Abstract
Cytoplasmic dynein and kinesin-1 are opposite-polarity, microtubule-based motors that transport a wide variety of cargo in eukaryotic cells. Many cellular cargos demonstrate bi-directional movement due to the presence of ensembles of dynein and kinesin, but are ultimately sorted with spatial and temporal precision. To investigate the mechanisms that coordinate motor ensemble behavior, we built a programmable synthetic cargo using three-dimensional DNA origami to which varying numbers of DNA oligonucleotide-linked motors could be attached, allowing control of motor type, number, spacing, and orientation in vitro. In ensembles of 1–7 identical-polarity motors, motor number had minimal affect on directional velocity, while ensembles of opposite-polarity motors engaged in a tug of war resolvable by disengaging one motor species.
Cytoplasmic dynein and kinesin-1 (referred to as “dynein” and “kinesin” here) are opposite-polarity, microtubule-based motors responsible for producing and maintaining subcellular organization via the transport of many cargos in eukaryotic cells (1, 2). Defects in these transport processes have been linked to neurological diseases (1, 3, 4). Microtubules contain inherent structural polarity, polymerizing rapidly at their “plus” end and more slowly at their “minus” end (5), with dynein and kinesin driving most minus- and plus-end-directed microtubule transport, respectively (2). Although some transport tasks require a single motor type, many cargos use both dynein and kinesin and move bidirectionally on microtubules (1, 6, 7). The mechanisms that allow ensembles of identical-polarity motors to coordinate their activity and ensembles of opposite-polarity motors to achieve both processive movement and rapid switches in direction are unknown.
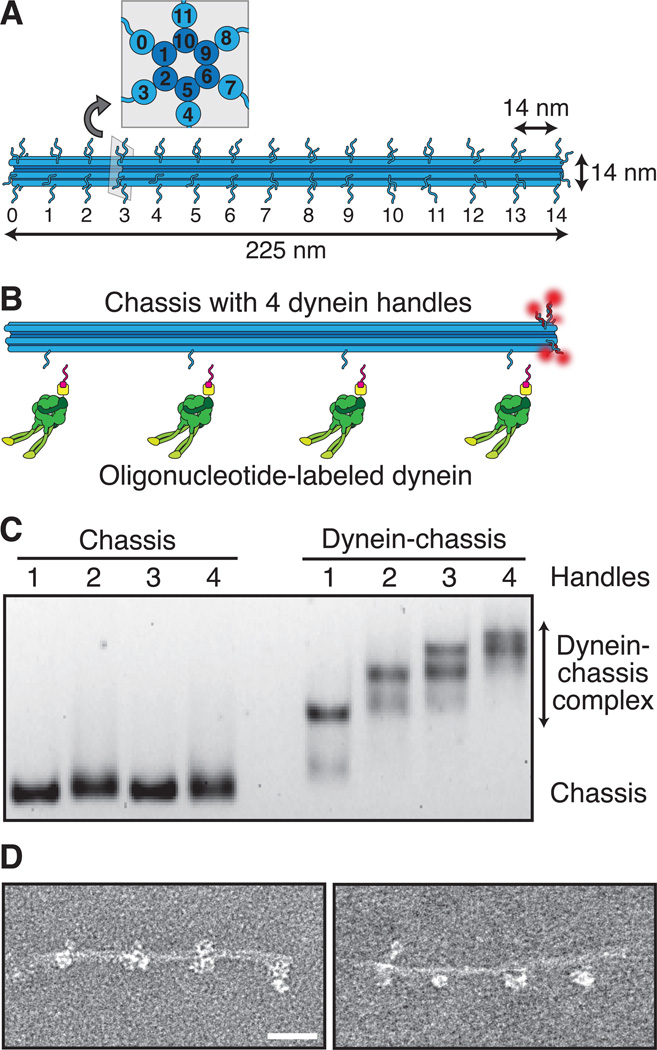
To dissect the biophysical mechanisms of motor-driven cargo transport, we designed a programmable, synthetic cargo using three-dimensional DNA origami (8, 9). The cargo consisted of a twelve-helix bundle with six inner and six outer helices (Fig. 1A, and fig. S1) (10). We refer to this structure as a “chassis,” akin to an automobile chassis that serves as a skeletal frame for the attachment of additional components. The origami chassis was made by rapidly heating and slowly cooling an 8064-nucleotide, single-strand DNA “scaffold” in the presence of 273 short, single-strand DNA "staples" (fig. S1A, and tables S1 to S3), which hybridize with discontinuous regions of the scaffold to fold it into a desired shape. Selective inclusion of staples with extra "handle" sequences that project out from the chassis provide site and sequence specific attachment points for motors, fluorophores, or other chemical moieties (Fig. 1B).
