Abstract
Background
Nontyphoidal Salmonella (NTS) species are important food-borne pathogens that cause gastroenteritis and bacteremia, and are responsible for a huge global burden of morbidity and mortality. The aim of this study was to investigate the prevalent serogroups and antibiotic resistance of NTS in our region.
Methods
We reviewed the serogroup distribution and antimicrobial susceptibility patterns of NTS strains obtained from 158 stool specimens of patients with acute diarrheal infection attending the outpatient and inpatient department at a university hospital in the Eastern Province of Saudi Arabia in the period from September, 2008 to April, 2011. A retrospective analysis of the 158 patients with NTS infection was conducted to determine the most prevalent NTS serogroups causing acute gastroenteritis and their antimicrobial susceptibility patterns.
Results
At this teaching hospital, a total of 17,436 fecal samples were analyzed during the 2008–2011 study period. Of these specimens, 158 tested positive for NTS, giving an overall prevalence of 9.06 per 1,000. Of 158 NTS cases, serogroup D1 (25.3%) was the most prevalent, followed by serogroup B (19.6%), and serogroup C1 (18.9). One third of all NTS serogroup strains tested were resistant to tetracycline. The NTS strains showed resistance to ampicillin (31.3%), amoxicillin/clavulanic acid (29.9%), trimethoprim/sulfamethoxazole (20.9%), and cefotaxime (14.93%).
Conclusion
The findings of this study support the concern that use of antibiotics in animal feeds may contribute to acquisition of resistance in food-borne bacteria, such as Salmonella. Our study also concludes that the prevalence of NTS in the Eastern Province of Saudi Arabia is very low compared with other studies worldwide.
Keywords: nontyphoidal Salmonella, serogroups, prevalence, antimicrobial resistance, Saudi Arabia
Introduction
Salmonella are motile Gram-negative facultative anaerobic bacteria in the family Enterobacteriaceae. The Salmonella genus consists of two species, ie, Salmonella enterica and Salmonella bongori.1 Most pathogenic species of Salmonella causing illness in humans belong to the S. enterica species.2 This species is further divided into six subspecies, ie, S. enterica subspecies enterica, S. enterica subspecies salamae, S. enterica subspecies arizonae, S. enterica subspecies diarizonae, S. enterica subspecies houtenae, and S. enterica subspecies indica. Nontyphoidal salmonellosis is caused by Salmonella species other than Salmonella typhi and Salmonella paratyphi. Nontyphoidal salmonellosis is one of the leading food-borne illnesses and accounts for considerable morbidity and mortality in both developed and developing countries. For instance, in the US, it was estimated that nontyphoidal salmonellosis accounted for 11% (rank number two) of food-borne illnesses, 35% (rank number one) of food borne-associated hospitalization, and 28% (rank number one) of foodborne-associated death.3,4
Nontyphoidal Salmonella (NTS) is recognized as one of the principal causes of food-borne infections worldwide.5 Among the more than 2,000 Salmonella serovars, Enteritidis is one of the top serovars reported in Saudi Arabia.6 Most cases of gastroenteritis caused by serovar Enteritidis occur sporadically or as limited outbreaks, but recent reports of large hospital-associated and nursing home-associated outbreaks emphasize the importance of serovar Enteritidis infection as a major public health problem.5,7,8 NTS is an important bacterial cause of diarrhea and community-acquired bloodstream infection.9,10 Globally, NTS gastroenteritis is estimated to cause 93.8 million illnesses and 155,000 deaths each year.11 Invasive NTS infection occurs when the organism spreads beyond the gastrointestinal mucosa to infect normally sterile sites, such as the bloodstream, meninges, bone, and joint spaces.12
Identification of NTS in fecal samples from patients is the first step in treating patients. Usually, the first line of treatment used by physicians is antibiotics. However, misuse of antibiotics inevitably leads to antibiotic resistance. The US Centers for Disease Control have discovered that, since the 1990s, NTS has shown increasing resistance to antibiotics, including ampicillin and chloramphenicol.13 Therefore, initial studies on NTS should not be restricted to prevalence, but should also include their antibiotic resistance. In the light of the above, the aim of the present study was to determine the prevalence and antimicrobial susceptibility of NTS serogroups at King Fahd Hospital of the University (KFHU), Al-Khobar, Saudi Arabia.
