Abstract
A Mediterranean diet increases intakes of n-3 and n-9 fatty acids and lowers intake of n-6 fatty acids. This can impact colon cancer risk since n-6 fatty acids are metabolized to pro-inflammatory eicosanoids. The purpose of this study was to evaluate interactions of polymorphisms in the fatty acid desaturase genes, FADS1 and FADS2, and changes in diet on fatty acid concentrations in serum and colon. A total of 108 individuals at increased risk of colon cancer were randomized to either a Mediterranean or a Healthy Eating diet. Fatty acids were measured in both serum and colonic mucosa at baseline and after 6 months. Each individual was genotyped for four single nucleotide polymorphisms in the FADS gene cluster. Linear regression was used to evaluate the effects of diet, genotype and the diet by genotype interaction on fatty acid concentrations in serum and colon. Genetic variation in the FADS genes was strongly associated with baseline serum arachidonic acid (n-6, AA) but serum eicosapentaenoic acid (n-3) and colonic fatty acid concentrations were not significantly associated with genotype. After intervention, there was a significant diet by genotype interaction for AA concentrations in colon. Subjects who had all major alleles for FADS1/2 and were following a Mediterranean diet had 16% lower AA concentrations in the colon after 6 months of intervention than subjects following the Healthy Eating diet. These results indicate that FADS genotype could modify the effects of changes in dietary fat intakes on AA concentrations in the colon.
Keywords: Fatty acid desaturase, diet-genotype interaction, colon cancer prevention, nutrition
Introduction
Many studies have suggested that a Mediterranean diet, as compared to a typical Western diet, may decrease the risk of various chronic diseases including colorectal cancer (1, 2). Rates of colorectal cancer were very low in Greece and have increased as diet has drifted away from the traditional eating pattern (3). The traditional Greek diet, relative to a Western diet, had lower intakes of n-6 polyunsaturated fatty acids (PUFA) and red meat, but higher intakes of plant-based foods, fish and monounsaturated fatty acids (MUFA) chiefly from olive oil (2). The fat content of the Mediterranean diet is of particular interest for colon cancer prevention since in intervention studies increasing fiber alone does not appear to be preventive, and increased intakes of fruit and vegetables have had modest preventive effects (4–6). In particular, we hypothesized lower intakes of n-6 linoleic acid and higher intakes of n-3 fatty acids have implications for preventing colon cancer since n-6 fatty acids are metabolized to eicosanoids such as prostaglandin E2 (PGE2) that is pro-inflammatory in the colon (7). PGE2 is formed from arachidonic acid (AA, 20:4 n-6) by cyclooxygenases in the colonic mucosa, and it plays an important role in colonic crypt cellular expansion and subsequent adenoma formation (8).
In addition to the possible effects of dietary intakes, genetic variation in fatty acid desaturase genes has been shown to influence serum and tissue AA concentrations (9–15). Delta-5 desaturase (FADS1) and delta-6 desaturase (FADS2) are key desaturase enzymes involved in the synthesis of AA and eicosapentaenoic acid (EPA, 20:5, n-3) from 18 carbon precursor fatty acids. Dietary intake of AA is low in humans; however, AA comprises between 5–10% of the phospholipids in cells due to elongation and desaturation of linoleic acid (18:2 n-6) to AA (16).
Polymorphisms in the FADS1 and FADS2 genes have been identified, and these significantly affect PUFA concentrations in serum. The minor alleles are associated with lower desaturase activity and lower concentrations of AA in blood (9–15). Analogous associations for EPA and docosahexaenoic acid (DHA) have not been consistent across studies, perhaps since certain types of fish can supply high amounts of pre-formed EPA and DHA. Dietary intakes are important to consider since conversion of dietary linolenic acid to longer chain n-3 fatty acids competes with the analogous process for n-6 fatty acids (17). (In addition to diet, desaturase activity appears to be important in cardiovascular health, and presence of the minor allele in FADS1/2 has been associated with improved measures of blood lipids, C-reactive protein, insulin and fasting glucose (18–21). This indicates that lower AA levels are associated with lower pro-inflammatory states. The prevalence of minor alleles appears to have evolved in response to Western diets that are plentiful in n-6 fatty acids, and they are more prevalent in persons of European descent than of African descent (11, 22).
Much less research is available on how FADS polymorphisms might affect changes in fatty acids in response to changes in diet, and the available studies have generally focused on n-3 fatty acid supplementation. Flaxseed supplementation, which provides linolenic acid (18:3, n-3), was less effective in increasing EPA concentrations in minor allele carriers of either FADS1 or FADS2, resulting in significant diet by genotype interactions on plasma concentrations of EPA and AA (23). Dietary n-3 fatty acids also may interact with FADS genotype in affecting concentrations of blood cholesterol and triglycerides, with significant beneficial effects for carriers of all minor alleles being found in some but not all studies (20, 24–26).
The goal of this present study was to assess potential interactions of polymorphisms in FADS1 and FADS2 with changes in diet on levels of arachidonic acid (AA) and eicosapentaenoic acid (EPA) in the serum and in the colonic mucosa of persons at increased risk for colon cancer. This was a secondary analysis of a randomized clinical trial that evaluated changes in fatty acids and carotenoids elicited by six months of intervention with either a Mediterranean or a standard Healthy Eating diet. In that study we observed that dietary changes had little effect on colon fatty acids, which led to the hypothesis that metabolic factors may be limiting for changes in fatty acids (27). The randomized study obtained both blood and colon biopsies. Here, the relationships of FADS polymorphisms with serum and colonic fatty acid concentrations were evaluated at baseline and after six months of dietary intervention.
Methods
Study Design and Eligibility
Details of recruitment and conduct of the Healthy Eating for Colon Cancer Prevention Study have been published previously (27, 28). The study was approved by the University of Michigan Medical Internal Review Board and was registered at the ClinicalTrials. org (NCT00475722). Briefly, 120 individuals at increased risk of colon cancer gave informed consent and were randomized to follow a modified Mediterranean diet or to Healthy People 2010 diet for 6 months. Blood and colonic mucosal tissue samples were collected at baseline and at 6 months by flexible sigmoidoscopy without prior preparation of the bowels. Blood was drawn after an overnight fast. At baseline, a Health Status Questionnaire was filled out by participants that included health and demographic data. Health information was asked again at 6 months. Dietary data was collected at 0 and 6 months using two days of food records and two 24-hour recalls.
The decision to genotype subjects with regard to fatty acid desaturases was made after the study began, and consent for genotyping could not be obtained from nine individuals, two of whom completed 6 months of study and seven of whom had dropped out after enrolling. Three samples were not genotyped successfully. The present analysis therefore included 108 of 120 subjects enrolled in the study and randomized to 6 months of counseling for either a Mediterranean or a Healthy Eating diet.