Fig. 1.
Design and validation of a three-dimensional DNA origami synthetic cargo. (A) Schematic of the twelve-helix bundle chassis structure with 6 inner and 6 outer helices. Each outer helix contains up to 15 optional handles, yielding 90 uniquely addressable sites. Each handle consists of an unpaired 21-bp (~7 nm) oligonucleotide sequence for hybridization to complementary anti-handle sequences covalently attached to motors or fluorophores. Inset shows an orthogonal cross-section. (B) Schematic of a chassis labeled with 5 fluorophores (red) at handle position 14 on each of 5 outer helices and dynein handles at positions 1, 5, 9, and 13 on a single outer helix. Oligonucleotide-labeled dynein is also shown. (C) Agarose gel shift assay of TAMRA-labeled chassis containing 1–4 handles in the absence (left lanes) or presence (right lanes) of dynein labeled with an anti-handle oligonucleotide. Chassis are visualized by TAMRA fluorescence. See fig. S2B for occupancy quantification. (D) Negative-stain TEM images of the 4 dyneinchassis complex. Scale bar, 40 nm.
Next, we purified well-characterized model dynein and kinesin motors and covalently linked them to DNA oligonucleotide “anti-handles” complementary to the handle sequences on the chassis. We used a minimal dimeric S. cerevisiae dynein (11, 12) and a minimal dimeric human kinesin-1 (13), both of which contained a SNAPf-tag at their cargo-binding domain for oligonucleotide anti-handle attachment.
We next assessed motor-chassis complex assembly. Gel shift assays of dynein chassis indicated an ~80% probability for individual dynein occupancy at each motor site on the chassis (Fig. 1C and fig. S2A). Due to kinesins small size relative to dynein, similar assays with kinesin chassis did not allow individual occupancy numbers to be resolved (fig. S2B). When the kinesin anti-handle was used with dynein, however, we again observed ~80% occupancy, indicating no handle sequence specific effects on motor-chassis linking (fig. S2C and S2D). Super-resolution fluorescence imaging by DNA-PAINT (14) revealed that sub-maximal handle incorporation into the folded chassis was probably responsible for incomplete motor occupancy (fig. S3), in agreement with previous reports (15, 16). Negative stain transmission electron microscopy (TEM) of fully assembled chassis structures showed dynein motors occupying sites on the chassis at the programmed locations (Fig. 1D).
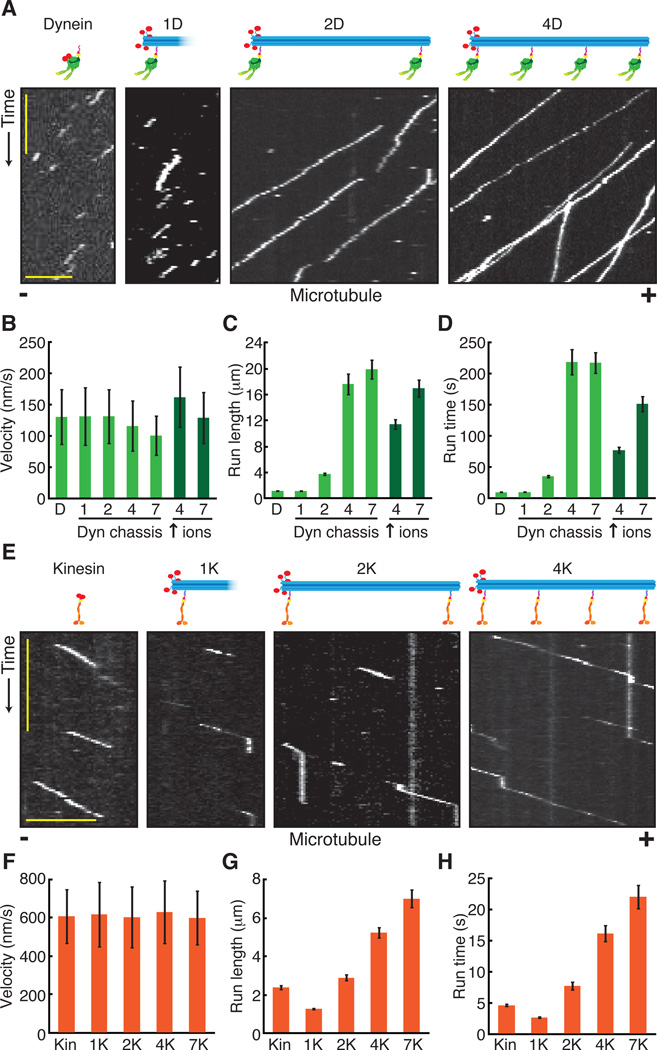
We quantified the motile properties of dynein alone or dynein ensembles on chassis with 1, 2, 4, or 7 motor attachment sites (1D, 2D, 4D, and 7D, respectively) on microtubules at the single-molecule level using total internal reflection fluorescence (TIRF) microscopy (Fig. 2A). The average velocity of a single dynein was similar to that of the 1D and 2D ensembles, while 4D and 7D ensembles moved slightly slower (Fig. 2B, and fig. S4A). The characteristic run length (total distance moved) and run time (total duration of the run) of dynein ensembles increased with the number of motor sites for the 1D, 2D, and 4D ensembles (Fig. 2C and D, and fig. S4B and S4C). The 4D and 7D ensembles were so processive that their run lengths and run times were similar to each other in standard assay buffer, where microtubule length and imaging duration become limiting (Fig. 2C and D). However, when assayed in high ionic strength buffer, which decreases dynein’s processivity (17), the 7D ensemble was more processive than the 4D ensemble (Fig. 2C and D, fig. S4, and fig. S5).
