
8 |
MDC020438.160: 35695 - 39116 |
IV |
| 14 |
MDP0000494270 |
9 |
MdCPK11 |
1489 |
498 |
0 |
15 |
MDC010082.361: 3158 - 4654 |
I |
| 15 |
MDP0000164868 |
4 |
MdCPK13a |
1757 |
585 |
8 |
4 |
MDC000306.525: 1570 - 6773 |
IV |
| 16 |
MDP0000649496 |
13 |
MdCPK13b |
1023 |
345 |
4 |
2 |
MDC000271.449: 354 - 2825 |
IV |
| 17 |
MDP0000802997 |
8 |
MdCPK17a |
1591 |
533 |
7 |
2 |
MDC040478.10: 1862 - 4930 |
II |
| 18 |
MDP0000138436 |
Unanchored |
MdCPK17b |
1605 |
534 |
7 |
3 |
MDC010071.376: 1022 - 3758 |
II |
| 19 |
MDP0000180811 |
11 |
MdCPK19 |
1496 |
504 |
9 |
1 |
MDC008434.490: 2551 - 5781 |
II |
| 20 |
MDP0000318339 |
14 |
MdCPK20a |
2994 |
1023 |
10 |
3 |
MDC031256.8: 21258 – 31086 |
I |
| 21 |
MDP0000513005 |
12 |
MdCPK20b |
1963 |
679 |
7 |
0 |
MDC008272.442: 6235 - 19231 |
I |
| 22 |
MDP0000232001 |
5 |
MdCPK21 |
1641 |
554 |
7 |
3 |
MDC002417.261: 24324 - 28052 |
II |
| 23 |
MDP0000262701 |
17 |
MdCPK24a |
1623 |
541 |
7 |
3 |
MDC020007.246: 24451 - 27032 |
IV |
| 24 |
MDP0000282003 |
9 |
MdCPK24b |
2860 |
954 |
12 |
2 |
MDC006465.419: 8202 - 16334 |
IV |
| 25 |
MDP0000297184 |
8 |
MdCPK26a |
1685 |
571 |
6 |
3 |
MDC012276.352: 7244 - 10346 |
I |
| 26 |
MDP0000457940 |
8 |
MdCPK26b |
4152 |
1403 |
8 |
0 |
MDC001323.383: 1559 - 7846 |
I |
| 27 |
MDP0000208913 |
2 |
MdCPK28 |
1861 |
626 |
13 |
10 |
MDC018730.149: 4526 - 9378 |
III |
| 28 |
MDP0000142398 |
12 |
MdCPK29 |
1584 |
527 |
7 |
4 |
MDC015573.110: 52421 - 55302 |
II |
| 29 |
MDP0000649508 |
15 |
MdCPK32a |
2081 |
709 |
10 |
4 |
MDC001801.279: 799 - 8644 |
IV |
| 30 | MDP0000179069 | 14 | MdCPK32b | 2011 | 676 | 10 | 3 | MDC006959.379: 1716 - 6520 | IV |
Figure 1.
Genomic distribution of MdCPK genes in Malus chromosomes. The number in brackets shows the position of the gene on the Malus chromosome pseudomolecules. The chromosome numbers are indicated at the top of each bar. Figure show, MdCPK genes are distributed evenly in different chromosome.
Phylogenetic analysis shows that MdCPKs are clustered into four clades
The Malus MdCPK amino acid sequence length ranged from 345 (MdCPK13b) to 1403 amino acids (MdCPK26b). Cluster analysis identified thirty MdCPKs nested into four distinct clades, as shown in Figure 2. A phylogenetic study of MdCPKs with orthologous A. thaliana, O. sativa, P. trichocarpa and Z. mays also clustered into four clades. MdCPKs are closely related to AtCPK and PtCPKs and that the proposed nomenclature for MdCPKs is consistent. The OsCPKs and ZmCPKs are less closely related to MdCPKs indicating their dicot-specific origin (Figure 2).
Figure 2.
Phylogenetic tress of Malus CPKs with orthologous CPKs of Arabidopsis thaliana (AtCPKs), Oryza sativa (OsCPKs), Populous trichocarpa (PtCPKs) and Zea mays (ZmCPKs). Phylogenetic trees show, all the CPKs are clustered into four different groups and MdCPKs genes are found to be much close to AtCPKs. Phylogenetic tree was constructed by MEGA5 software and statistical method used was Neighbor-joining, substitutition type: amino acid, Model: Jones-Taylor-Thornton (JTT) and no. of bootstrap replication was 500.
All MdCPKs have an EF-hand domain and palmitoylation sites
Ca2+ signals are decoded by many different protein kinases, and CPKs play significant roles in these signalling events [24,34,35]. The Ca2+ binding EF-hands are the predominant Ca2+ sensors, consisting of twelve residue loops, flanked on both sides by twelve alpha-helical domain residues [Additional file 2: Figure S2].
In response to E. amylovora and mechanical damage, MdCPKs are differentially expressed in resistant 'M.7' and susceptible 'G.D' M. x domestica cultivars
To clarify MdCPKs role in the resistance and susceptibility of M. × domestica to E. amylovora and mechanical damage (MD), we carried out a comparison between CPK gene expression patterns in the resistant Malling7 apple rootstock (M.7) and the susceptible golden delicious (GD) by using quantitative real time PCR analysis (qPCR) at 2, 6, 12 and 24 hours post inoculation (hpi) (Figure 3). These time points were selected based on previous analyses of the temporal transcriptional response of Malus to E. amylovora, indicating that basal defence to pathogen associated molecular patterns (PAMPs) occurred within 1–2 hpi, whereas expression of defence proteins occurred at 24–48 hpi [28]. These two genotypes were chosen based on their level of resistance and susceptibility to fire blight disease. M.7 is a highly resistant genotype whereas GD is a susceptible genotype to fire blight disease.
