Abstract
The gel-based proteomic analysis of plasma membranes from rat liver and chemically induced, malignant hepatocellular carcinoma Morris hepatoma 7777 was systematically optimized to yield the maximum number of proteins containing transmembrane domains (TMDs). Incorporation of plasma membrane proteins into polyacrylamide “tube gel” followed by in-gel digestion of “tube gel” pieces significantly improved detection by ESI-LC-MS/MS. Removal of less hydrophobic proteins by washing isolated plasma membranes with 0.1 M sodium carbonate enables detection of hydrophobic proteins with one or more TMDs in both tissues. Subsequent treatment of plasma membranes by a proteolytic enzyme (trypsin) causes the loss of some of the proteins that are detected after washing with sodium carbonate, but it enables the detection of other hydrophobic proteins containing TMDs. Introduction of mass spectrometers with higher sensitivity, higher mass resolution and mass accuracy, and a faster scan rate significantly improved detection of membrane proteins, but the improved sample preparation is still useful, and enables detection of additional hydrophobic proteins. Proteolytic pre-digestion of plasma membranes enables detection of additional hydrophobic proteins and better sequence coverage of TMD-containing proteins in plasma membranes from both tissues.
INTRODUCTION
The plasma membrane is the cell organelle that is involved in cell-cell and cell-matrix interactions, receptor binding, as well as the organization of the cytoskeleton. The components of the plasma membrane, mostly glyco-proteins and -lipids also determine the immunological identity of the cell [1-4]. The composition and antigenicity of the components of the cell surface are altered during malignant transformation and other pathological changes of the cell, and they are also the first ones that will initiate a reaction by the host immune system [2, 5]. The possibility that plasma membrane proteins, or fragments thereof, are the first ones to be secreted or shed into body fluids makes them likely candidates for disease and injury biomarkers [2, 5-7].
Depending on the type of interaction and level of hydrophobicity, integral membrane proteins can have one or more transmembrane domains (TMDs), or they are embedded or they are anchored by a lipid chain in the membrane bilayer. Peripheral membrane proteins are associated with the extra- or intracellular membrane surface, usually by ionic interactions or hydrogen bonds [5, 7]. Because of the microheterogeneity of plasma membrane proteins, caused primarily by differences in posttranslational modifications (PTMs), it has become necessary to develop new strategies for their proteomic analysis [3, 4]. The basic problem at the start of the analysis is how to prepare the plasma membrane with minimal levels of contaminating cytosolic proteins and proteins from other organelles. There are many different strategies for the removal of contaminating proteins and organelles, such as the use of either lectins or antibodies against integral membrane proteins immobilized on magnetic beads in order to isolate enriched plasma membrane vesicles [8, 9]. After appropriate chemical modification, cell surface proteins can also be isolated by the use of different methods, such as biotin-avidin affinity chromatography [10, 11]. However, such protocols are time consuming and less practical for incorporation into recently developed, high-throughput analyses and use of minimal sample amount for qualitative and, ultimately, quantitative proteomic analyses [6].
Methods for selective solubilization of membrane proteins according to their hydrophobicity and type of interaction with the lipid bilayer were developed over thirty years ago [12], and have been applied with gel-based or gel-free methodologies as a sample preparation technique for proteomic analyses of integral plasma membrane proteins [13, 14]. During stepwise solubilization, plasma membranes are frozen and thawed, and subsequently treated with salts, usually at higher pH, followed by chaotropic reagents, and then by different ionic or non-ionic detergents, or by combination of these substances [5, 8, 12]. We used this solubilization schema for qualitative and semi-quantitative comparison of proteomes of tissues and for identification of candidate biomarkers for hepatocellular carcinomas [13, 15]. Peng’s group recommended the use of a similar method for enrichment of membranes from the liver microsomal fraction, to be followed by SDS-PAGE, tryptic digestion of excised proteins and identification by LC-MS/MS [15]. The Peng’s group protocol was also discussed by the Asia Oceania Proteome Organization Membrane Proteomics Initiative as a standard protocol for sample preparation for further proteomic analysis of membrane proteins [16, 17].
The insolubility of integral membrane proteins in pure aqueous media interfere with proteolytic, most often tryptic, digestion of membane proteins and is the well-known and frequently discussed topic [2-7, 14, 18-20] of their underrepresentation in the lists of identified proteins. Protein identification and quantification with suitable sequence coverage is highly dependent on the generation of many unique tryptic peptides that can be separated by LC, most frequently by use of the reversed-phase chromatography, and detected by MS/MS. The low abundance of many of these proteins in cells and tissues, the presence of PTMs and the lack of charged residues (Arg/Lys in the case of trypsin digestion as preparation for LC-MS/MS) in the TMDs are additional reasons why the membrane proteins are frequently underrepresented in large scale proteomic investigations [6, 19]. As an alternative to gel-based membrane proteomics, a solution based multidimensional LC-MS strategies were developed for analysis of plasma membrane proteins [21-23], exemplified by solubilization and tryptic digestion of hydrophobic integral membrane proteins in a buffer containing 60% (v/v) methanol, representing a significant step on the path towards their improved detection by mass spectrometry [18, 24, 25]. Lu and Zhu [26] introduced the “tube-gel” digestion method for improved detection of very hydrophobic proteins, and Cao et al. [27] successfully applied this method for an improved analysis of rat liver plasma membrane proteome. By use of tube-gel digestion, proteins can be solubilized by different detergents and/or other reagents, and incorporated into a polyacrylamide gel matrix. The solubilization agents can then be washed out while the proteins are still immobilized in the gel matrix and can be subsequently digested by a protease, most frequently trypsin [26]. Using this approach, a major improvement in the detection of hydrophobic peptides from integral membrane proteins in the subsequent mass spectrometry step was achieved [28].