Fig. 2.
Single-molecule motile properties of chassis-motor complexes. (A) Kymographs of TMR-labeled dynein alone and TAMRA-labeled chassis with 1, 2, or 4 dyneins. Plus (+) and minus (−) denote microtubule polarity. Scale bars: 1 min (x), 5 μm (y). (B) Quantification of average segment velocities ± SD of dynein and dynein-chassis complexes. The 4D and 7D ensembles moved significantly slower than dynein alone, or the 1D or 2D ensembles (one-tailed t-test, P < 0.001; N ≥ 211). In higher ionic concentration (↑ ions), the 4D and 7D ensemble velocities were significantly different (one-tailed t-test, P < 0.001; N ≥ 208). (C) Quantification of run lengths ± SE of dynein and dynein-chassis ensembles (N ≥ 208). (D) Quantification of total run times ± SE of dynein and dynein-chassis ensembles (N ≥ 208). (E) Kymographs of TMR-labeled kinesin alone and TAMRA-labeled chassis with 1, 2, or 4 kinesins. Scale bars: 1 min (x), 5 μm (y). (F) Quantification of average segment velocities ± SD of kinesin and kinesinchassis ensembles. Comparison of velocities yielded no statistical differences (ANOVA test, P > 0.05; N ≥ 301). (G) Quantification of run lengths ± SE of kinesin and kinesinchassis ensembles (N ≥ 301). (H) Quantification of total run times ± SE of kinesin and kinesin-chassis ensembles (N ≥ 301). For additional statistical analysis see figs. S4–S6.
We performed a similar analysis of kinesin alone and kinesin ensembles on chassis with 1, 2, 4, or 7 motor attachment sites (1K, 2K, 4K, and 7K, respectively; Fig. 2E to H). The average velocities of kinesin ensembles remained constant (Fig. 2F, and fig. S6A), while run lengths and run times increased with increasing motor number (Fig. 2G and H, and fig. S6B and S6C).
Recent models of motor ensemble behavior using a transition state framework predict run lengths that are several orders of magnitude higher than we observed (18). In contrast, our data suggest that motor microtubule binding dynamics may be influenced by the presence and number of other motors on a shared cargo, similarly to previous work (19–22). For 1–7 kinesins or 1–2 dyneins, velocity was unaffected by motor number, however for 4D and 7D ensembles velocity was decreased, suggesting inter-motor interference can affect motor stepping rate. To test this hypothesis, we engineered chassis with locations for inactive mutant dyneins (denoted dI) incapable of binding ATP at dynein’s main site of ATP hydrolysis; this mutant binds microtubules tightly, but does not move (23). Dynein ensembles programmed to bind differing ratios of active and inactive motors (table S6) moved with reduced velocity (fig. S7), demonstrating that inter-motor negative interference decreases cargo velocity.