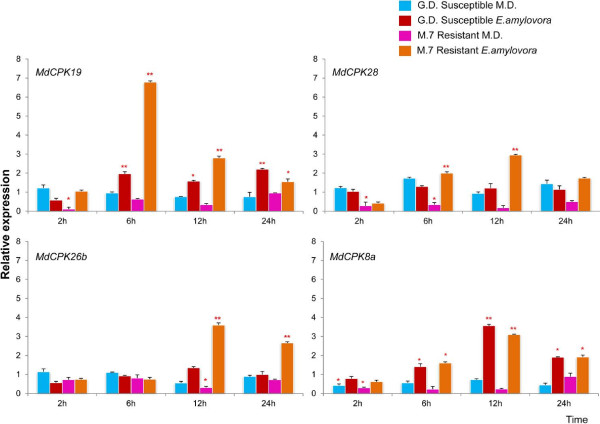
Figure 3.
Quantitative RT-PCR comparison of resistant and susceptible Malus cultivars after E. amylovora infection and mechanical damage at 2, 6 12 and 24 hpi. The transcript level of genes in resistant/susceptible cultivars at 2,6,12 and 24 hpi were normalised with those of EF1 and UB1 measured in the samples and expressed in relation to the normalised transcript level in the leaves of the respective uninfected plants (0 hrs). Metric bars represent the standard error (SE). Asterisks indicate significant differences: * P < 0.05, ** P < 0.01.
Few of the MdCPKs were up-regulated in the M.7 resistant genotype as compared to GD susceptible plants. Of the thirty MdCPKs analysed by qPCR, only eleven showed differential expression in susceptible and resistant apple genotypes following E. amylovora infection or MD (Figures 3, 4 and 5).
Figure 4.

Quantitative RT-PCR comparison of resistant and susceptible Malus cultivars after E. amylovora infection and mechanical damage at 2, 6, 12 and 24 hpi. The transcript level of genes in resistant/susceptible cultivars at 2,6,12 and 24 hpi were normalised with those of EF1 and UB1 measured in the samples and expressed in relation to the normalised transcript level in the leaves of the respective uninfected plants (0 hrs).Metric bars represent the standard error (SE). Asterisks indicate significant differences: * P < 0.05, ** P < 0.01.
Figure 5.
Quantitative RT-PCR comparison of resistant and susceptible Malus cultivars after E. amylovora infection and mechanical damage at 2, 6, 12 and 24 hpi. The transcript level of the genes in resistant/susceptible at 2,6,12 and 24 hpi were normalised with those of EF1 and UB1 measured in the samples and expressed in relation to the normalised transcript level in the leaves of the respective uninfected plants (0 hrs). Metric bars represent the standard error (SE). Asterisks indicate significant differences: * P < 0.05, ** P < 0.01.
Four MdCPKs were specifically induced after infection with E. amylovora (Figure 3). In the resistant genotype following E. amylovora infection, MdCPK19 and MdCPK28 were significantly up regulated at 6 and 12 hpi, whereas MdCPK26b was up regulated at 12 and 24 hpi. Following E. amylovora infection, MdCPK8a was similarly up regulated at 6, 12 and 24 hpi in both resistant and susceptible cultivars. None of the CPK genes were activated after MD (Figure 3). However, MdCPK8b was specifically induced in response to MD in the susceptible genotype at 2 and 6 hpi, whereas the resistant genotype showed no induction after either E. amylovora infection or MD (Figure 4).
Six other MdCPKs were differentially expressed in resistant and susceptible cultivars following E. amylovora infection and/or MD at different time points (Figure 5). In the resistant genotype, four CPKs (MdCPK1b, MdCPK1c, MdCPK9 and MdCPK29) were significantly up regulated at different time points following E. amylovora infection (Figure 5). It is interesting to note that in response to E. amylovora infection, MdCPK1b was up-regulated at later than MdCPK1c and that both genes were up-regulated later following MD than E. amylovora infection (24 hpi, Figure 5). The susceptible genotype showed a significant up regulation of MdCPK4b and MdCPK11 (except for 12 hpi) following MD (Figure 5).
E. amylovora induced differential intracellular cytosolic calcium variations in susceptible and resistant M. x domestica genotypes
CPK activity is often associated with variations in [Ca2+]cyt[3,35-38]. Having determined that some MdCPK genes are differentially expressed following E. amylovora infection in resistant and susceptible M. × domestica cultivars, we evaluated [Ca2+]cyt by Confocal Laser Scanning Microscopy (CLSM) with the calcium indicator, calcium orange. In the susceptible genotype, [Ca2+]cyt was found to decrease in both MD (Figure 6, A-C) and E. amylovora infected leaves (Figure 6, D-F) from 1 to 6 hpi. Conversely, the resistant genotype showed a consistent and significant (p < 0.05) increase in [Ca2+]cyt over the same time period. In particular, E. amylovora infected leaves (Figure 6, M-O) showed a higher [Ca2+]cyt level when compared to both infected (Figure 6, D-F) and MD resistant genotype leaves (Figure 6, H-L). A higher magnification of resistant genotype leaves at 6 hpi showed a clear cytosolic localization of Ca2+ (Figure 6, P), which is more evident than in the susceptible genotype infected leaves (Figure 6, G). Figure 6 also shows the relative percentage of calcium orange fluorescence in both resistant and susceptible apple cultivars upon MD and E. amylovora infection.