The frequency of counseling sessions was the same in both study arms. The Healthy Eating diet had dietary goals based on the Healthy People 2010 diet. The goals were to include 2 servings/day of fruit, 3 servings/day of vegetables with at least one of those servings being dark green or orange, 6 servings/day of grains with at least 3 from whole grains, less than 10% of calories from saturated fat and less than 30% of calories from total fat. The Mediterranean diet had goals for consumption of high n-3 foods such as fish or flax at least 2 times a week, consumption of foods in a manner to increase MUFA and decrease n-6 PUFA intakes, 6 servings/day of grains with at least 3 from whole grains, and 7–9 fruits and vegetable servings/day in specified variety.
Serum and Colonic Fatty Acids
Fatty acid analysis was performed by gas chromatography with mass spectral detection (GC-MS) of fatty acid methyl esters. Total lipids were extracted from serum using a 1:1 mixture of chloroform and methanol, and 17:0 (1,2-diheptadecanoyl-sn-glycero-3-phosphocholine) was used as the internal standard. For colon tissue, one biopsy of about 5 mg was sufficient for analysis of fatty acids. The biopsy was pulverized in liquid nitrogen, sonicated in 150 μl of ice-cold phosphate buffered saline containing 0.1% BHT and 1mM EDTA with an Ultrasonic processor (30 seconds twice), and then total lipids were extracted with 1 ml of chloroform and methanol (1:1). The organic layer in either case was used to prepare fatty acid methyl esters with METH-PREP II derivatization reagent (Alltech, Deerfield, IL). The GC-MS analysis was carried out with A SupelcoSP2330 column, 30m × 0.32mm × 0.2μm film thickness (Sigma-Aldrich, St. Louis, MO), a HP 7673/5971 GC-MS and helium as the carrier gas with a validated assay (29). The following fatty acids in serum and colon tissue were measured in 12 analytical different batches: 12:0, 14:0, 16:0, 16:1, 18:0, 18:1, 18:2 (n6), 18:3 (n3), 20:0, 20:1, 20:3 (n6), 20:4 (n6), 20:5 (n3) and 22:6 (n3).
DNA Extraction and Genotyping
Many polymorphisms have been identified in the FADS1/2 gene cluster. Haplotypes have been constructed using 3 to 18 single nucleotide polymorphisms, and AA concentrations were typically about 30% higher in carriers of all major alleles (9, 12, 16, 30). This literature has indicated that there was little additional benefit from genotyping more than three SNPs, we therefore chose to genotype the three SNPs used in the study of the Rzehak et al. (30). A subsequent genome-wide association study indicated that another polymorphism in the FADS1/2 region explained 18% of the inter-individual variation in AA concentrations, we therefore added rs174537 to the present analysis (21).
DNA was extracted from the buffy coat of heparinized blood samples. The buffy coat had been collected for each blood sample and mixed with 1% sodium dodecyl sulfate/1 mM EDTA prior to freezing at −80°C. After all the samples had been collected, they were treated with RNases A and heat-treated RNase T1 followed by digestion with protease K, solvent extraction and precipitation of DNA. The DNA was purified using MinElute Reaction Cleanup Kit (QIAGEN) to ensure high quality DNA for genotyping. Four SNPs were genotyped; one in the FADS2 gene (rs3834458), two SNPs located in FADS1 (rs174556 and rs174561), and one SNP located in the intragenic region between FADS1 and FADS2 (rs174537).
Three of the SNPs (rs174556, rs174561, and rs174537) were genotyped using TaqMan SNP Genotyping assays (Applied Biosystems). All assays were done both in real time and post read mode for allelic discrimination on an AB7900 system. The rs3834458 polymorphisms was detected by sequencing. For quality control 10% of all samples were re-genotyped. All plates incorporated positive and negative controls.
PCR reactions for rs3834458 included 5 μl (20 pmol/μl) of both forward and reverse primers, 12 μl AmpliTaq Gold master mix, 10 μg/μl genomic DNA, and Millipore water for a total volume of 25 μl. Primers used for rs3834458 were 5′-TCCACGATTCCCAAAGAGAC-3′ and 5′-TCTGCAACCTCCCTAGAGACA-3′. Samples were covered in mineral oil, denatured for 10 minutes at 95°C, were passed through 40 cycles of amplification consisting of 1 minute of denaturation at 95°C, 1 minute of primer annealing at 55°C, and 1 minute of elongation at 72°C. The PCR products were checked by running on a 2% agarose gel stained with ethidium bromide prior to sequencing. Sequencing was done on an ABI 3730 sequencer in the University of Michigan Sequencing Core Facility.
Statistical Analyses
The distributions of fatty acid variables were first checked for normality and transformed to approximate normality as needed prior to analyses. The transformations applied prior to analysis are given in the table footnotes, and variables were back transformed to calculate percent increases or differences. Untransformed means are shown in the Tables for ease of interpretation. Deviations from Hardy-Weinberg equilibrium for the genotypes of each SNP were tested using chi-square tests. Differences in baseline parameters between diet arms were assessed using independent t-tests or chi-square tests, as appropriate (Tables 1 and 2). Genotype data for the four SNPs were summarized to yield the count of minor alleles (the minimum and maximum counts were 0 and 8, respectively). Linear regression was used to evaluate the effect of number of minor alleles on fatty acid concentrations. Subsequently, a binary variable for genotype group was created by the presence/absence of minor alleles, i.e., all major alleles versus one or more minor alleles.
Table 1.
Characteristics of study subjects at baseline by diet arm. The data shown are mean (SD) or number (%).
| Characteristic | Healthy Eating arm (n=54) | Mediterranean arm (n=54) | P-value a |
|---|---|---|---|
| Gender, female | 38 (70%) | 40 (74%) | 0.67 |
| Age, years | 51.1 (13.3) | 54.9 (9.9) | 0.10 |
| Caucasian | 49 (91%) | 45 (83%) | 0.26 |
| BMI, kg/m2 | 26.9 (3.5) | 26.8 (3.9) | 0.88 |
| Completed 6 months of study | 43 (80%) | 47 (87%) | 0.31 |
| Minor Allele Frequency | |||
| FADS2 rs3834458 | 29 (54%) | 26 (48%) | 0.87 |
| FADS1 rs174556 | 28 (52%) | 24 (44%) | 0.68 |
| FADS1 rs174561 | 27 (50%) | 23 (43%) | 0.74 |
| FADS1 rs174537 | 30 (55%) | 27 (50%) | 0.73 |
| Number of minor alleles | 2.6 (2.2) | 2.2 (2.5) | 0.47 |
| Dietary fatty acids, % of energy b | |||
| n-6 PUFA | 6.55 (1.63) | 7.17 (2.14) | 0.09 |
| n-3 PUFA | 0.77 (0.26) | 0.88 (0.46) | 0.14 |
| Long chain n-3 PUFA c | 0.06 (0.11) | 0.07 (0.12) | 0.52 |
| Serum fatty acids, as % of total | |||
| 20:4, n-6 | 8.7 (1.9) | 9.2 (2.1) | 0.17 |
| Long chain n-3 PUFAc | 2.98 (1.20) | 3.04 (1.37) | 0.82 |
| Colon fatty acids, as % of total | |||
| 20:4, n-6 | 10.5 (3.1) | 10.1 (3.0) | 0.53 |
| Long chain n-3 PUFA c | 3.52 (1.53) | 3.18 (1.62) | 0.26 |
P-values for differences at baseline between diet arms are from two-sided t-tests for continuous variables or Chi-square tests for proportions. Natural log transformations were used to normalize the data before analysis except for dietary n-3 PUFA and serum 20:4, n-6, which did not require transformation. Data is shown untransformed for ease of interpretation.