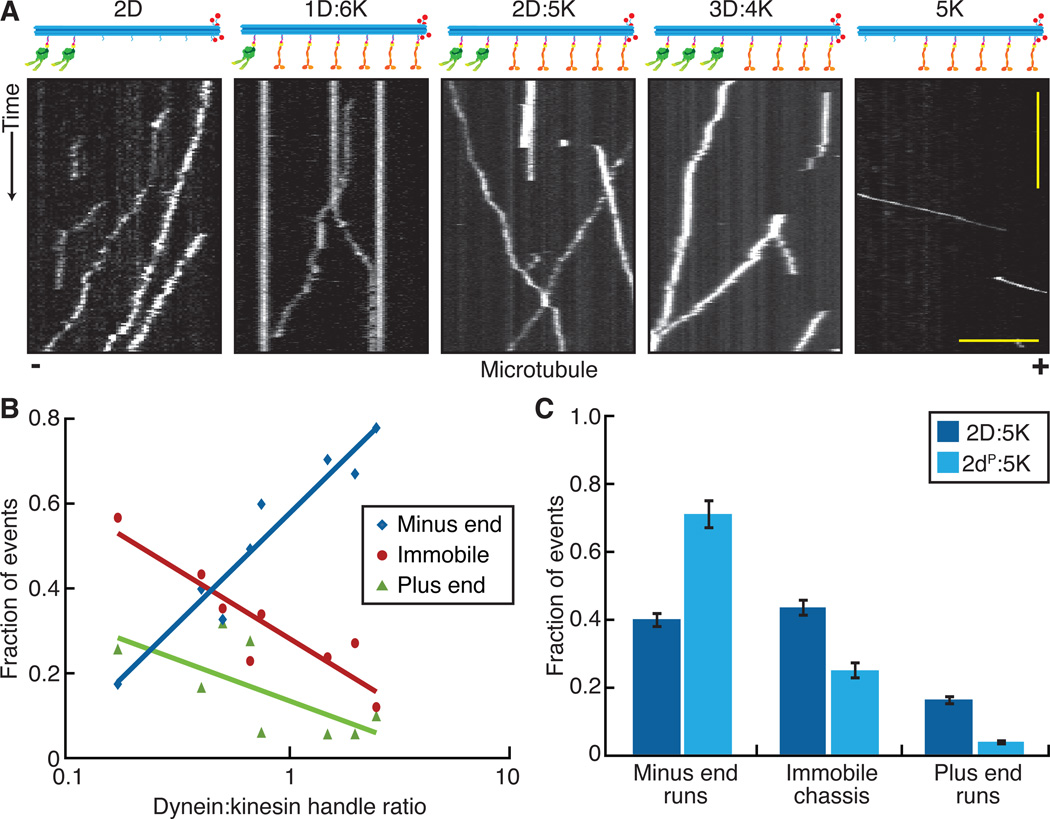
We next investigated the motility of chassis linked to “mixed” ensembles of opposite-polarity motors. We quantified the motility of chassis as a function of dynein to kinesin (D:K) ratio (table S6). All mixed-motor ensembles moved unidirectionally (Fig. 3A) with no reversals detected at a precision of ~10 nm. With the exception of the 1D:6K chassis, all ensembles were more likely to move toward the minus end of microtubules (Fig. 3B). Mixed-motor ensembles were relatively insensitive to increasing the number of kinesin motors compared to increasing the number of dynein motors, which could be due to kinesin ensembles operating predominantly through the actions of fewer motors at any given time (24). Based on the stall forces of dynein (~5 pN (25)) and kinesin (~7 pN (26)), we expected kinesin plus end runs would have been more dominant. In contrast, our results suggest that stall force was not the only parameter governing the behavior of opposite-polarity motor ensembles (27). Other parameters, such as microtubule affinity, detachment force, and velocity dependent on-rates could also be relevant (20–22, 28–31). Mixed-motor ensembles moved more slowly and for longer periods of time than equivalent single motor type ensembles (fig. S8A and S8B), with the magnitude of this effect more pronounced in the plus end direction. Notably, mixed ensembles of dynein and kinesin were more likely to be immobile than identical-motor ensembles, suggesting that opposite-polarity motors engage in a tug of war that prevents cargo movement (Fig. 3B).