Figure 6.
Intracellular Ca2+variations in GD and M7 leaf cells following pathogen infection and mechanical damage. Leaves were treated with calcium orange for 1 hr and then infected with E. amylovora. The cytosolic Ca2+ concentration of leaf cells was determined 1 hr, 2 hr and 6 hrs after infection. Mechanically damaged (MD) leaves served as a control for both genotypes. In the lower panel, the bar represents the relative percentage of calcium orange fluorescence in at least 3 replicates. Asterisk indicate significant (p <0.05) differences. Scale bars for Figures A-F and H-O = 100 μm, scale bars for Figures G and P = 400 μm.
Discussion
Regulation of Ca2+ homeostasis is important, particularly when Ca2+ is involved as a signalling ion. In plant cells, Ca2+-binding proteins also serve as regulators of internal free Ca2+ levels [4,5,38,39]. Since CPKs may be involved in the specificity and cross-talk of signal transduction in a variety of biotic and abiotic stresses, their possible involvement in active signalling cascades in response to pathogens deserves a thorough investigation. Recent expression profiles of M. × domestica blossom–Erwinia interaction revealed the involvement of CPKs in the signal transduction process [31]. However, a detailed study on the involvement of the MdCPK gene family in resistant and susceptible apple plants is lacking.
This work provides fundamental information on the phylogeny, gene structure, and gene expression of MdCPKs in response to pathogen and wound signalling in fire blight resistant and susceptible apple cultivars. The M. × domestica (GD) genome sequence is assembled in 21,554 scaffolds and different gene families reside within these scaffolds. The CPK gene family is one of them and is evenly distributed throughout the 17 pseudomolecules of the GD genome sequence. A global survey of the recent apple genome database reveals the presence of 30 MdCPKs from 57,386 annotated genes in the apple genome [40]. All the MdCPKs analysed here have the typical structures of the CPK family, including an N-terminal variable domain, a protein kinase domain, an auto-inhibitory domain, a calmodulin-like domain, an EF-hand like domain and a C-terminal domain. The calcium binding EF-hands are the predominant Ca2+ sensors.
Comparative plant genomics studies show that plant gene families are largely conserved over evolutionary timescales, including diversification of angiosperm and non-flowering plants [41]. Co-linearity resulting from the common ancestors of the angiosperms provides a powerful way of determining orthology, while comparative sequence analyses provides a wealth of information about the nature of sequence arrangement and evolution [42]. In this study, comparative sequence analysis of the orthologous protein sequences of Malus CPKs in relation to A. thaliana and P. trichocarpa CPKs showed a high level of conservation and significant orthology compared to O. sativa and Z. mays CPKs[12,13]. Improved orthologous gene detection is critically important for accurate functional annotation and study of comparative and evolutionary genomics. Besides this, all the 30 Malus CPKs are highly homologous to each other. Furthermore, the similarity found between MdCPK gene family with AtCPKs shows that Malus and Arabidopsis CPKs may derive from a common ancestor. Despite this evolutionary conservation of gene families, lineage-specific fluctuations in gene family size are frequent among taxa [41,43].
In this study we found that in group III there is only one Malus CPK (MdCPK28) present in the phylogenetic tree as compared to three from Arabidopsis, four from Zea, four from Oryza and two from Populous. The presence of MdCPK (MdCPK28) in group III was very divergent from other MdCPKs and may have evolved in Rosaceae following divergence with a distinct dicot specific function.
In the EF-hand loop, Ca2+ is coordinated in a pentagonal bi-pyramidal configuration [44]. The six residues involved in Ca2+ binding are 1, 3, 5, 7, 9 and 12. The invariant Glu (E) or Asp (D) amino acids at position 12 provide two O2 that can bind Ca2+ ions. The variable N-terminal domain contains myristoylation or palmitoylation sites. Palmitoylation is the reversible covalent attachment of palmitic acid to cysteine and less frequently to serine or threonine residues of proteins. Palmitoylation enhances the hydrophobicity of proteins and helps association with membranes (as well as sub-cellular trafficking between membrane compartments) and helps protein-protein interactions [45]. All MdCPKs here reported contain palmitoylation sites, usually present at the 4th or 5th position of the N-terminal end (Table 2).
Table 2.