In this study, an optimized gel-based method for sample preparation by pretreatment of plasma membranes with sodium carbonate, and predigestion with trypsin were combined with a modified “tube-gel” digestion [26] approach in order to detect the hydrophobic plasma membrane proteins. By use of this approach, liver plasma membranes and plasma membranes of the highly malignant type of chemically induced hepatocellular carcinoma, Morris hepatoma 7777 were analyzed. Starting with sample preparation and further procedure improvements, and ending with data analysis for plasma membrane protein identification, each step was analyzed and optimized in order to identify the maximal number of integral membrane proteins containing one of more TMDs. The application of this gel-based method for further identification of membrane proteins as possible disease biomarker candidates is also discussed.
EXPERIMENTAL SECTION
Isolation of plasma membranes
Buffalo rats bearing the chemically induced liver carcinoma Morris hepatoma 7777 were perfused with ice-cold PBS, liver and hepatoma were excised, minced, and dissociated using a Dounce type homogenizer in a buffer of 1 mM NaHCO3, 0.5 mM CaCl2 (pH 7.4, Lysis Buffer), and protease inhibitor cocktail (Calbiochem, San Diego, CA, USA). Plasma membranes were isolated as previously described [8, 13, 29]. Membrane fraction purity was routinely checked [29]. Ethical approval for these experiments was obtained from the Free University Berlin, Germany.
Pretreatment of rat liver plasma membrane fractions
1500 μg of isolated plasma membrane proteins from normal rat liver and Morris hepatoma 7777 were suspended in 10 mM Tris, pH 7.4, washed with ice-cold 100 mM Na2CO3, pH 11.0, and pelleted as described previously [13]. For sodium carbonate washed samples, membrane pellets were washed with cold deionized water, resuspended with 100 μL 25mM NH4HCO3, and stored at −80°C. For predigested samples, membrane pellets were washed with cold deionized water, resuspended with 1mL 25mM NH4HCO3, pH 8.6, and digested overnight at 37°C with 10 μg trypsin (Sigma, St. Louis, MO, USA). After predigestion, membranes were pelleted again by centrifugation at 40k RPM for 30 min and membrane pellets were then washed with cold deionized water, resuspended with 25 mM NH4HCO3, and stored at −80°C.
Incorporation of plasma membrane proteins into polyacrylamide gel and “tube gel” proteolytic digestion
For this procedure, the method developed by Lu and Zhu [26] was modified for trypsin digestion of differently pre-treated plasma membranes from rat liver and Morris hepatoma 7777. Pretreated membrane fractions from each condition were pelleted by centrifugation at 40k RPM for 30 min. Supernatants were removed and membrane pellets were solubilized with 40 μL of 2% SDS, 6M urea, 25 mM NH4HCO3, pH 8.0 and further incubated at 37°C for 30 min for better solubilization. Membrane proteins were reduced with 50 mM dithiothreitol (DTT) at 56°C for 1 h and alkylated with 40 mM iodoacetamide at room temperature in the dark for 45 min. The reduced and alkylated proteins were then incorporated into a polyacrylamide gel as described next. 50 μL of the protein solutions, 18.5 μL of acrylamide/bisacrylamide solution (40%, 29:1, w/v, ACROS Organics, Thermo Fisher Scientific), 2.5 μl of 10% (w/v) ammonium persulfate (Sigma), and 1 μL of TEMED (Bio-Rad, Hercules, CA) were mixed in an Eppendorf tube. After polymerization for 30 min at room temperature, the gel was cut into small pieces and washed twice with 25 mM NH4HCO3 and twice with 25 mM NH4HCO3, 50% (v/v) acetonitrile (ACN). The gel was further dehydrated with 100% ACN and then completely dried in vacuum centrifuge. Proteolytic digestion was performed with trypsin (Sigma) in 40 mM NH4HCO3, 10% ACN overnight at 37°C. Peptides were extracted from the gel using sequential extraction with 200 μL of 25 mM NH4HCO3, 200 μL of 0.1% (v/v) trifluoroacetic acid (TFA) in water, 200 μL of 0.1% TFA in ACN, and 200 μl of 100% ACN. The solutions were then combined and concentrated in a SpeedVac.
In-gel deglycosylation
In-gel deglycosylation was performed to facilitate the trypsin digestion of glycoproteins. After polymerization, washed and dried gels were rehydrated with freshly prepared digestion buffer containing 25 mU PNGase F (ProZyme, Inc., San Leandro, CA, USA.) in 25 mM NH4HCO3. Deglycosylation was performed at 37°C overnight. Gels were washed and sonicated three times with deionized water and once with ACN, and then completely dried in a speed vacuum. Trypsin digestion was then performed following the protocol described above.
Nano-LC-MS/MS Analysis (QSTAR XL)
Nano-flow reverse phase LC-MS/MS was performed using an Ultimate 3000 Nano LC system (LC Packings/Dionex, San Francisco, CA, USA) coupled a QSTAR XL quadrupole time-of-flight mass spectrometer (Applied Biosystems, Foster City, CA and Sciex, Concord, Ontario, Canada). Tryptic peptides were fractionated with a C18 reversed phase column (C18 PepMap 100, 75 μm ID × 15 mm, 3 μm particle size, LC Packings/Dionex) operating at a flow rate of 200 nL/min. Peptides were eluted into the QSTAR XL mass spectrometer directly using a reversed-phase gradient (10-130 min 10-35% (v/v) solvent B, 130-190 min 35-90% solvent B) via electrospary ionization. An electrospray voltage of 2.4 kV was applied. Spectra were collected in positive ion mode and in cycles of one full MS scan (m/z: 350-1500), followed by data-dependent MS/MS scans (m/z: 150-1500) sequentially of the three most abundant ions in each MS scan with charge state screening for +2 to +4 ions and dynamic exclusion time of 40 s.