Fig. 3.
Chassis attached to dynein and kinesin frequently engage in a stalled tug of war. (A) Kymographs of TAMRA-labeled chassis attached to dynein only (left most panel), kinesin only (right most panel), or varying ratios of dynein and kinesin motors (middle panels). Plus (+) and minus (−) denote microtubule polarity. Scale bars: 1 min (x), 5 μm (y). (B) Quantification of the fraction of events for each chassis observed as defined by their dynein to kinesin handle ratio. Chassis were immobile, moving toward the minus end, or moving toward the plus end (table S6, N ≥ 221). X-axis of dynein to kinesin ratios is a logarithmic scale and linear-log fits highlight the trends observed. (C) 10 Quantification of the fraction of events ± SE observed to be immobile, moving toward the minus end, or moving toward the plus end for mixed ensembles containing 2 dyneins and 5 kinesins (N ≥ 352). The dyneins were either wildtype (D) or a highly processive mutant (dP).
Based on the longer run lengths and run times of yeast dynein as compared to human kinesin, we hypothesized that dynein runs dominated in mixed-motor ensembles due to dynein’s higher microtubule affinity. To test this, we purified a mutant dynein with a higher processivity and affinity for microtubules (denoted dP) (17) and paired it with kinesins. The 2dP:5K ensemble was even more likely to move in the dynein direction and had fewer immobile chassis compared to the 2D:5K ensemble containing wildtype dynein (Fig. 3C). These results suggest that track affinity is a key motor property in governing opposite-polarity motor ensemble motility. Mixed ensembles containing the high affinity dynein mutant also produced slower plus end runs and longer run times in both directions compared to the equivalent wildtype system (fig. S8C and S8D).
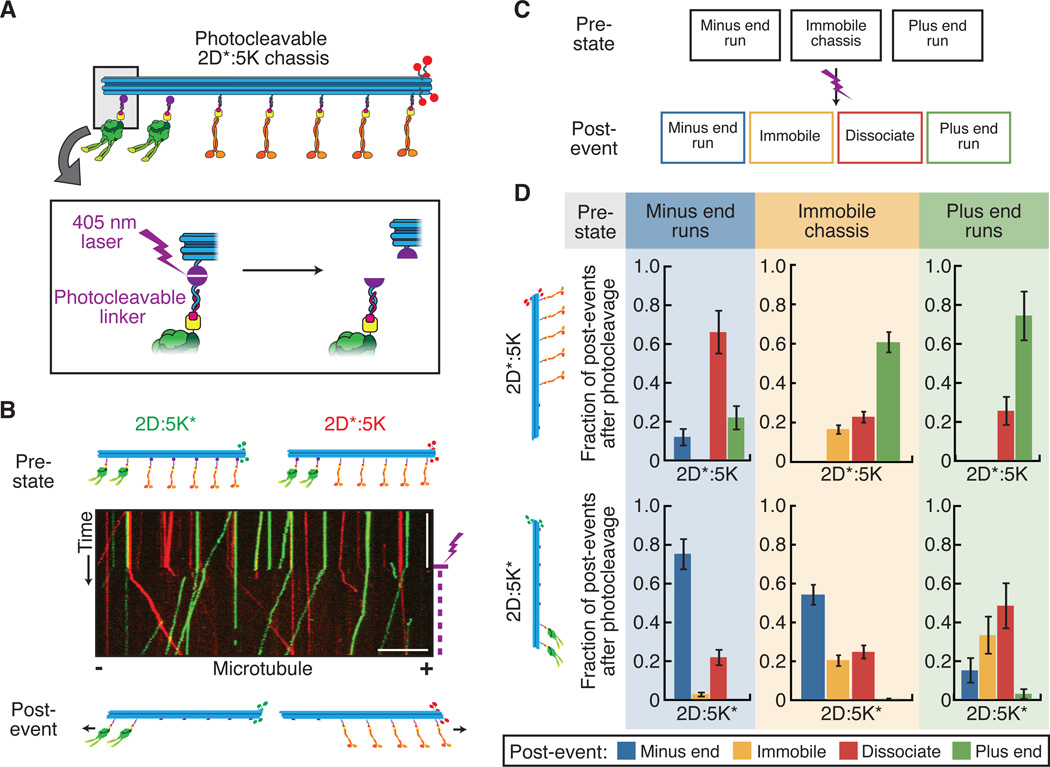
We wanted to determine if mixed-motor ensembles were non-motile due to a stalled tug of war. To regulate motor attachment to the chassis we introduced photocleavable linkers in selected handles such that illumination with a 405 nm laser released one motor type from the chassis (Fig. 4A). We designed two modified chassis: 2D:5K*, with photocleavable (*) kinesins, and 2D*:5K, with photocleavable dyneins. We monitored the motile properties of these chassis before and after laser-induced photocleavage (Fig. 4B). Cleavage was rapid (fig. S9); within seconds of photocleaving motors of one type, immobile chassis moved in the direction of the remaining motors (Fig. 4B). We classified the state of each chassis before and after photocleavage (Fig. 4C) and found that the majority of stalled tug of war events were resolved into active motility (Fig. 4D), indicating that disengagement of one motor type can resolve tug of war events between dynein and kinesin. Although we also observed rare events where ensembles switched directions after photocleavage, more commonly we observed that moving chassis would dissociate when moving in the direction of the cleaved motor (fig. S10).
Fig. 4.