Prediction of putative palmitoylation sites of MdCPKs using CSS-palm 3.0
| Gene | Position | Sequence | Score | Cutoff |
|---|---|---|---|---|
| MdCPK1a |
5 |
***MGNTCVGPSISK |
1.467 |
0.196 |
| MdCPK1b |
5 |
***MGNTCVGPSISK |
1.576 |
0.196 |
| MdCPK1c |
5 |
***MGNTCVGPSISK |
1.576 |
0.196 |
| MdCPK2 |
10 |
PRDDQIGCQXYLQLS |
2.645 |
1.225 |
| MdCPK4a |
37 |
QFGTTYLCTHKPTGA |
0.152 |
0 |
| MdCPK4b |
44 |
QFGTTYLCTHKPTGA |
0.157 |
0 |
| MdCPK8a |
93 |
EFGVTYLCTEASSNE |
0.224 |
0.196 |
| MdCPK8b |
4 |
****MGNCCVTLGAP |
3.132 |
1.225 |
| MdCPK8c |
4 |
****MGNCCATPQTG |
2.814 |
0.308 |
| MdCPK9 |
11 |
KATPSTICSTXASDL |
1.43 |
1.22 |
| MdCPK10a |
4 |
****MGNCNVCVRAD |
2.777 |
1.225 |
| MdCPK10b |
4 |
****MGNCNVCVRAD |
3.132 |
1.225 |
| MdCPK10c |
4 |
****MGNCNVCVRAD |
3.132 |
1.22 |
| MdCPK11 |
48 |
QFGTTYLCTEISSGH |
0.471 |
0 |
| MdCPK13a |
4 |
****MGNCCRSPAAV |
2.824 |
0.308 |
| MdCPK13b |
28 |
VILYILLCGVPPFWA |
0.219 |
0.196 |
| MdCPK17a |
4 |
****MGNCCSQCNTE |
3.567 |
0.308 |
| MdCPK17b |
4 |
****MGNCCSQRNTE |
4.248 |
0.308 |
| MdCPK19 |
139 |
RGQAVCPCLYGAGEL |
0.907 |
0.497 |
| MdCPK20a |
91 |
ITSRQFVCAHQGKHV |
0.357 |
0.308 |
| MdCPK20b |
198 |
QFGTTFLCVEKETNK |
0.31 |
0.308 |
| MdCPK21 |
3 |
*****MGCYSSKENA |
2.319 |
0.308 |
| MdCPK24a |
4 |
****MGSCLCTPANA |
0.943 |
0.308 |
| MdCPK24b |
4 |
****MGSCVCTPAKA |
4.019 |
0.497 |
| MdCPK26a |
5 |
***MGNTCRGSFRGK |
2.11 |
0.308 |
| MdCPK26b |
26 |
IGTPLYLCCRSLTFS |
1.657 |
0.308 |
| MdCPK28 |
4 |
****MGICFSAVKVS |
4.727 |
1.225 |
| MdCPK29 |
4 |
****MGLCFTKCQSH |
1.514 |
0.308 |
| MdCPK32a |
4 |
****MGNCCVTLGAP |
3.132 |
1.225 |
| MdCPK32b | 4 | ****MGNCCVTPQTG | 2.252 | 0.308 |
The prediction showed that all MdCPKs identified had palmitoylation sites characterised by the presence of cysteine residues at the N-terminal end, usually in positions 4 and 5.
Presence of “****” indicate palmitoylation site present at 4th position and “***” indicate palmitoylation site present at 5th position of respected CPK gene.
In eukaryotes and higher plants, some genes are spliced alternatively during various developmental stages or in response to stresses, creating multiple mRNA transcript for a single gene [46]. Spliced genes may encode proteins with different functions or different cellular or sub-cellular localizations [47]. In this study, MdCPKs were found to have several alternative spliced transcript variants (Table 1). The majority of plant alternative spliced transcripts have not yet been functionally characterised, but the evidence suggests that alternative splicing plays a major role in plant function, including stress response, and may impact domestication and trait selection [48]. Splicing variants play important roles within cells and increase proteome diversity and cellular function [49]. Thus, the presence of a significant number of alternative splicing variants in Malus might explain its domestication and resistance to stress response. Further studies are necessary to better understand their independent role in different stress responses.
Our study also provides information on the possible involvement of MdCPKs in regulating E. amylovora infection and wound response via Ca2+-mediated signalling. The differential expression of MdCPKs in fire blight resistant and susceptible M. × domestica cultivars shows the involvement of CPKs in the regulation of E. amylovora infection and/or to MD The selective expression of a few CPKs in the resistant cultivar in response to E. amylovora indicates the importance of these CPKs in modulating the resistance/susceptibility mechanisms by transducing the signal to downstream defence signalling pathways [3,4,38].
The early induction of a few CPKs observed, specifically, in the resistant cultivar, indicates they may play an important role in recognising pathogen infection and transducing the signals to downstream signalling cascades. These data show a divergent role for CPKs in response to various stimuli and their specific recognition [4,6,50,51].
[Ca2+]cyt variations occur in response to various biotic and abiotic stresses [3,4,52-55]. In our study we found that the M.7 resistant cultivar showed a significantly higher [Ca2+]cyt accumulation to E. amylovora infection and MD, whereas the GD susceptible cultivar showed a decreased [Ca2+]cyt accumulation. These [Ca2+]cyt differences between the M.7 and GD cultivars in response to E. amylovora infection show the ability of the resistant plant to recognise E. amylovora infection by significantly inducing [Ca2+]cyt accumulation and transducing downstream signalling cascades and are consistent with induction of MdCPKs genes. It has been shown that recognition of the pathogen or its effectors increases [Ca2+]cyt elevation in plant cells, which is a prerequisite for hypersensitive response development [56-58]. Despite a significant correlation between Ca2+ influxes and pathogen recognition, how the Ca2+ signal is transduced to downstream signalling events remains elusive. However, recent discoveries have identified six closely related CPKs in Arabidopsis (i.e. CPKs 1, 2, 4, 5, 6 and 11, all of them belonging to cluster I) as sensors and transducers of Ca2+ signalling triggered by recognition of pathogen effectors [6,24]. In our study, we found that most of the CPKs (such as MdCPK1b, 1c, 4b and 11) were differentially expressed in resistant and susceptible cultivars all belong to cluster I, indicating the importance of this cluster in the mechanism of resistance to the E. amylovora pathogen. Preliminary data has shown down regulation of some CPK genes in the flower of susceptible Malus after inoculation with E. amylovora[31].