Materials and methods
Study design
Clinical records of Salmonella strains from the clinical microbiology laboratory at KFHU for the period September 2008 to April 2011 were retrospectively reviewed. The KFHU routinely collects fecal samples from all patients admitted with acute gastroenteritis and diarrheal illness.
Culture procedure
All fecal samples were cultured directly on Hektoen agar and xylose lysine deoxycholate agar, inoculated in selenite F broth (Oxoid Ltd, Basingstoke, UK), subcultured, and identified further using an automated card system (bioMérieux Vitek Inc, Hazelwood, MO, USA) and the API 20E (bioMérieux, Marcy l’Etoile, France). All isolates were identified as Salmonella according to standard microbiologic techniques.13 The identified Salmonella isolates were then serogrouped using somatic group Salmonella A–G antisera (Murex Biotech Ltd, Dartford, UK).
Antibiotic susceptibility testing
Antibiotic susceptibility testing was determined using an automated Vitek machine with Gram-negative bacteria cards, which gave minimum inhibitory concentration results and interpretation of the results as resistant or susceptible according to the breakpoints for each antibiotic. Some antibiotic susceptibility testing was determined by means of the Kirby-Bauer disk diffusion method using the guidelines provided by the Clinical Laboratory Standards Institute (CLSI), formerly known as the National Committee for Clinical Laboratory Standards.14 Susceptibility tests were done on Mueller Hinton agar (Oxoid Ltd) using the following concentrations (μg/disc) of antibiotics (Oxoid Ltd): ampicillin 10 μg, amoxicillin/clavulanic acid 20/10 μg, amikacin 30 μg, aztreonam 30 μg, cefotaxime 5 μg, ceftriaxone 30 μg, cephalothin 30 μg, ciprofloxacin 5 μg, gentamicin 10 μg, imipenem 10 μg, piperacillin 100 μg, and trimethoprim/sulfamethoxazole (1.25 μg/23.75 μg). Results were scored as susceptible, moderately susceptible, or resistant, according to CLSI criteria. Escherichia coli 25922 (American Type Culture Collection, Manassas, VA, USA) was used as the reference strain.
Statistical analysis
The data were analyzed using bivariate tables and scatter graphs. The change in distribution over the years was analyzed using the chi-square test for a trend in situations where it is applicable (zeros not being adjacent to each other or cell frequencies not being less than two). The similarity of the distribution of the serogroups across the different years was tested using the Kruskal–Wallis test. The chi-square goodness of fit test was used to assess how a distribution compared with uniform distribution. For all tests, a P-value <0.05 was considered to be statistically significant. Analysis was done using MS Excel 2007 (Microsoft, Redmond, WA, USA) and PAST statistical software (version 2.04).15
Results
At this teaching hospital, a total of 17,436 fecal samples were analyzed during the 2008–2011 study period. Of these specimens, a total of 158 tested positive for NTS, giving an overall prevalence of 9.06 per 1,000. The figures for each year are shown in Table 1. As can be seen, the prevalence increased from 7.0 per 1,000 in 2008 to 9.89 per 1,000 in 2009; thereafter, the prevalence declined slightly between 2009 and 2011. Positive NTS samples during 2009 and 2010 remained constant at between 40 and 41 of total positive samples, but the trend varied among the different serogroups. Serogroup D remained the most frequent of the Salmonella serogroups. The annual prevalence of serogroup D increased by more than half in 2011 compared with 2008 (Table 2).
Table 1.