Disengagement of one motor species resolves stalled tug of war. (A) Schematic of a mixed-motor-chassis with dynein attached via photocleavable handles (purple circles). Photocleavage is induced by 405 nm laser pulses (inset). (B) Kymograph of 2D:5K*(green) and 2D*:5K (red) chassis. Purple lightening bolt indicates the start of laser pulses. Scale bars: 1 min (x), 10 μm (y). (C) Chassis classification scheme for data presented in panel D. Before (pre-state) and after (post-event) laser photocleavage the chassis were characterized as immobile, minus-end-directed, or plus-end-directed. Possible post-events also included dissociation from the microtubule. (D) Quantification of the post-photocleavage event motility of 2D*:5K (top) and 2D:5K* (bottom) chassis as a function of their pre-state (N ≥ 286). Each individual post-event fraction was calculated relative to the number of events within that given pre-state.
Using DNA origami, we built a versatile, synthetic cargo system that allowed us to determine the motile behavior of microtubule-based motor ensembles. In ensembles of identical-polarity motors, motor number had minimal affect on directional velocity, while ensembles of opposite-polarity motors engaged in a tug of war resolvable by disengaging one motor species. Yeast dynein’s high microtubule affinity allowed it to dominate in mixed ensembles while the ratio of dynein to kinesin dictated cargo directionality, supporting experiments performed in vivo or in cell-free lysates (32–34). The reduction in velocity reported here for opposite polarity motor ensembles also agrees with in vivo reports of dynein and kinesin tug of war (32, 35). The high probability with which mixed ensembles of active dynein and kinesin motors were immobile suggested that for this motor pair efficient bi-directional transport requires extrinsic regulation (36). Motors with comparable microtubule affinities and binding kinetics, such as those that coevolved in the same biological system, may produce bidirectional transport characteristics similar to those observed in vivo (6, 37, 38). The system we built provides a powerful platform to investigate the motile properties of any combination of identical- or opposite-polarity motors, and could also be used to investigate the role of motor regulation.
Supplementary Material
Acknowledgements
We thank C. Lin for assistance with electron microscopy, F. Aguet for assistance with data analysis, J. Huang, W. Qiu, W.B. Redwine, and A. Roberts for helpful advice and critical reading of the manuscript, members of the Reck-Peterson and Shih labs for advice and helpful discussions, and J. Iwasa for illustrations. EM data was collected at the Center for Nanoscale Systems, Harvard University. DNA-PAINT data was collected at the Nikon Imaging Center, Harvard Medical School. RJ is supported from the Alexander von Humboldt-Foundation through a Feodor Lynen fellowship. SRP is funded by the Rita Allen Foundation, the Harvard Armenise Foundation, and a NIH New Innovator award (1 DP2 OD004268-1). WMS is funded by NIH awards (1U54GM094608, 1DP2OD004641) and ONR awards (N000014091118, N000141010241).