Conclusions
Our data can be used to further extend our understanding of the downstream signalling network in fire blight resistant and susceptible apple cultivars by mutant and overexpressing candidate Malus CPKs analyses. Since Ca2+ and its binding proteins are involved in early recognition of pathogen infection and signal transduction to downstream target molecules [24], it would be interesting to understand downstream target genes and the possible role of phytohormones in regulating pathogen and wound defence mechanisms. We identified a few candidate CPKs which are specific to M7 and GD M. × domestica cultivars. Overexpression or silencing of these CPKs might modulate the resistance to E. amylovora infection. This study provides new tools for clarifying important signalling molecules in regulating the most devastating disease of Malus and other Rosaceae host plants.
Methods
Plant material and pathogen inoculation
One year old plants of Malus x domestica cv Golden Delicious (GD) and own-rooted M.7 rootstock, were grown in the greenhouse at 24°C. Erwinia amylovora strain Ea273 was grown overnight at 28°C in Kado medium [59] supplemented with 0.3 g/L MgSO4. The inoculum concentration was adjusted to 109 cfu ml-1 by dilution with sterile 0.05 M potassium phosphate buffer, pH 6.5. The youngest actively growing leaves of plants were transversally cut using scissors dipped in the bacteria suspension or phosphate buffer as a mechanical damage control [28]. Six plants were inoculated with Erwinia amylovora for each time point. Four to six mm wide leaf strips, parallel to the original cut, were collected according to the symptom progression at 0, 2, 6, 12, 24, and 48 hours post inoculation (hpi), frozen in liquid nitrogen and stored at -80°C.
Database search and identification of Malus CPKs
Calcium dependent protein kinase (CPK) genes from Malus x domestica were downloaded from the publicly available phytozome (http://www.phytozome.net, http://www.rosaceae.org) database using the hidden Markov model approach as well as the BLASTP protocol [32,33,60]. The BLASTP results are provided in supplementary Additional file 3: Table S1. CPK genes from Arabidopsis thaliana were used as query sequences to search Malus CPK genes. A. thaliana CPK genes were downloaded from “The Arabidopsis Information Resources” (TAIR) (http://www.arabidopsis.org) [44]. All sequences were confirmed by carrying out a BLASTP run against the TAIR database. Malus x domestica CPKs, which gave a BLASTP hit with Arabidopsis CPKs, were considered as Malus CPKs and the nomenclature was thus carried out accordingly. All the CPKs of M. x domestica were scanned using SCAN PROSITE software to confirm the presence of the EF-hands signature motif and hence CPK genes (http://prosite.expasy.org/scanprosite/) [61]. Identified Malus CPKs genes were aligned using CLUSTALW software, using BLSOUM62 software with gap open 10, gap extension 0.20, gap distance 5 and clustering neighbour joining [62] to find out the conserved EF-hand domains. Palmytoilation sites of CPKs were predicted using CSS palm software [63,64]. The protein sequences were carefully analysed for sequence redundancy followed by removal of alternatively spliced variants. In order to confirm the presence of alternatively spliced gene sequences, the genomic sequence of each candidate gene was also examined. Sequence similarity of Malus CPK genes was carried out using online software EMBOSS Needle (http://www.ebi.ac.uk/Tools/psa/emboss_needle/).
Chromosomal location
The phytozome (http://www.phytozome.net/, http://www.rosaceae.org) database was used for identification of putative MdCPKs. Each of the MdCPKs was positioned on the M. x domestica chromosome pseudo molecules using the apple genome browser (http://genomics.research.iasma.it/gb2/gbrowse/apple/).
Phylogenetic analysis of the MdCPK gene family
Multiple sequence alignment analysis carried out using CLUSTALW was used to construct the phylogenetic tree. The CPKs of Oryza sativa, A. thaliana, Populus trichocarpa, Zea mays and M. x domestica were used to construct the phylogenetic tree with MEGA software, version 5, using the neighbour joining statistical method and Jones-Taylor-Thornton (JTT) model [65].
RNA isolation and q-PCR
Total RNA from leaves was isolated using the Sigma Spectrum™ plant total RNA kit protocol. Before cDNA synthesis, RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA) according to the manufacturer’s instructions to ensure no DNA contamination, and first-strand cDNA synthesis was then carried out with approximately 1 μg RNA using an Invitrogen Superscript VILO™ First Strand cDNA Synthesis Kit and oligo-dT primers according to the manufacturer’s procedure. Primers were designed using Primer3 v. 0.4.0 (http://frodo.wi.mit.edu/) with melting temperatures at 58–60°C, primer lengths 20–24 bp and amplicon lengths 250–300 bp. All the primer sequences are listed in Additional file 4: Table S2. q-PCR was conducted on a Biorad iCycler® App 9001 Detection System using SYBR GreenER™ q-PCR supermix (Invitrogen). Reactions were prepared in a total volume of 20 μl containing: 10 μl of 2xSYBR Premix, 2 μl of cDNA template, 0.4 μl of each specific primer to a final concentration of 200 nM. The reactions were performed in the following conditions: initial denaturation step of 95°C for 10 s followed by two-step thermal cycling profile of denaturation at 95°C for 5 s, and combined primer annealing/extension at 60°C for 1 min for 40 cycles. No-template controls were included for each primer pair and each PCR reaction was performed in triplicate on 2 biological replicates. To verify the specificity of the amplicon for each primer pair, melting curve analysis was performed ranging from 60 to 95°C, with temperature increasing steps of 0.06°C/s (five acquisitions per °C) at the end of each run. Baseline and threshold cycles (Ct) were automatically determined using Biorad iCycler® IQ5 Software. Relative expression was calculated as described previously using EF1 and UB1 as the reference gene [66,67].