For dietary intakes, n-6 polyunsaturated fatty acids (n-6 PUFA) was the sum of 18:2, and 20:4, n-3 PUFA was the sum of 18:3, 20:5, 22:5, and 22:6, and ‘long chain n-3’ was the sum of 20:5, 22:5 and 22:6.
Long chain n-3 PUFA in serum and colon were the sum of 20:5, n-3 and 22:6, n-3.
Table 2.
Dietary intakes, serum fatty acid concentrations and colon fatty acid concentrations at baseline by genotype. The data shown are mean and SD.
| Fatty Acid | Number of Minor Alleles | P-Value a | |
|---|---|---|---|
| none n=46 | 1–8 n=62 | ||
| Dietary intakes (% of calories) | |||
| N-6 PUFA | 7.0 (2.2) | 6.8 (1.7) | 0.54 |
| N-3 PUFA | 0.8 (0.3) | 0.8 (0.4) | 0.98 |
| Long chain n-3 PUFA b | 0.08 (0.14) | 0.04 (0.06) | 0.22 |
| Serum fatty acids (% of fatty acids) | |||
| 20:4, n-6 | 10.2 (1.9) | 8.0 (1.7) | <0.001 |
| 20:5, n-3 | 0.8 (0.4) | 0.8 (0.6) | 0.19 |
| 22:6, n-3 | 2.18 (0.81) | 2.20 (1.00) | 0.78 |
| Long chain n-3 PUFA | 3.0 (1.1) | 3.0 (1.4) | 0.55 |
| 18:2 n-6 | 27.5 (6.1) | 26.7 (6.8) | 0.63 |
| 18:3 n-3 | 0.64 (0.23) | 0.87 (0.53) | 0.01 |
| N-3/N-6 ratio | 0.10 (0.03) | 0.12 (0.07) | 0.04 |
| Colon fatty acids (% of fatty acids) | |||
| 20:4, n-6 | 10.7 (3.1) | 10.0 (3.0) | 0.07 |
| 20:5, n-3 | 1.2 (0.9) | 1.2 (1.0) | 0.66 |
| 22:6, n-3 | 2.12 (0.77) | 2.20 (0.88) | 0.95 |
| Long chain n-3 PUFA | 3.3 (1.4) | 3.4 (1.7) | 0.94 |
| 18:2 n-6 | 19.3 (4.3) | 20.0 (7.6) | 0.97 |
| 18:3 n-6 | 1.14 (0.90) | 1.58 (1.41) | 0.19 |
| N-3/N-6 ratio | 0.15 (0.08) | 0.18 (0.11) | 0.15 |
P-values were obtained from linear mixed models with each of the baseline fatty acids regressed on genotype group with batch number as a random effect. The covariates of the model included age, baseline BMI, and baseline dietary intake measures of n-6 PUFA, n-3 PUFA and/or long chain n-3 PUFA as a percentage of energy. An inverse squared root transformation was applied to colon 18:2, n-6 and colon 22:6, n-3 to approximate normality, respectively. Natural logarithm transformations were used to normalize the rest fatty acid data except for serum 20:4, n-6 and 18:2, n-6, which did not require transformation. Means and SDs are shown untransformed for ease of interpretation.
Long chain n-3 PUFA in serum and colon were the sum of 20:5, n-3 and 22:6, n-3. In the diet, grams/day of the n-3 fatty acids 20:5, 22:5 and 22:6 were summed and expressed as a percentage of energy using 9 kcal/gram.
A linear mixed model was used to evaluate whether the presence of any FADS variant affects baseline fatty acid concentrations (AA, EPA). Each of the baseline fatty acids in both serum and colonic mucosa was regressed on genotype group (Table 2). Batch number was a random effect to account for heterogeneity since fatty acids were measured in different batches. The covariates in the model included age, gender, body mass index (BMI; in kg/m2), and dietary intake measures of n-6 PUFA, n-3 PUFA and long chain n-3 PUFA (sum of the n-3 fatty acids 20:5, 22:5 and 22:6) as a percentage of energy using 9 kcal/gram.
Next, we used linear mixed models to evaluate the changes in fatty acid concentrations after 6 months of diet intervention: dietary intake, serum, and colon fatty acid concentrations were regressed on time (baseline, 6 month) with a random intercept for each individual. For serum and colon fatty acids, batch number was included in the random effects. Separate analyses were performed for the two diet groups (Table 3). Finally, analyses were done to compare the changes in fatty acid composition over 6 months between the two diet arms and to assess if the changes were modified by the presence of minor alleles in FADS. For these analyses, each of the outcome variables (AA, EPA for both serum and colonic mucosa) at 6-month follow-up was regressed on genotype group, diet arm, and genotype group*diet assignment interaction by a linear mixed model (Table 4). The model was adjusted for age, BMI, and the concentration of each corresponding fatty acid at baseline. In all the models, batch number was incorporated as a random effect when appropriate. All reported P values were two-tailed. The statistical significance was set α = 0.05 level. Analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC).
Table 3.