Prevalence of nontyphoidal Salmonella, 2008–2011
| 2008 | 2009 | 2010 | 2011 | Total | |
|---|---|---|---|---|---|
| Positive samples | 27 | 41 | 40 | 50 | 158 |
| All samples | 3,854 | 4,145 | 4,142 | 5,295 | 17,436 |
| Prevalence (per 1,000) | 7.01 | 9.89 | 9.66 | 9.44 | 9.06 |
Table 2.
Prevalence of Salmonella serogroups isolated during 2008–2011
| Serogroup | 2008
|
2009
|
2010
|
2011
|
Total
|
P-value
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | Slope | Trend | |
| B | 7 | 25.9 | 11 | 26.8 | 3 | 7.5 | 10 | 20 | 31 | 19.6 | 0.255 | 0.107 |
| C1 | 5 | 18.5 | 11 | 26.8 | 10 | 25 | 4 | 8 | 30 | 19 | 0.114 | 0.135 |
| C2 | 5 | 18.5 | 4 | 9.8 | 3 | 7.5 | 4 | 8 | 16 | 10.1 | 0.186 | 0.638 |
| C3 | 0 | 0 | 1 | 2.4 | 5 | 12.5 | 1 | 2 | 7 | 4.4 | * | * |
| D1 | 1 | 3.7 | 7 | 17.1 | 15 | 37.5 | 17 | 34 | 40 | 25.3 | 0.001 | 0.2732 |
| D2 | 0 | 0 | 0 | 0 | 4 | 10.0 | 10 | 20 | 14 | 8.9 | * | * |
| E1 | 4 | 14.8 | 4 | 9.8 | 0 | 0 | 3 | 6 | 11 | 7 | 0.091 | 0.1945 |
| F | 0 | 0 | 1 | 2.4 | 0 | 0 | 0 | 0 | 1 | 0.6 | * | * |
| G1 | 2 | 7.4 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1.3 | * | * |
| H | 1 | 3.7 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 1.3 | * | * |
| K | 2 | 7.4 | 2 | 4.9 | 0 | 0 | 0 | 0 | 4 | 2.5 | * | * |
| Total¥ | 27 | 100 | 41 | 100 | 40 | 100 | 50 | 100 | 158 | 100 | ||
Notes:
chi-square trend test not applicable
total might be different from 100 because of rounding up.
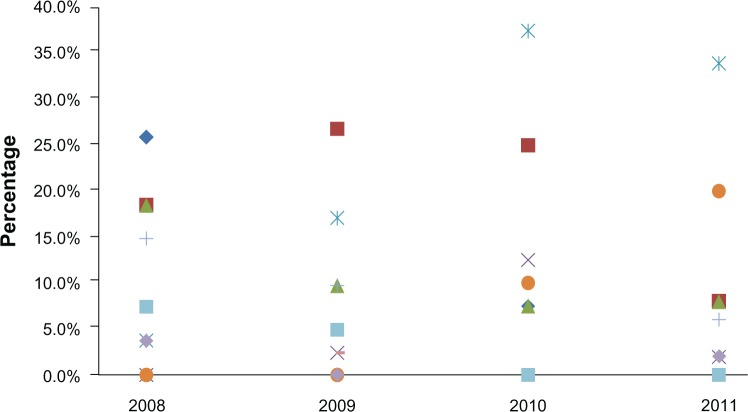
From 2008 to 2011, 158 strains of NTS were isolated from fecal samples of patients with gastroenteritis. Salmonella strains belonging to serogroup D1 had the highest prevalence (25.3%) followed by serogroup B (19.6%), and C1 serogroup (19%). Salmonella serogroup strains belonging to F (0.6%), G1 (1.3%), and H (1.3%) were the least prevalent (Table 2). The distribution of serogroups is shown in Figure 1. The only serogroup showing a clear increase in trend is D1. The chi-square test for trend confirms that the only significant change in slope over the study period occurred in serogroup D1. The trend for D1 is fairly linear. The Kruskal–Wallis test does not show any significant difference in year-to-year distribution between the different serogroups (H=0.413, P=0.941).