Nano-LC-MS/MS Analysis (LTQ Orbitrap Velos)
Tryptic peptides were fractionated on a 75 μm × 12 cm column containing 3 μm Monitor C18 resin (Orochem Technologies, Inc., Lombard, IL, USA) and having an integrated 10 μm ESI emitter tip (“Self-Pack” PicoFrit column; New Objective, Woburn, MA, USA). Solvent A was 0.1 M acetic acid in water and solvent B was 0.1 M acetic acid in acetonitrile. Peptides were eluted with a linear acetonitrile gradient (0-70% solvent B over 60 min), operated at 200 nL/min. using an Agilent 1200 HPLC (Agilent Technologies, Santa Clara, CA). The column eluate was introduced directly onto an LTQ Velos Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) with a 1.8 kV ESI voltage. Full MS scans in the m/z range of 300-1700 at a nominal resolution of 60,000 were collected in the Orbitrap, followed by data-dependent acquisition of MS/MS spectra for the ten most abundant ions in the LTQ ion trap. Only ions having a charge state ≥ 2 were considered for collision-induced dissociation. Repeated fragmentation of the same ion was minimized by employing a 30-second dynamic exclusion time.
MS Data Analysis using Mascot and ProteoIQ
MS/MS spectra were searched against the Uniprot rat protein database (downloaded April 2012) using the Mascot algorithm v.2.3.2 provided by Matrix Science [30]. The Uniprot rat database contained 74,212 protein entries (50% forward, 50% reversed). Mascot searches were performed with the following parameters: trypsin enzyme specificity, 2 possible missed cleavages, 0.2 Da (QSTAR XL) and 20 ppm (LTQ OrbiTrap Velos) mass tolerance for precursor ions, 0.5 Da mass tolerance for fragment ions. Search parameters specified a differential modification of oxidation on methionine and a static modification of carbamidomethylation (+57.0215 Da) on cysteine. Protein quantification was performed using ProteoIQ software v. 2.3.05 (BioInquire, Bogart, GA) with spectra count data. To provide high confidence on peptide sequence assignment and protein identification, data were filtered with following stringent criteria: Mowse score > 28 for all charge states, at least 2 peptides per protein, 1% peptide false discovery rate (FDR) and 1% protein FDR.
Bioinformatics Analysis
The transmembrane hidden Markov model (TMHMM) algorithm (TMHMM 2.0, http://www.cbs.dtu.dk/services/TMHMM) was used to predict the putative transmembrane domains (TMDs) in identified proteins [31]. The grand average hydrophobicity (GRAVY) values were calculated for each identified protein as the arithmetic mean of the sum of the hydropathy values of all the amino acids in a protein sequence. Proteins with positive GRAVY values are considered to be hydrophobic, and those with negative values, hydrophilic. To classify proteins identified from normal rat liver and Morris hepatoma 7777 plasma membranes, gene ontology terms were examined (www.geneontology.org) using ProteoIQ software based on the UniProt-GOA rat database [32].
RESULTS
Sample preparation
Using normal rat liver and Morris hepatoma 7777 as a model system, rat liver plasma membrane proteins were identified by LC-MS/MS after tryptic digestion of pre-treated samples that were solubilized with a SDS-urea mixture, using a modified protocol based on the tube-gel digestion, the method previously developed by Lu and Zhu [26]. Isolated plasma membranes from both tissues were firstly pretreated with either sodium carbonate [13] or predigested with trypsin [33] to remove soluble and other proteins that are associated with the plasma membrane.
Enzymatic digestion
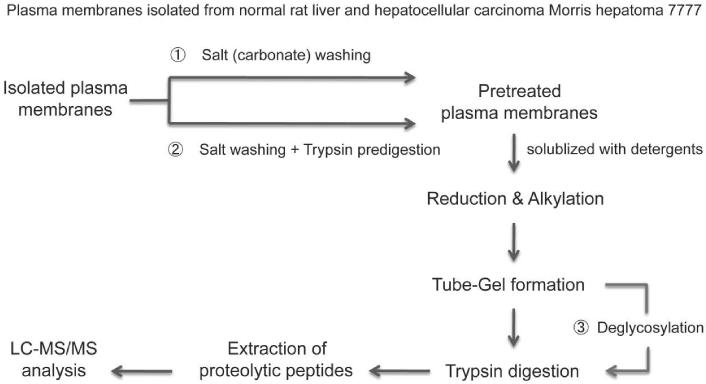
In order to make some highly glycosylated proteins more accessible to the proteolytic enzymes and to increase protein sequence coverage, deglycosylation was also performed after protein solubilization and formation of tube-gel (see Experimental Section), however, this additional treatment did not yield in increased detection of hydrophobic proteins. The complete scheme of the sample preparation and high-throughput analysis of isolated plasma membranes of both tissues is shown in Figure 1.
Figure 1. Experimental scheme of the high-throughput analyses of isolated plasma membranes of rat liver and hepatocellular carcinoma Morris hepatoma 7777.
Isolated plasma membranes from normal rat liver and hepatocellular carcinoma Morris hepatoma 7777 were pretreated with either 1) Salt (sodium carbonate) washing only or 2) sodium carbonate washing followed by trypsin predigestion to remove the soluble and peripheral membrane proteins. Subsequently, the resulting plasma membrane pellets were solubilized with detergents. Proteins were reduced, alkylated, and then directly incorporated into a polyacrylamide gel. The enhanced gel-assisted digestion was then performed. Peptides extracted from the gel were applied to LC-MS/MS analysis for protein identification and quantification. 3) Deglycosylation was performed after the formation of tube-gel to increase the protein sequence coverage for glycoprotein identification.