Effects of dietary intervention on changes in dietary intakes and fatty acid concentrations.
| Fatty Acid | Healthy Eating Baseline (n=54) | 6 Months (n=43) | Mediterranean Baseline (n=54) | 6 Months (n=47) |
|---|---|---|---|---|
| Dietary Fatty Acids (% of energy) | ||||
| SFA a | 12.3 (2.7) | 7.9 (2.3) b | 11.2 (2.6) | 8.3 (2.3) b |
| MUFA | 13.0 (2.2) | 10.6 (3.3) b | 13.0 (3.4) | 16.4 (3.7) b |
| n-6 PUFA | 6.5 (1.6) | 6.3 (2.2) | 7.2 (2.1) | 6.2 (1.9) b |
| 18:3, n-3 | 0.72 (0.22) | 0.78 (0.32) | 0.81 (0.43) | 0.90 (0.67) |
| Long-chain n-3 PUFA | 0.06 (0.11) | 0.12 (0.16) b | 0.07 (0.12) | 0.17 (0.22) b |
| Serum Fatty Acids (% of total fatty acids) | ||||
| SFA | 34.0 (5.3) | 33.9 (5.0) | 34.0 (5.4) | 33.5 (5.3) |
| MUFA | 24.7 (6.0) | 24.2 (5.2) | 24.1 (4.6) | 26.6 (4.1) b |
| 18:2, n-6 | 27.1 (6.0) | 27.6 (5.9) | 27.1 (6.9) | 25.1 (5.3) b |
| 20:4, n-6 | 8.67 (1.92) | 8.54 (2.30) | 9.2 (2.1) | 8.9 (2.2) |
| 18:3, n-3 | 0.79 (0.28) | 0.81 (0.32) | 0.75 (0.55) | 0.74 (0.43) |
| Long-chain n-3 PUFA | 2.97 (1.20) | 3.26 (1.45) | 3.04 (1.38) | 3.42 (1.39) b |
| Colon Fatty Acids (% of total fatty acids) | ||||
| SFA | 32.4 (4.9) | 32.4 (3.6) | 32.4 (3.3) | 31.9 (3.4) |
| MUFA | 30.5 (4.0) | 31.4 (3.8) | 32.3 (5.0) | 32.8 (4.7) |
| 18:2, n-6 | 20.2 (7.9) | 19.1 (4.8) | 19.3 (4.4) | 18.9 (5.3) |
| 20:4, n-6 | 10.5 (3.1) | 10.5 (2.6) | 10.1 (3.0) | 10.3 (2.7) |
| 18:3, n-3 | 1.39 (1.24) | 1.26 (1.03) | 1.39 (1.24) | 1.16 (1.07) |
| Long-chain n-3 PUFA | 3.52 (1.53) | 3.86 (1.47) b | 3.18 (1.62) | 3.47 (1.60) b |
The transformations used for dietary variables were natural log both for 18:3, n-3 and for long chain n-3 fatty acids (sum of 20:5 and 22:6). The transformations used for serum concentrations were squared for saturated fat (SFA), and natural log both for 18:3, n-3 and long chain n-3 fatty acids. The transformations used for colon fatty acids were: squared for SFA, inverse squared root for 18:2, n-6 and natural log for MUFA, 20:4, n-6, 18:3, n-3, and long chain n-3 fatty acids. Means and standard deviations are shown un-transformed for ease of interpretation.
Significant different from baseline for that diet arm (p<0.05).
Table 4.
Interactions of diet group assignment with genotype on fatty acid concentrations in serum and colonic mucosa at 6 months a.
| Sample | Fatty Acid (at 6 months) | Effect | Estimate | SE | p-Value |
|---|---|---|---|---|---|
| Serum | 20:4, n-6 | Diet | 0.368 | 0.458 | 0.42 |
| Genotype | −0.286 | 0.483 | 0.56 | ||
| Diet*Genotype | −0.630 | 0.602 | 0.30 | ||
| Baseline 20:4, n-6 | 0.804 | 0.085 | <0.001 | ||
| Age | −0.016 | 0.014 | 0.25 | ||
| BMI | 0.011 | 0.039 | 0.78 | ||
|
| |||||
| 20:5, n-3 b | Diet | 0.092 | 0.149 | 0.54 | |
| Genotype | −0.019 | 0.145 | 0.90 | ||
| Diet*Genotype | −0.015 | 0.199 | 0.94 | ||
| Baseline 20:4, n-6 | 0.442 | 0.097 | <0.001 | ||
| Age | 0.009 | 0.005 | 0.04 | ||
| BMI | −0.013 | 0.013 | 0.31 | ||
|
| |||||
| Colon | 20:4, n-6 b | Diet | −0.178 | 0.066 | 0.01 |
| Genotype | −0.226 | 0.066 | 0.001 | ||
| Diet*Genotype | 0.266 | 0.089 | 0.004 | ||
| Baseline 20:4, n-6 | 0.293 | 0.071 | <0.001 | ||
| Age | <0.001 | 0.002 | 0.85 | ||
| BMI | 0.001 | 0.006 | 0.82 | ||
|
| |||||
| 20:5, n-3 b | Diet | −0.014 | 0.120 | 0.91 | |
| Genotype | −0.027 | 0.116 | 0.82 | ||
| Diet*Genotype | 0.063 | 0.158 | 0.69 | ||
| Baseline 20:4, n-6 | 0.721 | 0.057 | <0.001 | ||
| Age | 0.004 | 0.004 | 0.28 | ||
| BMI | −0.003 | 0.011 | 0.79 | ||
Analyses were performed by linear mixed models by including each fatty acid concentration at 6 months as the response variable and covariates: diet arm (Mediterranean versus Healthy Eating), genotype (presence of any minor alleles versus all major alleles at the four SNPs in the FADS gene), interaction of diet and genotype, baseline age, baseline BMI and baseline concentration of each fatty acid. Batch of sample analysis was treated as a random effect.
Natural log transformation was applied to fatty acid variables approximate normality in analysis.
Results
Baseline Characteristics and Genotyping
The overall study consisted of 108 study participants after exclusions for lack of genotyping consent (n=9) and incomplete genotype data (n=3). Genotyping success rate of the 4 SNPs chosen to define the FADS1/2 haplotype as described in Methods, was between 96.7% and 98.3%. Minor allele frequencies were in the range of 25.0% to 32.9%. The genotype distribution for each SNP did not deviate from Hardy-Weinberg equilibrium (p >0.05).
Baseline characteristics for the Healthy Eating diet group (n = 54) and the Mediterranean diet group (n = 54) were summarized in Table 1. No significant differences were found in minor allele frequency of any SNP, gender, race, age, or BMI between the two diet groups at baseline. Likewise, baseline measurements of AA, EPA, and long chain n-3 fatty acids (the sum of EPA and DHA) did not differ significantly in the serum or the colonic mucosa between the two diet groups (Table 1).
Baseline measures
Linear regression analysis indicated that the number of minor alleles was a significant predictor of baseline serum AA concentration (p < 0.001) and almost significant for colonic AA concentration (p = 0.058). A greater number of minor alleles was significantly associated with lower AA concentration in serum. Dietary AA intakes were not a significant predictor of either serum of colon concentrations. For long chain n-3 fatty acids, however, the situation was the reverse. Dietary intake of long chain n-3 fatty acids was a significant predictor of baseline serum long chain n-3 concentration (p < 0.001) and colonic long chain n-3 concentration (p = 0.044) while the number of minor alleles was not a significant predictor of either.