Footnotes
Supplementary Materials
Materials and Methods
Figure S1–S10
Tables S1–S6
Scaffold sequence
caDNAno file of chassis structure
References and Notes
- 1.Hirokawa N, Niwa S, Tanaka Y. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Vale RD. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 3.Vallee RB, Tsai J-W. Genes Dev. 2006;20:1384–1393. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- 4.Banks GT, Fisher EMC. Genome Biol. 2008;9:214. doi: 10.1186/gb-2008-9-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai A, Mitchison TJ. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 6.Welte MA. Curr Biol. 2004;14:R525–37. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 7.Bryantseva SA, Zhapparova ON. Cell Biol. Int. 2012;36:1–6. doi: 10.1042/CBI20110413. [DOI] [PubMed] [Google Scholar]
- 8.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 9.Douglas SM, et al. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas SM, et al. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reck-Peterson SL, et al. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu W, et al. Nat Struct Mol Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- 14.Jungmann R, et al. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 15.Ke Y, Voigt NV, Gothelf KV, Shih WM. J Am Chem Soc. 2012;134:1770–1774. doi: 10.1021/ja209719k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko SH, Gallatin GM, Liddle JA. Adv. Funct. Mater. 2012;22:1015–1023. [Google Scholar]
- 17.Redwine WB, et al. Science. 2012;337 doi: 10.1126/science.1224151. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumpp S, Lipowsky R. Proc Natl Acad Sci USA. 2005;102:17284–17289. doi: 10.1073/pnas.0507363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leduc C, Pavin N, Jüicher F, Diez S. Phys. Rev. Lett. 2010;105:128103. doi: 10.1103/PhysRevLett.105.128103. [DOI] [PubMed] [Google Scholar]
- 20.Rogers AR, Driver JW, Constantinou PE, Kenneth Jamison D, Diehl MR. Physical chemistry chemical physics : PCCP. 2009;11:4882–4889. doi: 10.1039/b900964g. [DOI] [PubMed] [Google Scholar]
- 21.Lu H, et al. J of Biol Chem. 2012 [Google Scholar]
- 22.Xu J, Shu Z, King SJ, Gross SP. Traffic. 2012;9999 doi: 10.1111/j.1600-0854.2012.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 24.Jamison DK, Driver JW, Rogers AR, Constantinou PE, Diehl MR. Biophys J. 2010;99:2967–2977. doi: 10.1016/j.bpj.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yildiz A, Tomishige M, Gennerich A, Vale RD. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Constantinou PE, Diehl MR. J Biomech. 2010;43:31–37. doi: 10.1016/j.jbiomech.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Ali MY, et al. Proc Natl Acad Sci USA. 2011;108:E535–41. doi: 10.1073/pnas.1104298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vale RD, Malik F, Brown D. J Cell Biol. 1992;119:1589–1596. doi: 10.1083/jcb.119.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunwar A, Vershinin M, Xu J, Gross SP. Curr Biol. 2008;18:1173–1183. doi: 10.1016/j.cub.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamison DK, Driver JW, Diehl MR. J. of Biol. Chem. 2012;287:3357–3365. doi: 10.1074/jbc.M111.296582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levi V, Serpinskaya AS, Gratton E, Gelfand V. Biophys J. 2006;90:318–327. doi: 10.1529/biophysj.105.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster M, Lipowsky R, Assmann M-A, Lenz P, Steinberg G. Proc Natl Acad Sci USA. 2011;108:3618–3623. doi: 10.1073/pnas.1015839108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amrute-Nayak M, Bullock SL. Nat Cell Biol. 2012;14:416–423. doi: 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soppina V, Rai AK, Ramaiya AJ, Barak P, Mallik R. Proc Natl Acad Sci USA. 2009;106:19381–19386. doi: 10.1073/pnas.0906524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunwar A, et al. Proc Natl Acad Sci. 2011;108:18960–18965. doi: 10.1073/pnas.1107841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Encalada SE, Szpankowski L, Xia C-H, Goldstein LSB. Cell. 2011;144:551–565. doi: 10.1016/j.cell.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egan MJ, Tan K, Reck-Peterson SL. J Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin C, et al. Nature Chemistry. 2012 in press. [Google Scholar]
- 40.Yildiz A, et al. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.