Protein identification
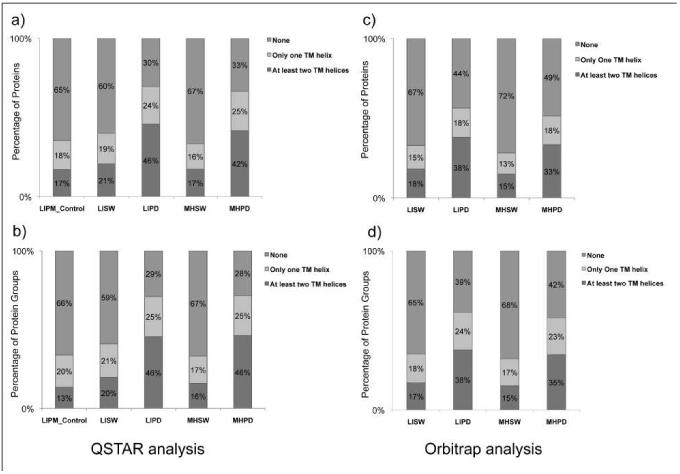
After use of different methods for sample preparation, membrane proteins were identified by LC-MS/MS either by use of a QSTAR or Orbitrap mass spectrometer (see Experimental Section). As shown in Table 1 and Figure 2, results regarding the numbers of identification of both proteins and protein groups for both tissues, liver and Morris hepatoma 7777, were comparable. After pretreatment with sodium carbonate, 625 proteins in liver plasma membranes (LISW) and 603 proteins in plasma membranes of Morris hepatoma (MHSW) were detected after ESI-MS/MS with the QSTAR mass spectrometer. 375 (60%) of LISW and 401 (63%) of MHSW proteins did not have any TMD. In liver plasma membranes, 40% of proteins have TMDs, and roughly half of them (21%) have two or more. The number of proteins with TMDs in Morris hepatoma 7777 plasma membranes is slightly lower (37%), and 18% of them have two or more TMDs. Interestingly, more proteins with three or more TMDs were detected in liver plasma membranes than in Morris hepatoma 7777 (13.6% versus 11.3%, respectively). After tryptic predigestion prior to the final gel-tube digestion, some membrane-associated proteins were removed, and only 375 proteins in liver plasma membranes (LIPD) and 355 proteins in the corresponding Morris hepatoma 7777 samples (MHPD) were detected. However, only 30.5% of detected proteins in liver and 33.5% in Morris hepatoma did not have any TMD, and the number of detected proteins with TMDs raised to 59.5% (liver) and 56.5% (Morris hepatoma). The absolute number of proteins containing TMDs was also higher, especially the number of proteins with 3 or more TMDs, which increased approximately 40% (85 to 138 for liver, and 68 to 108 for Morris hepatoma plasma membranes). As a control, we also performed the analysis of a liver plasma membrane sample after solubilization, without any pretreatment (“LIPM Control” in the Table 1). As shown in the Table 1, the number of identified proteins was significantly lower (403 compared with 625 after sodium carbonate treatment), and this procedure was not further investigated.
Table 1. Proteins and protein groups identified from normal rat liver plasma membranes and hepatocellular carcinoma Morris hepatoma 7777 plasma membranes using different treatments.
Mass spec data were acquired by a) a QSTAR XL mass spectrometer and b) an Orbitrap Velos mass spectrometer. MS/MS spectra were searched against the Uniprot rat protein database using the Mascot algorithm v.2.3.2 provided with Matrix Science as described in methods. Mascot results were filtered using ProteoIQ software with the following stringent criteria: Mowse score > 28 for all charge states, at least 2 peptides per protein, 1% peptide false discovery rate (FDR) and 1% protein FDR.
| a) QSTAR analysis | |||||
|---|---|---|---|---|---|
| Samples | Total (Proteins) | None | TMD=1 | TMD=2 | TMD>=3 |
| LIPM_Control | 473 | 306 | 86 | 34 | 47 |
| LISW | 625 | 375 | 121 | 44 | 85 |
| LIPD | 375 | 114 | 89 | 34 | 138 |
| MHSW | 603 | 401 | 105 | 37 | 68 |
| MHPD | 355 | 119 | 148 | 40 | 108 |
| Samples | Total (Protein Groups) | None | TMD=1 | TMD=2 | TMD>=3 |
| LIPM_Control | 215 | 142 | 73 | 12 | 17 |
| LISW | 268 | 158 | 110 | 17 | 36 |
| LIPD | 169 | 49 | 120 | 13 | 64 |
| MHSW | 252 | 168 | 84 | 12 | 28 |
| MHPD | 169 | 48 | 121 | 16 | 62 |
| b) Orbitrap analysis | |||||
|---|---|---|---|---|---|
| Samples | Total (Proteins) | None | TMD=1 | TMD=2 | TMD>=3 |
| LISW | 2248 | 1508 | 331 | 103 | 306 |
| LIPD | 955 | 416 | 176 | 82 | 281 |
| MHSW | 2154 | 1544 | 289 | 95 | 226 |
| MHPD | 1036 | 503 | 187 | 83 | 263 |
| Samples | Total (Protein Groups) | None | TMD=1 | TMD=2 | TMD>=3 |
| LISW | 967 | 626 | 175 | 45 | 121 |
| LIPD | 434 | 168 | 102 | 36 | 128 |
| MHSW | 952 | 642 | 161 | 44 | 101 |
| MHPD | 477 | 201 | 110 | 38 | 128 |
Figure 2. Representation of the distributions of proteins or protein groups with predicted TM helices identified from normal rat liver plasma membranes (LIPM) or hepatocellular carcinoma Morris hepatoma 7777 plasma membranes (MHPM) using different treatments.