Subsequent analyses were done categorizing subjects into two groups by presence or absence of any minor alleles in the FADS gene cluster. The only dietary or demographic factor to differ by genotype at baseline was BMI, which was lower in carriers of any minor alleles (mean of 27.8, SD 3.7, in all major allele carriers and mean of 26.1, SD 3.6, in carriers of any minor alleles, p=0.02 by the 2-sided t-test). Age was not significantly different (p=0.11) but was retained as a covariate. No significant difference was found for other demographic characteristics (race, gender, smoking, common medication use) between minor allele and all major allele carriers. Results were similar when using any one SNP individually versus all minor SNPS (not shown).
Serum and colon fatty acid concentrations at baseline by genotype group are shown in Table 2. Linear mixed models were used to evaluate differences between genotype groups. The presence of any minor alleles was highly significantly associated with baseline serum 20:4, n-6 concentrations (p<0.0001) and 18:3, n-3 concentration (p=0.01), and marginally significant for colonic 20:4, n-6 concentration (p = 0.07), with adjustment for age, BMI, and dietary intakes of n-6 PUFA, n-3 PUFA and/or long chain n-3 PUFA as a percentage of energy. Specifically, mean serum 20:4, n-6 concentration (% of total fatty acids) for minor allele carriers were estimated to be 2% (95% CI = [1%, 3%]) lower, whereas mean serum 18:3, n-3 concentration for minor allele carriers were estimated to be 21% (95% CI = [4%, 41%]) higher, compared to those individuals with all major alleles in the four SNPs in FADS. There was no significant association of genotype with EPA nor with long chain n-3 fatty acids (the sum of EPA and DHA). Genotype group also had no significant effects on total cholesterol, LDL, HDL, triglycerides, insulin, glucose, and CRP with p>0.11 in each case (not shown).
Effects of Dietary Intervention on Fatty Acid Intakes and Fatty Acid Concentrations in Serum and Colon
We first evaluated changes in fatty acids by diet group assignment alone without considering the genotype groups. Table 3 displays dietary intakes, serum, and colon fatty acid concentrations for the two diet arms at baseline and after 6 months of intervention. Based on data from food records and 24-hour recalls, dietary intakes of saturated fats (SFA) and monounsaturated fats (MUFA) were significantly reduced (p<0.0001) and long chain n-3 PUFA was significantly increased (p=0.004) in the Healthy Eating group after 6 months. The decrease in mean SFA resulted in an increased polyunsaturated fat: saturated fat ratio from 0.60 to 0.92 in the Healthy Eating group (p=0.008 from mixed linear regression models controlling for age). In the Mediterranean group, dietary intakes of SFA and n-6 PUFA both significantly decreased (p<0.0001), while MUFA and long chain n-3 PUFA significantly increased (p<0.0001), in accord with the counseling goals. The mean polyunsaturated fat: saturated fat ratio increased non-significantly from 0.72 to 0.77 in the Mediterranean group.
Serum 18:2 n-6 significantly decreased (p=0.02), and both MUFA and n-3 PUFA significantly increased (p=0.0005 and p=0.01, respectively) in the Mediterranean arm only (Table 3). There was little change in colon fatty acid concentrations. The only significant change was for long chain n-3 PUFA that significantly increased in both Healthy Eating (p=0.01) and Mediterranean groups (p=0.01).
Interactions of Genotype and Diet Intervention
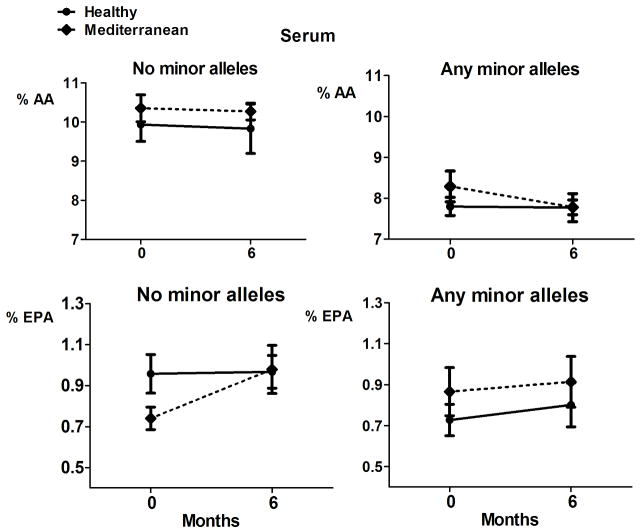
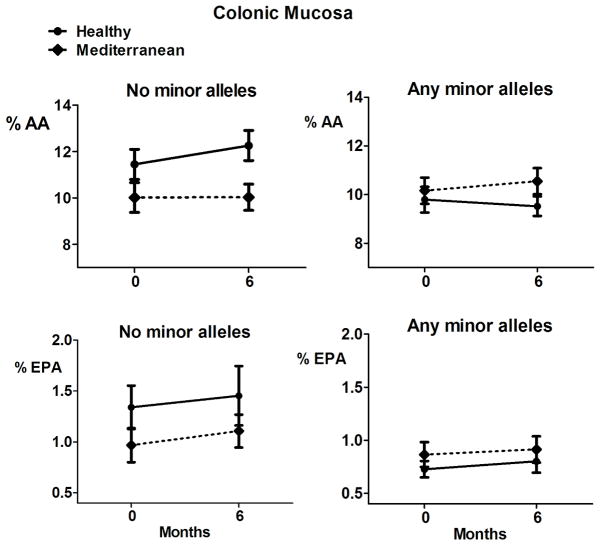
Figures 1 and 2 show the raw means in each group over time. Table 4 shows the linear mixed model results for the analysis of the genotype by diet interaction. There was a significant interaction of genotype by diet for 20:4, n-6 (AA) concentrations in the colon (p=0.004). No significant genotype-by-diet interactions were found for AA in serum nor for EPA. Among subjects with no minor alleles, mean colon AA concentrations were estimated to be 16% (95% CI = [5%, 26%]) lower for the Mediterranean arm than the Healthy Eating arm at 6 months. These results indicate that after adjusting for baseline AA concentrations, mean colon AA concentrations at 6 months were significantly different between diet arms only in persons with no minor alleles in the FADS1/2 gene cluster. This was mainly due to an increase in colon AA in the Healthy Eating diet arm while colon AA concentrations remained fairly constant in the Mediterranean group.
Figure 1.
Serum concentrations of arachidonic acid (AA) and eicosapentaenoic acid (EPA) at baseline and after 6 months of intervention. Subjects were grouped by presence/absence of any minor alleles in the FADS1/FADS2 gene cluster (rs3834458, rs174556, rs174561, and rs174537). The data shown are untransformed mean and SE.