Figure 1.
Distribution of Salmonella serogroups isolated during 2008–2011.
Resistance to ampicillin (31.3%) and amoxicillin/clavulanic acid (29.9%) was observed in most NTS strains. Resistance to sulfamethoxazole, trimethoprim, and cefotaxime was observed in 20.9% and 14.93% of strains (Table 3). Of all strains, the lowest resistance was noted with ciprofloxacin (3.0%, Table 3). The Kruskal–Wallis test did not show any significant difference in year-to-year distribution between the different antibiotics (H=1.803, P=0.6205). All strains were susceptible to amikacin, aztreonam, ceftriaxone, cephalothin, gentamicin, imipenem, and piperacillin.
Table 3.
Antibiotic resistance in Salmonella isolated during 2008–2011
| Antibiotic | 2008
|
2009
|
2010
|
2011
|
Total
|
P-value
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | Slope | Trend | |
| AM | 13 | 33.3 | 10 | 35.7 | 9 | 37.5 | 10 | 23.3 | 42 | 31.3 | 0.337 | 0.569 |
| AMC | 13 | 33.3 | 10 | 35.7 | 9 | 37.5 | 8 | 18.6 | 40 | 29.9 | 0.153 | 0.167 |
| CXM | 4 | 10.3 | 0 | 0 | 0 | 0 | 16 | 37.21 | 20 | 14.93 | * | * |
| CIP | 3 | 7.7 | 0 | 0 | 0 | 0 | 1 | 2.3 | 4 | 3 | * | * |
| SXT | 6 | 15.4 | 8 | 28.6 | 6 | 25 | 8 | 18.6 | 28 | 20.9 | 0.838 | 0.152 |
| Total¥ | 39 | 100 | 28 | 100 | 24 | 100 | 43 | 100 | 134 | 100 | ||
Notes:
chi-square trend test not applicable
total might be different from 100 because of rounding up.
Abbreviations: AM, ampicillin; AMC, amoxicillin/clavulanic acid; CXM, cefotaxime; CIP, ciprofloxacin; SXT, sulfamethoxazole.
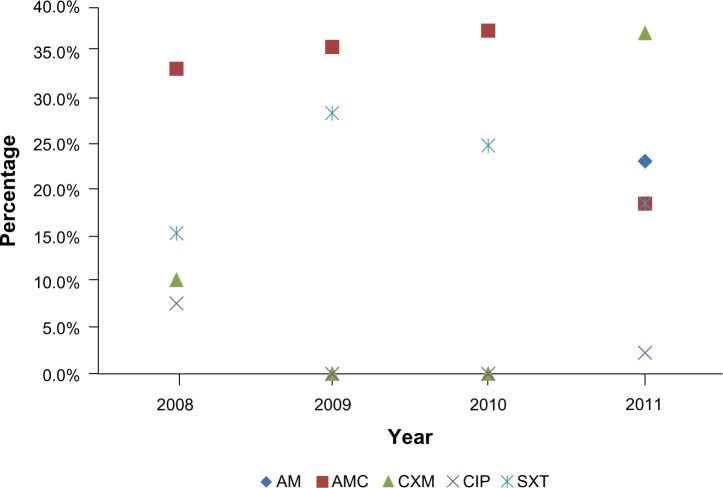
Overall, antibiotic resistance in the NTS serogroup strains increased from 29.1% in 2008 to 32.1% in 2011. According to the chi-square goodness-of-fit test, this increase was not significantly different from the uniform distribution during the study period (P=0.065). Resistance to cefotaxime increased from 10.3% of all isolates in 2008 to 14.93% of all isolates in 2011 (Figure 2). The number of isolates showing resistance to trimethoprim/sulfamethoxazole fluctuated between six and eight throughout the study period. There was an increase in the number of isolates showing resistance to trimethoprim/sulfamethoxazole from four in 2008 to 16 in 2011 (Table 3). The highest overall resistance (40.2%) was observed among strains belonging to serogroup B, followed by serogroups C2 (18.6%), C1 and D1 (10.4%), and C3 (8.2%, Table 4).