Isolated plasma membranes were 1) LIPM Control: LIPM directly analyzed using the nonelectrophoretic gel-assisted digestion approach, 2) LISW or MHSW: LIPM or MHPM were pretreated with carbonate washing, 3) LIPD or MHPD: LIPM or MHPM treated with carbonate washing followed by predigestion with trypsin. After the enhanced gel-assisted digestion, tryptic peptides were subjected to LC-MS/MS analysis for protein identification and quantification. Both data acquired by a QSTAR XL mass spectrometer (left) and an Orbitrap Velos mass spectometer (right) are shown for comparison. The putative transmembrane helices were predicted using the transmembrane hidden Markov model (TMHMM) algorithm for each identified protein. Distributions of proteins (above) or protein groups (below) with their predicted TM helices are both presented.
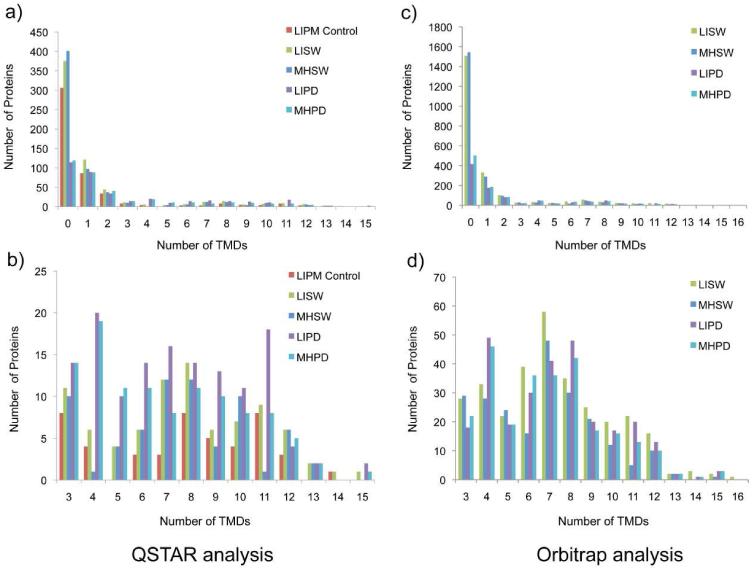
As expected, significantly higher numbers of proteins were identified when the more sensitive Orbitrap mass spectrometer was used. After sodium carbonate treatment, 2248 proteins were identified in liver plasma membranes, and 2154 in Morris hepatoma samples (about 3.6× more in both tissues). In liver plasma membranes, 1508 proteins (67%) have no TMD, and 740 (33%) have 1 or more predicted TMDs. After the same pretreatment, a comparable number of proteins (2154) was identified in Morris hepatoma plasma membranes, and an even higher percentage of them (72%) without any TMD. Consequently, only 28% of the proteins have 1 or more TMDs. This time, roughly the same percentage of proteins containing three or more TMDs were detected in both plasma membrane samples (13.6% for liver comapared to 13.0% in Morris hepatoma). After tryptic predigestion (LIPD and MHPD in Table 1), 955 proteins were identified in liver and 1036 in Morris hepatoma 7777 plasma membranes. More than 50% of membrane-associated proteins were removed or lost during this treatment. As shown in Figures 2 and 3, the percentage of identified proteins with TMDs significantly raised (56% for the liver and 52% for Morris hepatoma) with this sample preparation technique. However, the number of identified proteins for both liver and Morris hepatoma plasma membranes also decreased by more than 50%, and the absolute number of detected proteins that contain PTMs was also lower (see Table 1). However, the tryptic pre-digestion resulted in detection of a very different hydrophobic membrane protein pattern in both liver and Morris hepatoma 7777 (see Figure 4 and Table 2).
Figure 3. Histogram illustrations of the distributions of proteins with predicted TM helices identified from normal rat liver and hepatocellular carcinoma Morris hepatoma plasma membrane using different treatments.
Both data acquired by a QSTAR XL mass spectrometer (left) and an Orbitrap Velos mass spectometer (right) are shown for comparison. Identified proteins predicted with more than three TM helices are also shown in a more detailed view (below). Isolated plasma membranes were 1) LIPM Control: normal rat liver plasma membrane directly analyzed using the nonelectrophoretic gel-assisted digestion approach, 2) LISW or MHSW: LIPM or MHPM treated with carbonate washing, 3) LIPD or MHPD: LIPM or MHPM treated with carbonate washing followed by predigestion with trypsin. After the enhanced gel-assisted digestion, tryptic peptides were analyzed by LC-MS/MS for protein identification and quantification.
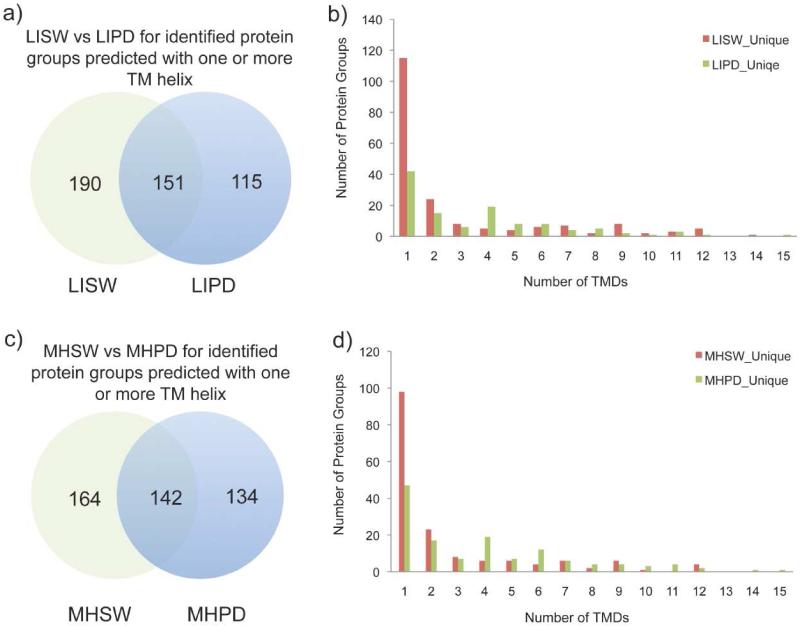
Figure 4. Comparisons of proteins predicted with one or more TM helix identified from normal rat liver and hepatocellular carcinoma Morris hepatoma plasma membrane using different treatments.