Determination of intracellular calcium variations using confocal laser scanning microscopy (CLSM) and calcium orange
Calcium orange dye (stock solution in DMSO, Molecular Probes) was diluted in 5 mM MES-Na buffer (pH 6.0) to a final concentration of 5 μM. This solution was applied to intact M. x domestica leaves as detailed in [68]. Five μM calcium orange solution was applied and after 60 min the leaf was mounted on a Nikon Eclipse C1 spectral CLSM stage, without separating the leaf from the plant, to assess basic fluorescence levels as a control. The microscope operated with a Krypton/Argon laser at 488 nm with a BP of 500–540 nm and a LP of 650 nm. Digital images were analysed using NIH image software as described earlier [53]. After pathogen inoculation (see above) or mechanical damage performed with scissors, leaves were perfused with calcium orange and analysed using CLSM as described above. Controls were represented by application of 5 μM calcium orange solution to intact leaves. At least 5 biological replicates were performed and several images taken for each biological replicate.
Data and statistical analysis
At least 2 biological replications and 3 technical replication sets were used for the statistical treatment of data. The data are expressed as mean values; error bars indicate the standard error. To evaluate the significance of differences in data, ANOVA followed by Fisher’s PLSD test was performed.
Abbreviations
CPKs: Calcium-dependent protein kinases; MdCPK: Malus x domestica CPK; Ca2+: Calcium; [Ca2+]cyt: Cytosolic calcium concentration; Hpi: Hours post inoculation; CaM: Clamodulins; CBL: Calcineurin B-like proteins; MAPK: Mitogen activated protein kinase; MD: mechanical damage.
Competing interests
The authors declare that there are no competing interests.
Authors’ contributions
Conception and design of the experiments: CNK, TKM, MM, MEM. Carrying out of experiments: CNK, TKM, AC, AO, FV. Analysis of data: CNK, MEM, MM. Provision of reagents/materials/analysis tools: CNK, MM, MEM. Writing of the paper: CNK, TKM, MEM, MM. All authors read and approved the final manuscript.
Supplementary Material
Multiple sequence alignment of MdCPK genes. Amino acid sequence alignment of MdCPK genes show presence of kinase domain and four calcium binding EF-hands in regulatory domain. In EF-hands, Ca2+ ion are co-ordinated in a pentagonal bipyramidal configuration. Ca2+ binding amino acid residue are present at position 1, 3, 5, 7, 9 and 12. The conserved Glu (E) or Asp (D) provides two oxygen for liganding Ca2+. Multiple sequence alignment of MdCPK genes were carried out using multalin (http://multalin.toulouse.inra.fr/multalin) software using statistical programme BLOSUM. Red and blue color indicate high and low conserved domains/motifs respectively, whereas black indicate neutral.
Schematic representation of transcript of MdCDPK genes. Box mark represents the exon and line represents the intron of specific CDPK gene. The name to the right of the gene structure indicates the gene name.
Q PCR Primer list of all MdCDPK genes used in this study.
The BLASTP score of MdCPKs found during their identification. The E- value found during BLASTP search show very significant similarity.
Contributor Information
Chidananda Nagamangala Kanchiswamy, Email: chidananda.nagamangala@fmach.it.
Tapan Kumar Mohanta, Email: nostoc.tapan@gmail.com.
Andrea Capuzzo, Email: andrea.capuzzo@unito.it.
Andrea Occhipinti, Email: andrea.occhipinti@unito.it.
Francesca Verrillo, Email: francesca.verrillo@unito.it.
Massimo E Maffei, Email: massimo.maffei@unito.it.
Mickael Malnoy, Email: mickael.malnoy@fmach.it.
Acknowledgements
The research leading to these results has received funding from the Autonomous Province of Trento, with reference to the Call 1 - post-doc 2012 – Incoming, approved with provincial government resolution no. 1023 of 5/7/10.
References
- Harper JF, Harmon A. Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol. 2005;6(7):555–566. doi: 10.1038/nrm1679. [DOI] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 1991;352(6335):524–526. doi: 10.1038/352524a0. [DOI] [PubMed] [Google Scholar]
- Kanchiswamy CN, Takahashi H, Quadro S, Maffei ME, Bossi S, Bertea C, Zebelo SA, Muroi A, Ishihama N, Yoshioka H. et al. Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 2010;10:97. doi: 10.1186/1471-2229-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura G, Maffei ME. Calcium and secondary CPK signaling in plants in response to herbivore attack. Biochem Biophys Res Commun. 2010;400(4):455–460. doi: 10.1016/j.bbrc.2010.08.134. [DOI] [PubMed] [Google Scholar]
- Ma W. Roles of Ca2+ and cyclic nucleotide gated channel in plant innate immunity. Plant Sci: Int J Exp Plant Biol. 2011;181(4):342–346. doi: 10.1016/j.plantsci.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J. et al. Bifurcation of Arabidopsis NLR immune signaling via Ca(2)(+)-dependent protein kinases. PLoS Pathog. 2013;9(1):e1003127. doi: 10.1371/journal.ppat.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF. Communicating with calcium. The Plant cell. 1999;11(4):691–706. doi: 10.1105/tpc.11.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Sebastia C, Hardin SC, Clouse SD, Kieber JJ, Huber SC. Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch Biochem Biophys. 2004;428(1):81–91. doi: 10.1016/j.abb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR. et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132(2):666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell. 2002;14(Suppl):S389–400. doi: 10.1105/tpc.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithofer A. CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 2012;159(3):1159–1175. doi: 10.1104/pp.112.198150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Liu J, Yang X, Ma R. Genome-wide identification of the maize calcium-dependent protein kinase gene family. Appl Biochem Biotechnol. 2013;169(7):2111–2125. doi: 10.1007/s12010-013-0125-2. [DOI] [PubMed] [Google Scholar]
- Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, Zhou G. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa) Mol Biol Rep. 2013;40(3):2645–2662. doi: 10.1007/s11033-012-2351-z. [DOI] [PubMed] [Google Scholar]
- Li AL, Zhu YF, Tan XM, Wang X, Wei B, Guo HZ, Zhang ZL, Chen XB, Zhao GY, Kong XY. et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.) Plant Mol Biol. 2008;66(4):429–443. doi: 10.1007/s11103-007-9281-5. [DOI] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129(2):469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol. 2004;55:263–288. doi: 10.1146/annurev.arplant.55.031903.141627. [DOI] [PubMed] [Google Scholar]
- Harmon AC, Yoo BC, McCaffery C. Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33(23):7278–7287. doi: 10.1021/bi00189a032. [DOI] [PubMed] [Google Scholar]
- Ito T, Nakata M, Fukazawa J, Ishida S, Takahashi Y. Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca(2+)-dependent protein kinase is important for substrate recognition. Plant Cell. 2010;22(5):1592–1604. doi: 10.1105/tpc.109.073577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camoni L, Harper JF, Palmgren MG. 14-3-3 proteins activate a plant calcium-dependent protein kinase (CDPK) FEBS Lett. 1998;430(3):381–384. doi: 10.1016/S0014-5793(98)00696-6. [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR. et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 2006;4(10):e327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ. et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19(10):3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Sun HL, Mei C, Wang XJ, Yan L, Liu R, Zhang XF, Wang XF, Zhang DP. The Arabidopsis Ca(2+) -dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 2011;192(1):61–73. doi: 10.1111/j.1469-8137.2011.03793.x. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464(7287):418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, He P. Nuclear dynamics of Arabidopsis calcium-dependent protein kinases in effector-triggered immunity. Plant Signal Behav. 2013;8(4):e23868. doi: 10.4161/psb.23868. PMID: 23425856; http://dx.doi.org/10.4161/psb.23868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Korban SS, Sosinski B, Abbott AG, Aldwinckle HS, Folta KM, Iezzoni A, Main D, Arus P, Dandekar AM. et al. Multiple models for Rosaceae genomics. Plant Physiol. 2008;147(3):985–1003. doi: 10.1104/pp.107.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoy M, Martens S, Norelli JL, Barny MA, Sundin GW, Smits TH, Duffy B. Fire blight: applied genomic insights of the pathogen and host. Annu Rev Phytopathol. 2012;50:475–494. doi: 10.1146/annurev-phyto-081211-172931. [DOI] [PubMed] [Google Scholar]
- Venisse JS, Malnoy M, Faize M, Paulin JP, Brisset MN. Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora. Mol Plant. 2002;15(12):1204–1212. doi: 10.1094/MPMI.2002.15.12.1204. [DOI] [PubMed] [Google Scholar]
- Baldo A, Norelli JL, Farrell RE Jr, Bassett CL, Aldwinckle HS, Malnoy M. Identification of genes differentially expressed during interaction of resistant and susceptible apple cultivars (Malus x domestica) with Erwinia amylovora. BMC Plant Biol. 2010;10:1. doi: 10.1186/1471-2229-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner SE, Norelli JL, De Silva N, Fazio G, Peil A, Malnoy M, Horner M, Bowatte D, Carlisle C, Wiedow C. et al. Putative resistance gene markers associated with quantitative trait loci for fire blight resistance in Malus 'Robusta 5' accessions. BMC Genet. 2012;13:25. doi: 10.1186/1471-2156-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Halbrendt N, Fazio G, Makalowska I, Altman N, Praul C, Maximova SN, Ngugi HK, Crassweller RM, Travis JW. et al. Rootstock-regulated gene expression patterns associated with fire blight resistance in apple. BMC Genomics. 2012;13:9. doi: 10.1186/1471-2164-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarowar S, Zhao Y, Soria-Guerra RE, Ali S, Zheng D, Wang D, Korban SS. Expression profiles of differentially regulated genes during the early stages of apple flower infection with Erwinia amylovora. J Exp Bot. 2011;62(14):4851–4861. doi: 10.1093/jxb/err147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(D1):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D. The genome of the domesticated apple (Malus x domestica Borkh.) Nat Genet. 2010;42(10):833-+. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- Zou J-J, Wei F-J, Wang C, Wu J-J, Ratnasekera D, Liu W-X, Wu W-H. Arabidopsis Calcium-Dependent Protein Kinase CPK10 Functions in Abscisic Acid- and Ca2 + -Mediated Stomatal Regulation in Response to Drought Stress. Plant Physiol. 2010;154(3):1232–1243. doi: 10.1104/pp.110.157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Hayashi N, Kikuchi S, Ohsugi R. CDPK-mediated abiotic stress signaling. Plant Signal Behav. 2012;7(7):817–821. doi: 10.4161/psb.20351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Regad L, Lauriere C. Characterization of Arabidopsis calcium-dependent protein kinases: activated or not by calcium? Biochem J. 2012;447(2):291–299. doi: 10.1042/BJ20112072. [DOI] [PubMed] [Google Scholar]
- Holder AA, Mohd Ridzuan MA, Green JL. Calcium dependent protein kinase 1 and calcium fluxes in the malaria parasite. Microbes Infect. 2012;14(10):825–830. doi: 10.1016/j.micinf.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Szczegielniak J, Borkiewicz L, Szurmak B, Lewandowska-Gnatowska E, Statkiewicz M, Klimecka M, Ciesla J, Muszynska G. Maize calcium-dependent protein kinase (ZmCPK11): local and systemic response to wounding, regulation by touch and components of jasmonate signaling. Physiol Plant. 2012;146(1):1–14. doi: 10.1111/j.1399-3054.2012.01587.x. [DOI] [PubMed] [Google Scholar]