Figure 2.
Distribution of antibiotic resistance in Salmonella isolated during 2008–2011.
Abbreviations: AM, ampicillin; AMC, amoxicillin/clavulanic acid; CXM, cefotaxime; CIP, ciprofloxacin; SXT, sulfamethoxazole.
Table 4.
Overall antimicrobial resistance in Salmonella by serogroup during 2008–2011
| Serogroup | Year
|
Total
|
||||
|---|---|---|---|---|---|---|
| 2008 | 2009 | 2010 | 2011 | n | % | |
| B | 15 | 20 | 3 | 16 | 54 | 40.0 |
| C1 | 1 | 0 | 4 | 9 | 14 | 10.4 |
| C2 | 14 | 3 | 3 | 5 | 25 | 18.7 |
| C3 | 0 | 0 | 11 | 0 | 11 | 8.2 |
| D1 | 1 | 3 | 3 | 7 | 14 | 10.4 |
| D2 | 0 | 0 | 0 | 3 | 3 | 2.2 |
| E1 | 2 | 2 | 0 | 2 | 6 | 4.5 |
| G1 | 2 | 0 | 0 | 0 | 2 | 1.5 |
| H | 4 | 0 | 0 | 1 | 5 | 3.7 |
| Total¥ | 39 | 28 | 24 | 43 | 134 | 100.0 |
Note:
Total might be different from 100 because of rounding up.
Discussion
NTS infections are a leading cause of food poisoning and enteric infection, and are an important public health problem worldwide.3,9,10,16,17Salmonella may cause gastroenteritis in people of all ages, and is responsible for severe invasive disease in infants, the elderly, and the immunocompromised.18,19 The frequency of antimicrobial resistance and number of resistance determinants in Salmonella has risen markedly.20 Antimicrobial susceptibility to ampicillin, trimethoprim/sulfamethoxazole, and quinolones for Salmonella isolated from fecal specimens should be routinely tested and reported using the CLSI guidelines.
A surveillance study by Su et al demonstrated an obvious increase in overall antimicrobial resistance among Salmonella from 20%–30% in the early 1990s to as high as 70% in some countries at the turn of the century.21 Conventional antibiotics, such as ampicillin, chloramphenicol, and trimethoprim/sulfamethoxazole, are no longer an appropriate choice for the treatment of invasive salmonellosis.21 Increasing antimicrobial resistance in NTS is a global public health problem that complicates antimicrobial therapy, and is increasingly due to the overuse and misuse of antimicrobial agents in animal feeds.22–24 Several studies worldwide have reported increased morbidity and mortality in patients infected with resistant Salmonella strains.25,26 Resistance to antibiotics is posing a serious problem in the treatment of salmonellosis. In the present study, serogroup D1 (25.3%) was the most frequent and prevalent serogroup, followed by serogroup B (19.6%) and serogroup C1 (18.9%). There was an increase in gastroenteritis caused by serogroup D1 from 2008 to 2011 (Figure 1 and Table 2). Our study also confirms the emergence and rapid increase in cases of NTS infection with serogroup D1, particularly serovar Enteritidis. A similar study conducted by Chiu et al at a university hospital in Taiwan reported that the incidence of serogroup D Salmonella has been increasing in Taiwan.5 The present study indicates that serogroup D was more prevalent than serogroup B and other serogroups, and if this trend were to continue, the incidence of NTS serogroup D would soon surpass that of serogroup B infection. Such a trend has been reported in the US, Europe, Taiwan, Malawi, Thailand, and Malaysia.5,8,20,27–29 Serovar Enteritidis is known to be closely associated with layer and broiler flocks, and infection is generally believed to be derived from poultry and poultry products, including eggs.8,30
The current study findings are consistent with those of other studies reporting that most of the NTS (serovar Enteritidis) are resistant to a wide range of antimicrobial agents.7,30–32 However, in our study, we found a higher rate of resistance to trimethoprim/sulfamethoxazole (17.7% of isolates during the period from September, 2008 to April, 2011, Table 3). These findings are in agreement with those of a similar study on the emergence of Salmonella with extended spectrum β-lactamase enzymes that hydrolyze and confer resistance to cefotaxime in several European countries.33,34 In the present study, some of the strains (14.93%) were reported to be resistant to cefotaxime and ciprofloxacin (3%). Ciprofloxacin and cefotaxime are the antimicrobial agents recommended for treatment of invasive infections due to Salmonella.35,36