The data shown here were acquired on an Orbitrap Velos mass spectrometer. Venn diagram representations (left) and histogram representations (right) of identified proteins predicted with one or more TM helix were represented for a) b) LISW and LIPD, and c) d) MHSW and MHPD.
Table 2. Comparisons of total protein groups or protein groups predicted with one or more TM helix identified from normal rat liver plasma membrane (LIPM) and hepatocellular carcinoma Morris hepatoma plasma membrane (MHPM) using different treatments.
Data shown here was acquired by an Orbitrap Velos mass spectrometer as described in Table 1.
| Samples | Total Protein Groups | Protein Groups with TMD>=1 |
|---|---|---|
| Common(LIPDvsLISW) | 263 | 151 |
| LIPD_Unique | 171 | 115 |
| LISW_Unique | 704 | 190 |
| Common(MHPDvsMHSW) | 264 | 142 |
| MHPD_Unique | 213 | 134 |
| MHSW_Unique | 688 | 164 |
| Common(LIPDvsMHPD) | 392 | 245 |
| LIPD_Unique | 42 | 21 |
| MHPD_Unique | 85 | 31 |
| Common(LISWvsMHSW) | 760 | 261 |
| LISW_Unique | 207 | 80 |
| MHSW_Unique | 192 | 45 |
DISCUSSION
As a basic analytical method for protein detection, mass spectrometry is constantly improving in the direction of higher sensitivity and accuracy. However, the analysis of membrane proteins, especially ones from plasma membranes, has always been hampered by their diversity of PTMs, high hydrophobicity caused by the TMDs, and their relatively low abundance. To enable the analysis of complete plasma membrane proteomes, especially by use of high-throughput methods, it is essential that the sample preparation protocols are combined with the workflows that are tuned to cope with the above mentioned characteristics of these proteins, such as the high number of PTMs and the hydrophobic peptides that are parts of their TMDs or other structures that interact with hydrophobic plasma membrane domains. Additionally, protein solubilization techniques, protein and peptide separation and (mostly proteolytic) cleavage procedures have to be optimized in order to insure optimal yield of hydrophobic peptides [5, 34].
The Asia Oceania Human Proteome Organization (AOHUPO) started the Membrane Proteomics Initiative for the analysis of membrane proteomes. As a common standard, the mouse liver membrane fractions were analyzed in 17 different laboratories. The membrane fraction containing plasma membranes and membranes of intracellular organelles was pre-treated with 0.1M sodium carbonate, pH 11.5, in order to remove membrane associated proteins. After this treatment, proteins were immediately digested (in-solution digestion) or differently separated by use of SDS-PAGE, isoelectric focussing, 2-D blue native PAGE or chromatographic separation, followed by tryptic digestion and LC-MS/MS [17]. In all participating laboratories, the LC-MS/MS for protein identification was performed by a Dionex UltiMate™ 3000 LC system and a Thermo Finningan LTQ mass spectrometer equipped with a nanospray ion source. In the subsequent publication, Peng et. al. [16] recommended a “Standard Using SDS-PAGE Shotgun Proteomics”. According to this protocol, the plasma membrane fraction is treated with 0.1M sodium carbonate, pH 11.5, and subsequently separated with SDS-PAGE prior to the tryptic digestion and LC-ESI-MS/MS. Following this procedure, roughly 1000 proteins were identified, and 47% of them had TMDs. We used a stepwise solubilization of plasma membranes (freezing-thawing followed by alkaline treatment (at pH 11) and solubilization by use of detergents) as a sample preparation protocol for the identification of liver and Morris hepatoma 7777 plasma membrane proteins. After the in-solution digestion, approximately 600 proteins in three fractions of both liver and Morris hepatoma 7777 plasma membranes were detected. However, we detected a significantly lower number of proteins with TMDs. Our less favorable result was presumably due to the lower sensitivity of the mass spectrometer that was used [13]. Although it yields in detection of a relatively high number of membrane proteins with TMDs, the protocol that was recommended by the Asia Oceania Human Proteome Organization for the analysis of liver membrane proteins, and that was published by Peng et al. [16], can be considered neither as high-throughput method nor as shotgun proteomics. Due to the sample preparation steps that are generated by excising slices after SDS-PAGE, this procedure is time-consuming. According to the previously published results obtained by in-solution/gel-free 2D-LC-MS methods, the term “Shotgun Proteomics” is exclusively related to a gel-free procedure [21, 24].
Wu et al. [33] recommended proteolytic pre-digestion of membranes to remove membrane-associated proteins. As shown in Figure 1, in a parallel experiment, proteolytic (tryptic) pre-digestion of membranes after treatment with sodium carbonate was also performed. In the protocols described here, different sample preparation techniques were combined with the so-called “tube gel” digestion method that was developed by Lu and Zhu [26]. Not only sample preparation and the difficulties in detecting hydrophobic peptides by mass spectrometry [35], but also the limited accessibility of hydrophobic TMDs of plasma membrane proteins to proteolytic digestion are the primary reasons for lack of their identification [26]. As demonstrated by Cao et al. [27] and Han et al. [28], the introduction of the “tube in-gel” digestion method has made a significant contribution to better identification of hydrophobic proteins, presumably because of their enhanced access to the proteolytic enzymes.