- Zebelo SA, Matsui K, Ozawa R, Maffei ME. Plasma membrane potential depolarization and cytosolic calcium flux are early events involved in tomato (Solanum lycopersicon) plant-to-plant communication. Plant Sci: Int J Exp Plant Biol. 2012;196:93–100. doi: 10.1016/j.plantsci.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D. et al. The genome of the domesticated apple (Malus x domestica Borkh.) Nat Genet. 2010;42(10):833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist EA, Kamisugi Y. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319(5859):64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Ramakrishna W, Dubcovsky J, Park YJ, Busso C, Emberton J, SanMiguel P, Bennetzen JL. Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics. 2002;162(3):1389–1400. doi: 10.1093/genetics/162.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J. et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One. 2007;2(12):e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40(D1):D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PIH. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307(5716):1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- Asano T, Tanaka N, Yang GX, Hayashi N, Komatsu S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: Comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 2005;46(2):356–366. doi: 10.1093/pcp/pci035. [DOI] [PubMed] [Google Scholar]
- Eckardt NA. Alternative splicing and the control of flowering time. The Plant cell. 2002;14(4):743–747. doi: 10.1105/tpc.000000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18(9):1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- Furnham N, Ruffle S, Southan C. Splice variants: A homology modeling approach. Proteins-Structure Function and Genetics. 2004;54(3):596–608. doi: 10.1002/prot.10568. [DOI] [PubMed] [Google Scholar]
- Wurzinger B, Mair A, Pfister B, Teige M. Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal Behav. 2011;6(1):8–12. doi: 10.4161/psb.6.1.14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM. The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol Biol. 2013;82(3):267–278. doi: 10.1007/s11103-013-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa R, Bertea CM, Foti M, Narayana R, Arimura G, Muroi A, Horiuchi J, Nishioka T, Maffei ME, Takabayashi J. Exogenous polyamines elicit herbivore-induced volatiles in lima bean leaves: involvement of calcium, H2O2 and Jasmonic acid. Plant Cell Physiol. 2009;50(12):2183–2199. doi: 10.1093/pcp/pcp153. [DOI] [PubMed] [Google Scholar]
- Mithofer A, Mazars C, Maffei ME. Probing Spatio-temporal Intracellular Calcium Variations in Plants. Methods Mol Biol. 2009;479:79–92. doi: 10.1007/978-1-59745-289-2_5. [DOI] [PubMed] [Google Scholar]
- Maffei ME, Mithofer A, Arimura G, Uchtenhagen H, Bossi S, Bertea CM, Cucuzza LS, Novero M, Volpe V, Quadro S. et al. Effects of feeding Spodoptera littoralis on lima bean leaves. III. Membrane depolarization and involvement of hydrogen peroxide. Plant Physiol. 2006;140(3):1022–1035. doi: 10.1104/pp.105.071993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei M, Bossi S, Spiteller D, Mithofer A, Boland W. Effects of feeding Spodoptera littoralis on lima bean leaves. I. Membrane potentials, intracellular calcium variations, oral secretions, and regurgitate components. Plant Physiol. 2004;134(4):1752–1762. doi: 10.1104/pp.103.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, Mansfield J. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J: Cell Mol Biol. 2000;23(4):441–450. doi: 10.1046/j.1365-313x.2000.00804.x. [DOI] [PubMed] [Google Scholar]
- Ma W, Smigel A, Tsai YC, Braam J, Berkowitz GA. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 2008;148(2):818–828. doi: 10.1104/pp.108.125104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, Von Bodman S, Berkowitz GA. Death don't have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell. 2007;19(3):1081–1095. doi: 10.1105/tpc.106.045096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado CI, Heskett MG. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology. 1970;60(6):969–976. doi: 10.1094/Phyto-60-969. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJA, De Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41(D1):E344–E347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel. 2008;21(11):639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):research0034.0031 - research0034.0011. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricchi I, Occhipinti A, Bertea CM, Zebelo SA, Brillada C, Verrillo F, De Castro C, Molinaro A, Faulkner C, Maule AJ. et al. Separation of early and late responses to herbivory in Arabidopsis by changing plasmodesmal function. Plant J. 2013;73(1):14–25. doi: 10.1111/j.1365-313X.2012.05103.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of MdCPK genes. Amino acid sequence alignment of MdCPK genes show presence of kinase domain and four calcium binding EF-hands in regulatory domain. In EF-hands, Ca2+ ion are co-ordinated in a pentagonal bipyramidal configuration. Ca2+ binding amino acid residue are present at position 1, 3, 5, 7, 9 and 12. The conserved Glu (E) or Asp (D) provides two oxygen for liganding Ca2+. Multiple sequence alignment of MdCPK genes were carried out using multalin (http://multalin.toulouse.inra.fr/multalin) software using statistical programme BLOSUM. Red and blue color indicate high and low conserved domains/motifs respectively, whereas black indicate neutral.
Schematic representation of transcript of MdCDPK genes. Box mark represents the exon and line represents the intron of specific CDPK gene. The name to the right of the gene structure indicates the gene name.
Q PCR Primer list of all MdCDPK genes used in this study.
The BLASTP score of MdCPKs found during their identification. The E- value found during BLASTP search show very significant similarity.