NTS species are important food-borne pathogens, with acute gastroenteritis being the most common clinical manifestation. However, invasion beyond the gastrointestinal tract occurs in approximately 5% of patients with NTS gastroenteritis, resulting in bacteremia.12 In industrialized countries, NTS constitutes a well recognized public health problem that, in healthy subjects, is usually encountered clinically as self-limited gastroenteritis.37,38 In immunocompromised and debilitated hosts, NTS can become invasive, leading to bacteremia, sepsis, and focal infections (eg, meningitis).38,39 Invasiveness is also observed in infants younger than 3 months of age who become infected with NTS in industrialized countries, resulting in bacteremia and focal infections.40 In sub-Saharan Africa, studies have documented the important role of NTS as invasive bacterial pathogens.41–44
We conclude that there is a need to establish standard first-line therapy for salmonellosis. Clinicians should be aware of the local epidemiology of NTS and carefully review the results of antimicrobial susceptibility testing once available from the clinical laboratory. Active monitoring of NTS serogroups for antibiotic resistance patterns is essential because of potential acquisition of resistance genes by pre-existing serovar Enteritidis strains via horizontal gene transfer.5,19,20 Knowledge of the distribution of prevalence of Salmonella serogroups is potentially of epidemiologic and public health importance.
Acknowledgments
The authors thank their laboratory specialists (Lauro Bartolome, Piodennis Dasal, Bader Sager, Abdulrahim Osman) for contributing to this research.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Grimont PAD, Weill FX.Antigenic formulae of the Salmonellae serovars 9th edGeneva, Switzerland: World Health Organization; 2007Available from: http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-000036-089Accessed September 1, 2013 [Google Scholar]
- 2.Crum-Cianfone NF. Salmonellosis and the gastrointestinal tract: more than just peanut butter. Curr Gastroenterol Rep. 2008;10:424–431. doi: 10.1007/s11894-008-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States – major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention CDC Estimates of foodborne illness in the United States 2011Available from: http://www.cdc.gov/features/dsfoodborneestimates/Accessed September 1, 2013
- 5.Chiu CH, Su LH, Hung CC, Chen KL, Chu C. Prevalence and antimicrobial susceptibility of serogroup D nontyphoidal Salmonella in a university hospital in Taiwan. J Clin Microbiol. 2004;42:415–441. doi: 10.1128/JCM.42.1.415-417.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panhotra BR, Saxena AK, Al-Ghamdi AM. Emerging nalidixic acid and ciprofloxacin resistance in non-typhoidal Salmonella isolated from patients having acute diarrheal disease. Ann Saudi Med. 2004;24:332–336. doi: 10.5144/0256-4947.2004.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling JM, Koo IC, Kam KM, et al. Antimicrobial susceptibility and molecular epidemiology of Salmonella enterica serotype Enteritidis strains isolated in Hong Kong from 1986 to 1996. J Clin Microbiol. 1998;36:1693–1699. doi: 10.1128/jcm.36.6.1693-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigue DC, Tauxe RV, Rowe B, et al. International increase in Salmonella Enteritidis: a new pandemic? Epidemiol Infect. 1990;105:21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crump JA, Medalla FM, Joyce KW, et al. Emerging Infections Program NARMS Working Group Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother. 