An additional problem is that in MS, especially in less sensitive mass spectrometers, membrane proteins are usually detected by means of their hydrophilic regions, and short peptides are detected more frequently than long ones [35]. As shown in Figures 2-4 and Tables 1 and 2, when protein detection was performed by the less sensitive QSTAR mass spectrometer, membrane pre-digestion after a wash with sodium carbonate gives better results, regarding detection of more hydrophobic proteins with one or more TMDs, than a wash with sodium hydroxide alone. This type of sample preparation enables detection of more hydrophobic proteins in both liver and Morris hepatoma 7777 plasma membranes (see Figures 2a and 2b, and Figure S1a and S1b). When a more sensitive, up-to-date Orbitrap mass spectrometer was used, the relative amount of proteins with TMDs was still higher in proteolytically pre-treated samples (see Figure 2 c and d, and Figure 3), but the differences in GRAVY score were reduced (see Figure 5 c and d). Obviously, this mass spectrometer is able to detect additional hydrophobic peptides and also larger peptides. As shown in Figure 4, tryptic pre-digestion as a sample preparation method is still useful for the identification of additional membrane proteins. Although a significant number of membrane proteins has been lost after this treatment (see Table 1), additional, mostly hydrophobic proteins were detected (see Figure 4). Membrane pre-digestion by proteolytic enzymes also enables higher sequence coverage of some hydrophobic proteins containing one or more TMDs (see Table 3). Interestingly, deglycosylation of plasma membranes by PGNase F before their proteolytic digestion (see the Experimental Section and the scheme in Figure 1) did not increase the number of detected hydrophobic proteins (not shown here).
Table 3. Sequence coverage of three membrane proteins with 1 or more TMDs after different sample pre-treatment.
Untreated liver plasma membranes were directly digested with trypsin (“tube gel” digestion) without any sample preparation (LIPM Control), or the samples were pre-treated with 0.1M sodium carbonate (LISW), or with sodium carbonate followed by tryptic pre-digestion (LIPD) before digestion. Protein identification was performed by ESI-LC-MS/MS by use of a QSTAR mass spectrometer (see Experimental Section).
| Gene Name |
Protein Name | TMDs | Total % Seq Coverage (LIPM Control) |
Total % Seq Coverage (LISW) |
Total % Seq Coverage (LIPD) |
|---|---|---|---|---|---|
| Ugt2b15 | UDP glucuronosyltransferase 2B15 | 1 | 15.1% | 17.2% | 20.0% |
| Itgbl | Integrin beta 1 | 1 | 4.8% | 4.6% | 12.4% |
| Slco1b2 | Solute carrier organic anion transporter family member 1B2 |
12 | 4.8% | 11.1% | 14.7% |
After proteolytic pre-treatment, all three marker proteins for liver plasma membranes containing one TMD could be detected, namely the enzymes dipeptidyl peptidase IV, 5′nucleotidase and CEACAM 1, the molecule involved in cell-cell adhesion (not shown here). MS/MS detection of the latter, CEACAM 1, causes problems because of its high level of glycosylation, and requires additional steps for its targeted enrichment [36].
CONCLUSIONS
Incorporation of plasma membrane proteins of rat liver and Morris hepatomas 7777 into polyacrylamide gel and “tube gel” proteolytic digestion significantly improves their tryptic digestion and detection by ESI-LC-MS/MS.
Removal of membrane-associated proteins by washing with 0.1 M sodium carbonate enables the detection of hydrophobic proteins with one or more TMD in both tissues.
Subsequent treatment of plasma membranes by a proteolytic enzyme (trypsin) causes the loss of some proteins that are detected after washing by sodium carbonate only, but it enables the detection of additional hydrophobic proteins containing TMDs.
Deglycosylaton of plasma membrane proteins by treatment with PGNase F did not yield in detection of additional hydrophobic proteins.
The introduction of the Orbitrap mass spectrometer with its high sensitivity, high mass resolving power and mass accuracy, and a faster scan rate, significantly improved detection of membrane proteins without the necessity of SDS-PAGE as a sample preparation step. Other sample preparation steps are still useful, and they enable detection of a higher number of hydrophobic proteins also by the more modern LC-MS/MS system. Proteolytic pre-digestion of plasma membranes also enables detection of additional hydrophobic proteins and better coverage of other proteins containing TMDs in plasma membranes of both tissues.
Supplementary Material
ACKNOWLEDGEMENT
We thank Professor Jasminka Giacometti from the Department of Biotechnology, University of Rijeka, Croatia, for the editing of the manuscript, and technical support, and Professor Arthur Salomon, Department of Molecular Pharmacology, Physiology and Biotechnology, Brown University, Providence, USA, for the advices for optimization of mass spectrometry analyzes.
This work was supported by the National Institutes of Health, Centers of Biochemical Research Excellence (COBRE), Grant No. P20RR017695 and by Sonnenfeld-Stiftung, Berlin, Germany.