Figure 2.
Colon mucosa concentrations of arachidonic acid (AA) and eicosapentaenoic acid (EPA) at baseline and after 6 months of intervention. Subjects were grouped by presence/absence of any minor alleles in the FADS1/FADS2 gene cluster (rs3834458, rs174556, rs174561, and rs174537). There was a significant gene*diet interaction for AA in persons with no minor alleles in any of the single nucleotide polymorphisms in linear mixed models. The data shown are untransformed mean and SE.
Discussion
This randomized, dietary intervention study afforded the opportunity to evaluate the impact of FADS genotype and diet on fatty acid concentrations in both serum and colonic mucosa of individuals at increased risk for colon cancer. The number of minor alleles in the FADS gene cluster, but not diet, predicted serum AA concentrations. This agrees well with results of previous studies, namely that carriers of minor alleles have lower AA concentrations (9–15). For EPA concentrations in serum, genotype had no effect while diet did have a significant effect, likely because n3 fatty acid intakes were fairly low and limiting in this study population. It should, however, be noted that diet in this study was assessed using self-report on four separate days. In addition to the possibility of mis-reporting of intakes, those four days might not represent usual intakes over the last month of study and therefore will weaken any apparent associations with diet.
In epidemiological studies, relatively higher dietary intakes of both n-3 and n-9 fatty acids are thought to be protective while high intakes of n-6 fatty acids increase risk of several cancers including that of the colon (31). This has been confirmed in experimental models of colon cancer, and low versus high n6 fatty acid diets are associated with decreased tumors and lower production of certain eicosanoids such as prostaglandin E2 (PGE2) (32, 33). In the colon, prostaglandin E2 (PGE2) has been tightly linked with colon cancer risk (34). Increased n-3 fatty acid intakes also reduce PGE2 production (35–39). Interestingly, a reduction in n-6 fatty acid intakes can augment increases in EPA after n-3 fatty acid supplementation (40). Bartoli et al. observed inhibition of aberrant crypt foci, adenocarcinomas, decreased mucosal arachidonate (20:4) and decreased PGE2 in rats fed either n-9 or n-3 diets relative to rats fed diets high in n-6 fatty acids (41). The levels of colon mucosal PGE2 were directly proportional to arachidonate levels in the colon in that study (41). This data makes it important to better understand factors that could affect AA and EPA levels in the human colon.
Unlike serum fatty acids, genotype had no significant effects on fatty acid concentrations in the colon at baseline (Table 2). It may be the case that serum concentrations of fatty acids are affected by first pass liver metabolism more so than tissues. After absorption of fatty acids, mainly in the small intestine, the liver is the initial site of fatty acid metabolism. The subsequent distribution of fatty acids from the circulation to tissues will be dependent on lipoprotein lipase activity in each tissue site and on tissue-specific metabolic conversions. In a well-controlled study in pigs, increased dietary intakes of linolenic acid and/or linoleic acid significantly affected metabolism of each other to longer chain fatty acids in the liver, but the effect was minimal in brain cortex (42). In a human lipodomic study, fatty acid desaturase activity of blood reflected activity in the liver but not in adipose tissue (43). Serum and colon fatty acid concentrations therefore not only diet and genotype, but any tissue-specific regulation of fatty acid metabolism.
Since the present study was a randomized clinical trial, we then evaluated the effects of the two dietary interventions on changes in fatty acid intakes and levels over time. Both dietary interventions decreased SFA intakes and increased n-3 PUFA intakes. Only the Mediterranean intervention resulted in increased MUFA and decreased n-6 PUFA intakes. Serum fatty acids in the Mediterranean arm reflected these changes in diet (Table 3). In the colon, however, the only significant change was an increase in n-3 PUFA. This indicates that tissue-specific processes may limit the impact of dietary changes in colon fatty acids. The increase in colon n-3 PUFA is interesting, however, since the increases in dietary n-3 PUFA were modest in each diet arm.
The effect of FADS genotype on fatty acid concentrations in colon was only evident after intervention (Table 4). Study subjects who were carriers of all major alleles and randomized to the Healthy Eating intervention had higher colon AA concentrations after 6 months than subjects with all major alleles in the Mediterranean group. It is not entirely clear why this should be the case, but the Healthy Eating intervention did result in a higher relative amount of n-6 PUFA to other dietary fats. This could have helped increase the percentage of AA in the colon fatty acids after the Healthy Eating intervention. In addition to polymorphisms in FADS, other factors could be operative to affect fatty acid desaturation such as diet-induced changes in the expression and the activity of FADS, and to changes in substrate competition (44). In carriers of all major alleles randomized to the Mediterranean intervention, AA levels stayed relatively low at both time points and were estimated to be 16% lower than in the Healthy arm after 6 months of intervention.
Limitations of this study include the small sample size, the relatively short intervention length, and the self-report of diet which is known to be subject to biases. It may take longer for a change in diet to be fully manifest, especially in tissues. In addition, the measurement of fatty acids was done as a percentage of total fatty acids such that increases in one fatty acid on a volume basis would result in decreases in other fatty acids. An additional consideration is that AA concentrations are not easily modifiable by changes in n6 fatty acids in the diet, especially if AA is not elevated at the outset (45). Strengths of the study include that it was a randomized study, and measures were available before and after diet change in both serum and colonic mucosa of individuals at increased risk for colon cancer.
In conclusion, this study showed that those subjects with no minor alleles in the FADS1/2 cluster had higher concentrations of AA in serum. Polymorphism in FADS1/2 had no effect on concentrations of EPA, perhaps because concentrations of this fatty acid are more highly driven by dietary intakes. The trends were similar in colon tissue fatty acids but not significant. After randomization to Mediterranean or Healthy Eating intervention for 6 months, there was a significant gene*diet interaction for colon AA concentrations. Subjects who had all major alleles for FADS1/2 had significantly lower AA concentrations in the colon after 6 months if they were in the Mediterranean diet arm. Since AA is the substrate for prostaglandin E2 production, these results indicate that a Mediterranean diet could be especially favorable for reducing colon cancer risk in the subset of subjects with all major alleles in FADS1/2. Future work should evaluate the effects of these FADS polymorphisms on colonic pro-inflammatory states.
Acknowledgments
We thank all the individuals who volunteered for the Healthy Eating Study for Colon Cancer Prevention. The parent study was designed and conducted in collaboration with Drs. Dean E. Brenner, Mack T. Ruffin, D. Kim Turgeon and Ananda Sen. Mary Rapai was the coordinator for the study and Maria Cornellier was the study dietitian.