2011;55:1148–1154. doi: 10.1128/AAC.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voetsch AC, Van Gilder TJ, Angulo FJ, et al. Emerging Infections Program FoodNet Working Group FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38:S127–S134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- 11.Majowicz SE, Musto J, Scallan E, et al. International Collaboration on Enteric Disease ‘Burden of Illness’ Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 12.Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32:263–269. doi: 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- 13.Farmer JJ., III . Enterobacteriaceae: introduction and identification. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. Washington, DC: American Society for Microbiology Press; 1995. [Google Scholar]
- 14.Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document 2010; M100-S20 Wayne, PA: Clinical and Laboratory Standards Institute; 2010Available from: http://www.techstreet.com/products/1662846Accessed September 1, 2013 [Google Scholar]
- 15.Hammer O, Harper D, Ryan P. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica. 2001;4(1):9. [Google Scholar]
- 16.Sun S, Negrea A, Rhen M, Andersson DI. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2009;53:2298–2305. doi: 10.1128/AAC.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majowicz SE. The global burden of nontyphoidal Salmonella gasteroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 18.Rocourt J, Moy G, Vierk K, et al. The present state of foodborne disease in OECD countries Geneva, Switzerland: World Health Organization; 2003Available from: http://www.who.int/foodsafety/publications/foodborne_disease/oecd_fbd.pdfAccessed September 1, 2013 [Google Scholar]
- 19.Goldberg MB, Rubin RH. The spectrum of Salmonella infection. Infect Dis Clin North Am. 1988;2:571–598. [PubMed] [Google Scholar]
- 20.World Health Organization The medical impact of the use of antimicrobials in food animals: report and proceedings of a WHO meeting, Berlin, Germany, October 13–17, 1997 Geneva, Switzerland: World Health Organization; 1997Available from: http://whqlibdoc.who.int/hq/1997/WHO_EMC_ZOO_97.4.pdfAccessed September 1, 2013 [Google Scholar]
- 21.Su LH, Chiu CH, Chu C, Ou JT. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis. 2004;39:546–551. doi: 10.1086/422726. [DOI] [PubMed] [Google Scholar]
- 22.Tsai MH, Wu SR, Lee HY, et al. Recognition of mechanisms involved in bile resistance important to halting antimicrobial resistance in non-typhoidal Salmonella. Int J Antimicrob Agents. 2012;40:151–157. doi: 10.1016/j.ijantimicag.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 23.Swartz MN. Human diseases caused by foodborne pathogens of animal origin. Clin Infect Dis. 2002;34:111–122. doi: 10.1086/340248. [DOI] [PubMed] [Google Scholar]
- 24.McDonald LC, Chen MT, Lauderdale TL, Ho M. The use of antibiotics critical to human medicine in food-producing animals in Taiwan. J Microbiol Immunol Infect. 2001;34:97–102. [PubMed] [Google Scholar]
- 25.Helms M, Vastrup P, Gerner-Smidt P, Mølbak K. Excess mortality associated with antimicrobial drug-resistant Salmonella Typhimurium. Emerg Infect Dis. 2002;8:490–495. doi: 10.3201/eid0805.010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varma JK, Molbak K, Barrett TJ, et al. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis. 2005;191:554–561. doi: 10.1086/427263. [DOI] [PubMed] [Google Scholar]