REFERENCES
- 1.Polanski M, Anderson MD. Biomarker Insights. 2006;2:1–48. [PMC free article] [PubMed] [Google Scholar]
- 2.Rucevic M, Hixson D, Josic Dj. Electrophoresis. 2011;32:1549–1564. doi: 10.1002/elps.201100212. [DOI] [PubMed] [Google Scholar]
- 3.Cordwell SJ, Thingholm TE. Proteomics. 2010;10:611–627. doi: 10.1002/pmic.200900521. [DOI] [PubMed] [Google Scholar]
- 4.Gilmore JM, Washburn MP. J. Proteomics. 2010;73:2078–2091. doi: 10.1016/j.jprot.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Josic Dj., Clifton J. Proteomics. 2007;7:3010–3029. doi: 10.1002/pmic.200700139. [DOI] [PubMed] [Google Scholar]
- 6.Helbig AO, Heck AJR, Slijper M. J. Proteomics. 2010;73:868–878. doi: 10.1016/j.jprot.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Sprenger R, Jansen ON. Proteomics. 2010;10:3997–4011. doi: 10.1002/pmic.201000312. [DOI] [PubMed] [Google Scholar]
- 8.Lawson EL, Clifton JG, Huang F, Li X, Hixson DC, Josic Dj. Electrophoresis. 2006;27:2747–2758. doi: 10.1002/elps.200600059. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y-C, Block G, Chen H, Fulch-Puy E, Foronjy R, Jalili R, Jendersen CB, Kimura M, Kraft E, Lindemose S, Lu J, McLain T, Nutt L, Ramon-Garcia S, Smith J, Spivak A, Wang ML, Zanic M, Lin S-H. Prot. Expr. Purif. 2008;62:233–229. doi: 10.1016/j.pep.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Nature Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Zhang W, White MA, Zhao Y. Anal. Chem. 2003;75:3751–3757. doi: 10.1021/ac034184m. [DOI] [PubMed] [Google Scholar]
- 12.Fujuki Y, Hubbard AN, Fowler S, Lazarow PD. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clifton JG, Li X, Reutter W, Hixson DC, Josic Dj. J. Chromatogr. B. 2007;849:293–301. doi: 10.1016/j.jchromb.2006.08.047. [DOI] [PubMed] [Google Scholar]
- 14.Speers AE, Wu CC. Chem. Rev. 2007;107:3687–3714. doi: 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- 15.Rucevic M, Rosenquist T, Breen L, Cao L, Clifton J, Hixson D, Josic Dj. J. Proteomics. 2012;76:79–90. doi: 10.1016/j.jprot.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Peng L, Kapp EA, McLauchlan D, Jordan W. Proteomics. 2011;11:4376–4384. doi: 10.1002/pmic.201100169. [DOI] [PubMed] [Google Scholar]
- 17.Peng L, Kapp EA, Fenyo D, Kwon M-S, Jiang P, Wu S, Jiang Y, Aguilar M-I, Ahmed N, Baker MS. The Asia Oceania Human Proteome Organisation Membrane Proteomics Initiative. Proteomics. 2010;10:4142–4148. doi: 10.1002/pmic.201000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blonder J, Chan KC, Issaq HJ, Veenstra TD. Nature Protocols. 2007;1:2784–2790. doi: 10.1038/nprot.2006.359. [DOI] [PubMed] [Google Scholar]
- 19.Jamsson M, Wårell C, Lavander F, James P. J. Proteome Res. 2008;7:659–665. doi: 10.1021/pr070545t. [DOI] [PubMed] [Google Scholar]
- 20.Blonder J, Conrads TP, Veenstra TD. Expert Rev. Proteomics. 2004;1:153–163. doi: 10.1586/14789450.1.2.153. [DOI] [PubMed] [Google Scholar]
- 21.Zhang N, Chen R, Young N, Wishart D, Winter P, Weiner JH, Li L. Proteomics. 2007;7:484–493. doi: 10.1002/pmic.200600518. [DOI] [PubMed] [Google Scholar]
- 22.Chick JM, Haynes PA, Molloy MP, Bjellqvist B, Baker MS, Len AC. J. Proteome Res. 2008;7:1036–1045. doi: 10.1021/pr700611w. [DOI] [PubMed] [Google Scholar]
- 23.Blackler AR, Speers AE, Ladinsky MS, Wu CC. J. Proteome Res. 2008;7:3028–3034. doi: 10.1021/pr700795f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blonder J, Hale ML, Chan KC, Yu LR, Lucas DA, Conrads TP, Zhou M, Popoff MR, Issaq HJ, Stiles BG, Veenstra TD. J. Proteome Res. 2005;4:523–531. doi: 10.1021/pr049790s. [DOI] [PubMed] [Google Scholar]
- 25.Wigelsworth DJ, Ruthel G, Schnell L, Herrlich P, Blonder J, Veenstra TD, Carman RJ, Wilkins TD, Van Nhieu GT, Pauillac S, Gibert M, Sauvonnet N, Stiles BG, Popoff MR, Barth H. PLoS One. 2012;7:e51356. doi: 10.1371/journal.pone.0051356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X, Zhu H. Mol. Cell. Proteomics. 2005;4:1948–1958. doi: 10.1074/mcp.M500138-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao R, He QY, Zhou H, He QZ, Liu Z, Wang XC, Chen P, Xie H, Liang SP. J. Proteome Res. 2008;7:535–545. doi: 10.1021/pr070411f. [DOI] [PubMed] [Google Scholar]
- 28.Han C-L, Chien C-W, Chen W-C, Chen Y-R, Wu C-P, Li H, Chen Y-J. Mol. Cell. Proteomics. 2008;7:1983–1997. doi: 10.1074/mcp.M800068-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Tauber R, Reutter W. Eur. J. Biochem. 1978;83:37–45. doi: 10.1111/j.1432-1033.1978.tb12065.x. [DOI] [PubMed] [Google Scholar]
- 30.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 32.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweoler R. Nucleic Acids Res. 2009;37:D396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd Nature Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 34.Helbig AO, Heck AJR, Slijper M. J. Proteomics. 2010;73:868–878. doi: 10.1016/j.jprot.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Eichacker LA, Granvogl B, Mirus O, Muller BC, Mies C, Schleiff H. J. Biol. Chem. 2004;279:50915–50922. doi: 10.1074/jbc.M405875200. [DOI] [PubMed] [Google Scholar]
- 36.Rucevic M, Clifton JG, Huang F, Li X, Callanan H, Hixson D, Josic Dj. J. Chromatogr. A. 2006;1123:199–204. doi: 10.1016/j.chroma.2006.02.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.