This study was supported by a grant from the Ronald P. and Joan M. Nordgren Cancer Research Fund, NIH grant RO1 CA120381, and Cancer Center Support Grant P30 CA046592. The study used core resources supported by a Clinical Translational Science Award, NIH grant UL1RR024986 (the Michigan Clinical Research Unit), the Michigan Diabetes Research Center NIH grant 5P60 DK020572 (Chemistry Laboratory), and the Michigan Nutrition and Obesity Research Center NIH grant P30 DK089503.
Abbreviations
- FADS1
Fatty Acid Desaturase 1(Delta-5 desaturase)
- FADS2
Fatty Acid Desaturase 2 (Delta-6 desaturase)
- AA
Arachidonic Acid (20:4, n-6)
- EPA
Eicosapentaenoic Acid (20:5, n-3)
- DHA
Docosahexaenoic Acid
- MUFA
Monounsaturated Fatty Acids
- PUFA
Polyunsaturated Fatty Acids
- SFA
Saturated Fatty Acids
References
- 1.Verberne L, Bach-Faig A, Buckland G, Serra-Majem L. Association between the Mediterranean diet and cancer risk: a review of observational studies. Nutr Cancer. 2010;62:860–70. doi: 10.1080/01635581.2010.509834. [DOI] [PubMed] [Google Scholar]
- 2.Giacosa A, Barale R, Bavaresco L, Gatenby P, Gerbi V, Janssens J, et al. Cancer prevention in Europe: the Mediterranean diet as a protective choice. Eur J Cancer Prev. 2013;22:90–5. doi: 10.1097/CEJ.0b013e328354d2d7. [DOI] [PubMed] [Google Scholar]
- 3.Frenandez E, Vecchia CL, Gonzales JR, Lucchini F, Negri E, Levi F. Coverging patterns of colorectal cancer mortality in Europe. European Journal of Cancer. 2005;41:430–7. doi: 10.1016/j.ejca.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Koushik A, Hunter DJ, Spiegelman D, Beeson WL, van den Brandt PA, Buring JE, et al. Fruits, vegetables, and colon cancer risk in a pooled analysis of 14 cohort studies. J Natl Cancer Inst. 2007;99:1471–83. doi: 10.1093/jnci/djm155. [DOI] [PubMed] [Google Scholar]
- 5.Bingham SA, Day NE, Luben R, Ferrari P, Slimani N, Norat T, et al. Dietary fibre in food and protection against colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC): an observational study. Lancet. 2003;361:1496–501. doi: 10.1016/s0140-6736(03)13174-1. [DOI] [PubMed] [Google Scholar]
- 6.Alberts DS, Einspahr J, Ritenbaugh C, Aickin M, Rees-McGee S, Atwood J, et al. The effect of wheat bran fiber and calcium supplementation on rectal mucosal proliferation rates in patients with resected adenomatous colorectal polyps. Cancer Epidemiol Biomarkers Prev. 1997;6:161–9. [PubMed] [Google Scholar]
- 7.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–93. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu WK, Sung JJ, Lee CW, Yu J, Cho CH. Cyclooxygenase-2 in tumorigenesis of gastrointestinal cancers: an update on the molecular mechanisms. Cancer Lett. 2010;295:7–16. doi: 10.1016/j.canlet.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Malerba G, Schaeffer L, Xumerle L, Klopp N, Trabetti E, Biscuola M, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–99. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 10.Glaser C, Heinrich J, Koletzko B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism. 2010;59:993–9. doi: 10.1016/j.metabol.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Sergeant S, Hugenschmidt CE, Rudock ME, Ziegler JT, Ivester P, Ainsworth HC, et al. Differences in arachidonic acid levels and fatty acid desaturase (FADS) gene variants in African Americans and European Americans with diabetes or the metabolic syndrome. Br J Nutr. 2012;107:547–55. doi: 10.1017/S0007114511003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, Kompauer I, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–56. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 13.Aslibekyan S, Jensen MK, Campos H, Linkletter CD, Loucks EB, Ordovas JM, et al. Fatty Acid desaturase gene variants, cardiovascular risk factors, and myocardial infarction in the costa rica study. Front Genet. 2012;3:72. doi: 10.3389/fgene.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baylin A, Ruiz-Narvaez E, Kraft P, Campos H. alpha-Linolenic acid, Delta6-desaturase gene polymorphism, and the risk of nonfatal myocardial infarction. Am J Clin Nutr. 2007;85:554–60. doi: 10.1093/ajcn/85.2.554. [DOI] [PubMed] [Google Scholar]
- 15.Mathias RA, Fu W, Akey JM, Ainsworth HC, Torgerson DG, Ruczinski I, et al. Adaptive evolution of the FADS gene cluster within Africa. PLoS One. 2012;7:e44926. doi: 10.1371/journal.pone.0044926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lattka E, Illig T, Koletzko B, Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol. 2010;21:64–9. doi: 10.1097/MOL.0b013e3283327ca8. [DOI] [PubMed] [Google Scholar]
- 17.Hussein N, Ah-Sing E, Wilkinson P, Leach C, Griffin BA, Millward DJ. Long-chain conversion of [13C]linoleic acid and alpha-linolenic acid in response to marked changes in their dietary intake in men. J Lipid Res. 2005;46:269–80. doi: 10.1194/jlr.M400225-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, Trabetti E, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr. 2008;88:941–9. doi: 10.1093/ajcn/88.4.941. [DOI] [PubMed] [Google Scholar]
- 20.Standl M, Lattka E, Stach B, Koletzko S, Bauer CP, von Berg A, et al. FADS1 FADS2 gene cluster, PUFA intake and blood lipids in children: results from the GINIplus and LISAplus studies. PLoS One. 2012;7:e37780. doi: 10.1371/journal.pone.0037780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, Guralnik JM, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathias RA, Sergeant S, Ruczinski I, Torgerson DG, Hugenschmidt CE, Kubala M, et al. The impact of FADS genetic variants on omega6 polyunsaturated fatty acid metabolism in African Americans. BMC Genet. 2011;12:50. doi: 10.1186/1471-2156-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillingham LG, Harding SV, Rideout TC, Yurkova N, Cunnane SC, Eck PK, et al. Dietary oils and FADS1–FADS2 genetic variants modulate [13C]alpha-linolenic acid metabolism and plasma fatty acid composition. Am J Clin Nutr. 2013;97:195–207. doi: 10.3945/ajcn.112.043117. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Feskens EJ, Dolle ME, Imholz S, Verschuren WM, Muller M, et al. Dietary n-3 and n-6 polyunsaturated fatty acid intake interacts with FADS1 genetic variation to affect total and HDL-cholesterol concentrations in the Doetinchem Cohort Study. Am J Clin Nutr. 2010;92:258–65. doi: 10.3945/ajcn.2009.29130. [DOI] [PubMed] [Google Scholar]