- 27.Gordon MA, Graham SM, Walsh AL, et al. Epidemics of invasive Salmonella enterica serovar Enteritidis and S. enterica serovar Typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008;46:963–969. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 28.Kiratisin P. Bacteraemia due to non-typhoidal Salmonella in Thailand: clinical and microbiological analysis. Trans R Soc Trop Med Hyg. 2008;102:384–388. doi: 10.1016/j.trstmh.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Dhanoa A, Fatt QK. Non-typhoidal Salmonella bacteraemia: epidemiology, clinical characteristics and its association with severe immunosuppression. Ann Clin Microbiol Antimicrob. 2009;8:15. doi: 10.1186/1476-0711-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen SJ, Bishop R, Brenner FW, et al. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987–1997. J Infect Dis. 2001;183:753–761. doi: 10.1086/318832. [DOI] [PubMed] [Google Scholar]
- 31.Lee LA, Puhr ND, Maloney EK, Bean NH, Tauxe RV. Increase in antimicrobial-resistant Salmonella infections in the United States, 1989–1990. J Infect Dis. 1994;170:128–134. doi: 10.1093/infdis/170.1.128. [DOI] [PubMed] [Google Scholar]
- 32.Threlfall EJ, Ward LR, Skinner JA, Rowe B. Increase in multiple antibiotic resistance in nontyphoidal salmonellas from humans in England and Wales: a comparison of data for 1994 and 1996. Microb Drug Resist. 1997;3:263–266. doi: 10.1089/mdr.1997.3.263. [DOI] [PubMed] [Google Scholar]
- 33.Batchelor M, Threlfall EJ, Liebana E. Cephalosporin resistance among animal-associated Enterobacteria: a current perspective. Expert Rev Anti Infect Ther. 2005;3:403–417. doi: 10.1586/14787210.3.3.403. [DOI] [PubMed] [Google Scholar]
- 34.Meakins S, Fisher IS, Berghold C, et al. Enter-net Participants Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000–2004: a report from the Enter-net International Surveillance Network. Microb Drug Resist. 2008;14:31–35. doi: 10.1089/mdr.2008.0777. [DOI] [PubMed] [Google Scholar]
- 35.Threlfall EJ. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infection. FEMS Microbiol Rev. 2002;26:141–148. doi: 10.1111/j.1574-6976.2002.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 36.Angulo FJ, Johnson KR, Tauxe TW, et al. Origins and consequences of antimicrobial-resistent nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb Drug Resist. 2000;6:77–83. doi: 10.1089/mdr.2000.6.77. [DOI] [PubMed] [Google Scholar]
- 37.Voetsch AC, Van Gilder TJ, Angulo FJ, et al. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38:S127–S134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- 38.Adak GK, Long SM, O’Brien SJ, et al. Trends in indigenous foodborne disease and deaths, England and Wales: 1992 to 2000. Gut. 2002;51:832–841. doi: 10.1136/gut.51.6.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy M, Villar R, Vugia DJ, et al. Emerging Infections Program FoodNetWorking Group Hospitalizations and deaths due to Salmonella infections, FoodNet, 1996–1999. Clin Infect Dis. 2004;38:S142–S148. doi: 10.1086/381580. [DOI] [PubMed] [Google Scholar]
- 40.Vugia DJ, Samuel M, Farley MM, et al. Emerging Infections Program FoodNetWorking Group Invasive Salmonella infections in the United States, FoodNet, 1996–1999: incidence, serotype distribution, and outcome. Clin Infect Dis. 2004;38:149–156. doi: 10.1086/381581. [DOI] [PubMed] [Google Scholar]
- 41.Berkley JA, Lowe BS, Mwangi I, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 42.Ikumapayi UN, Antonio M, Sonne-Hansen J, et al. Molecular epidemiology of community-acquired invasive non-typhoidal Salmonella among children aged 2–29 months in rural Gambia and discovery of a new serovar, Salmonella enterica Dingiri. J Med Microbiol. 2007;56:1479–1484. doi: 10.1099/jmm.0.47416-0. [DOI] [PubMed] [Google Scholar]
- 43.Sigaúque B, Roca A, Mandomando I, et al. Community-acquired bacteremia among children admitted to a rural hospital in Mozambique. Pediatr Infect Dis J. 2009;28:108–113. doi: 10.1097/INF.0b013e318187a87d. [DOI] [PubMed] [Google Scholar]
- 44.Mandomando I, Macete E, Sigaúque B, et al. Invasive non-typhoidal Salmonella in Mozambican children. Trop Med Int Health. 2009;14:1467–1474. doi: 10.1111/j.1365-3156.2009.02399.x. [DOI] [PubMed] [Google Scholar]