- 25.Hellstrand S, Sonestedt E, Ericson U, Gullberg B, Wirfalt E, Hedblad B, et al. Intake levels of dietary long-chain PUFAs modify the association between genetic variation in FADS and LDL-C. J Lipid Res. 2012;53:1183–9. doi: 10.1194/jlr.P023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cormier H, Rudkowska I, Paradis AM, Thifault E, Garneau V, Lemieux S, et al. Association between polymorphisms in the fatty acid desaturase gene cluster and the plasma triacylglycerol response to an n-3 PUFA supplementation. Nutrients. 2012;4:1026–41. doi: 10.3390/nu4081026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen A, Ren J, Ruffin MT, Turgeon DK, Brenner DE, Sidahmed E, et al. Relationships Between Serum and Colon Concentrations of Carotenoids and Fatty Acids in a Randomized Dietary Intervention Trial. Cancer Prevention Research. 2013 doi: 10.1158/1940-6207.CAPR-13-0019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djuric Z, Ruffin MTt, Rapai ME, Cornellier ML, Ren J, Ferreri TG, et al. A Mediterranean dietary intervention in persons at high risk of colon cancer: recruitment and retention to an intensive study requiring biopsies. Contemp Clin Trials. 2012;33:881–8. doi: 10.1016/j.cct.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren J, Mozurkewich EL, Sen A, Vahratian AM, Ferreri TG, Morse AN, et al. Total Serum Fatty Acid Analysis by GC-MS: Assay Validation and Serum Sample Stability Current Pharmaceutical Analysis. 2013. Epub March 8, 2013:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rzehak P, Heinrich J, Klopp N, Schaeffer L, Hoff S, Wolfram G, et al. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br J Nutr. 2008:1–7. doi: 10.1017/S0007114508992564. [DOI] [PubMed] [Google Scholar]
- 31.Bartsch H, Nair J, Owen RW. Dietary polyunsaturated fatty acids and cancers of the breast and colorectum: emerging evidence for their role as risk modifiers. Carcinogenesis. 1999;20:2209–18. doi: 10.1093/carcin/20.12.2209. [DOI] [PubMed] [Google Scholar]
- 32.Rao CV, Hirose Y, Indranie C, Reddy BS. Modulation of experimental colon tumorigenesis by types and amounts of dietary fatty acids. Cancer Res. 2001;61:1927–33. [PubMed] [Google Scholar]
- 33.Rao CV, Simi B, Wynn TT, Garr K, Reddy BS. Modulating effect of amount and types of dietary fat on colonic mucosal phospholipase A2, phosphatidylinositol-specific phospholipase C activities, and cyclooxygenase metabolite formation during different stages of colon tumor promotion in male F344 rats. Cancer Res. 1996;56:532–7. [PubMed] [Google Scholar]
- 34.Backlund MG, Mann JR, Dubois RN. Mechanisms for the prevention of gastrointestinal cancer: the role of prostaglandin E2. Oncology. 2005;69 (Suppl 1):28–32. doi: 10.1159/000086629. [DOI] [PubMed] [Google Scholar]
- 35.Dommels YE, Haring MM, Keestra NG, Alink GM, van Bladeren PJ, van Ommen B. The role of cyclooxygenase in n-6 and n-3 polyunsaturated fatty acid mediated effects on cell proliferation, PGE(2) synthesis and cytotoxicity in human colorectal carcinoma cell lines. Carcinogenesis. 2003;24:385–92. doi: 10.1093/carcin/24.3.385. [DOI] [PubMed] [Google Scholar]
- 36.Bartram HP, Gostner A, Scheppach W, Reddy BS, Rao CV, Dusel G, et al. Effects of fish oil on rectal cell proliferation, mucosal fatty acids, and prostaglandin E2 release in healthy subjects. Gastroenterology. 1993;105:1317–22. doi: 10.1016/0016-5085(93)90135-y. [DOI] [PubMed] [Google Scholar]
- 37.Nieto N, Fernandez MI, Torres MI, Rios A, Suarez MD, Gil A. Dietary monounsaturated n-3 and n-6 long-chain polyunsaturated fatty acids affect cellular antioxidant defense system in rats with experimental ulcerative colitis induced by trinitrobenzene sulfonic acid. Dig Dis Sci. 1998;43:2676–87. doi: 10.1023/a:1026655311878. [DOI] [PubMed] [Google Scholar]
- 38.Kuratko CN, Becker SA. Dietary lipids alter fatty acid composition and PGE2 production in colonic lymphocytes. Nutrition and Cancer. 1998;31:56–61. doi: 10.1080/01635589809514678. [DOI] [PubMed] [Google Scholar]
- 39.Broughton KS, Wade JW. Total fat and (n-3):(n-6) fat ratios influence eicosanoid production in mice. J Nutr. 2002;132:88–94. doi: 10.1093/jn/132.1.88. [DOI] [PubMed] [Google Scholar]
- 40.James MJ, Gibson RA, Cleland LG. Dietary polyunsaturated fatty acids and inflammatory mediator production. Am J Clin Nutr. 2000;71:343S–8S. doi: 10.1093/ajcn/71.1.343s. [DOI] [PubMed] [Google Scholar]
- 41.Bartoli R, Fernandez-Banares F, Navarro E, Castella E, Mane J, Alvarez M, et al. Effect of olive oil on early and late events of colon carcinogenesis in rats: modulation of arachidonic acid metabolism and local prostaglandin E(2) synthesis. Gut. 2000;46:191–9. doi: 10.1136/gut.46.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smink W, Gerrits WJ, Gloaguen M, Ruiter A, van Baal J. Linoleic and alpha-linolenic acid as precursor and inhibitor for the synthesis of long-chain polyunsaturated fatty acids in liver and brain of growing pigs. Animal. 2012;6:262–70. doi: 10.1017/S1751731111001479. [DOI] [PubMed] [Google Scholar]
- 43.Kotronen A, Seppanen-Laakso T, Westerbacka J, Kiviluoto T, Arola J, Ruskeepaa AL, et al. Comparison of lipid and fatty acid composition of the liver, subcutaneous and intra-abdominal adipose tissue, and serum. Obesity (Silver Spring) 2010;18:937–44. doi: 10.1038/oby.2009.326. [DOI] [PubMed] [Google Scholar]
- 44.Tu WC, Cook-Johnson RJ, James MJ, Muhlhausler BS, Gibson RA. Omega-3 long chain fatty acid synthesis is regulated more by substrate levels than gene expression. Prostaglandins Leukot Essent Fatty Acids. 2010;83:61–8. doi: 10.1016/j.plefa.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Rett BS, Whelan J. Increasing dietary linoleic acid does not increase tissue arachidonic acid content in adults consuming Western-type diets: a systematic review. Nutr Metab (Lond) 2011;8:36. doi: 10.1186/1743-7075-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]