Abstract
The bacterial potassium channel KcsA is gated open by the binding of protons to amino acids on the intracellular side of the channel. We have identified, via channel mutagenesis and x-ray crystallography, two pH-sensing amino acids and a set of nearby residues involved in molecular interactions that influence gating. We found that the minimal mutation of one histidine (H25) and one glutamate (E118) near the cytoplasmic gate completely abolished pH-dependent gating. Mutation of nearby residues either alone or in pairs altered the channel’s response to pH. In addition, mutations of certain pairs of residues dramatically increased the energy barriers between the closed and open states. We proposed a Monod–Wyman–Changeux model for proton binding and pH-dependent gating in KcsA, where H25 is a “strong” sensor displaying a large shift in pKa between closed and open states, and E118 is a “weak” pH sensor. Modifying model parameters that are involved in either the intrinsic gating equilibrium or the pKa values of the pH-sensing residues was sufficient to capture the effects of all mutations.
INTRODUCTION
Ligand-gated ion channels convert the energy of ligand binding into the gating of ion-conducting pores, allowing the passage of current across biological membranes. In this way, electrical activity under the explicit control of signaling molecules underlies many physiological processes such as postsynaptic neurotransmission in nerve and muscle, pain sensation, and sensory transduction (Hille, 2001).
The proton-gated prokaryotic K+ channel KcsA (Schrempf et al., 1995; Cuello et al., 1998; Heginbotham et al., 1999) was the first ion channel modeled with high resolution x-ray crystallography (Doyle et al., 1998; Zhou et al., 2001). The pore appeared closed, as the inner TM2 helices formed a tight hydrophobic constriction called the “bundle crossing.” Spectroscopic studies confirmed that the conformation of the bundle-crossing gate was pH dependent (Perozo et al., 1998; Liu et al., 2001). Structural studies of MthK (Jiang et al., 2002a), a Ca2+-gated K+ channel (Zadek and Nimigean, 2006; Li et al., 2007), provided a model for the open conformation (Jiang et al., 2002b), later found to be consistent with the presumed open-state structure of a mutant KcsA pore (Cuello et al., 2010b). These open versus closed structural snapshots represent the conformational states of KcsA that equilibrate in a proton-dependent manner.
In a previous report, we made several mutations near the bundle-crossing gate of KcsA that dramatically altered pH-dependent gating (Thompson et al., 2008). In particular, we confirmed a contribution from a key histidine (H25) behind the bundle crossing that had been proposed as the sole pH sensor in a nuclear magnetic resonance (NMR) study (Takeuchi et al., 2007). Despite a significant effort by us and others to modify or ablate pH sensing using mutagenesis (Thompson et al., 2008; Cuello et al., 2010a,c; Hirano et al., 2011), the energetic mechanism underlying proton-dependent gating remained obscure partly because of ambiguity of interpretation after channel modification. Mutations may alter the energy of ligand binding, the ligand-independent intrinsic energy of gating, or both, and these outcomes are often not distinguishable by analysis of channel activity alone (Colquhoun, 1998). Furthermore, there is a high density of arginines and ionizable residues at this location, and mutations of these residues may change proton binding (pKa) to nearby pH-sensing sites by alteration of local electrostatics or solvent accessibility, making the determination of pH-sensor contributions difficult.
In this study, we investigated the specific role of each ionizable residue located at the bundle crossing of KcsA using channel recordings in lipid bilayers and x-ray crystallography of two bundle-crossing mutant channels. We propose that proton binding to H25 and E118 is energetically linked to proton-dependent gating in KcsA, and that R121, R122, and E120 play a supportive role. We propose that specific intersubunit and intrasubunit interactions alter the intrinsic equilibrium between closed and open states, whereas proton binding gates the channel according to a Monod–Wyman–Changeux (MWC) kinetic scheme. In our model, H25 is a “strong” pH sensor, characterized by a large pKa shift between closed and open conformations, enabling this residue to change the gating equilibrium to a large degree. On the other hand, E118 is modeled as a “weak” pH sensor, suggesting a smaller energetic contribution from this residue.
MATERIALS AND METHODS
Protein expression, purification, and reconstitution in lipid bilayers
KcsA mutations were introduced with the QuikChange site-directed DNA mutagenesis kit (Agilent Technologies) and verified with sequencing. All pH-sensor mutations were made on the background of the E71A mutant, shown to remove selectivity filter inactivation (Cordero-Morales et al., 2006). KcsA protein was expressed and purified as described previously (Heginbotham et al., 1999; Thompson et al., 2008). All reagents were from Sigma-Aldrich unless otherwise noted. In brief, either N-terminal 6His-tagged KcsA pASK90 or C-terminal 6His-tagged KcsA pQE60 constructs were transformed into JM83 (ATCC) or BL21 (DE3; Invitrogen) Escherichia coli cells, respectively, and grown at 37°C in Terrific Broth or Luria-Bertani media to OD600 = 1. For pASK90, protein expression was induced for 90 min with 0.2 mg/ml anhydrotetracycline (Acros Organics), and for pQE60, protein expression was induced for 3 h with 500 µM IPTG. Because of poor expression at 37°C, H25R/E118A expression was induced at 20°C overnight. Cells were harvested via centrifugation at 5,000 g for 15 min at 4°C, resuspended in 100 mM KCl and 50 mM Tris, pH 7.5, and disrupted with probe sonication (Thermo Fisher Scientific). Membranes were extracted for 2 h at room temperature with 50 mM n-decyl maltoside (DM; Affymetrix) followed by centrifugation at 35,000 g for 45 min. Protein was purified from the soluble fraction in buffer (100 mM KCl, 20 mM Tris, and 5 mM DM, pH 7.5) with either a Ni2+-affinity column (EMD Millipore) or a HiTrap Co2+-affinity column (GE Healthcare), eluted with the addition of 300 mM imidazole, and finished with gel filtration (Superdex 200; GE Healthcare).
Purified channel was reconstituted into liposomes at protein to lipid ratios of 0.1–10 µg protein per milligram of lipid (3:1 POPE/POPG; Avanti Polar Lipids, Inc.). Detergent was removed using a G50 fine (GE Healthcare) gel-filtration column in buffer containing 400 mM KCl, 5 mM NMG, and 20 mM Tris, pH 7.5. Liposome aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
Single-channel recording and analysis
KcsA pH-sensor mutants were recorded and analyzed as described previously (Thompson et al., 2008). In brief, channels were recorded in a horizontal planar lipid bilayer system consisting of two chambers separated by a plastic partition (transparency slide; IKON) containing a 50–100-µm hole, across which bilayers were formed using a mixture of POPE (7.5 mg/ml) and POPG (2.5 mg/ml) dissolved in n-decane. Recording solutions were: 70 mM KCl, 30 mM KOH, 10 mM MOPS, 10 mM succinate, and 10 mM Tris, pH adjusted to 4–9. Proteoliposomes were applied to the cis (upper) chamber, which contained recording solution at pH 7. Initially, the trans (lower) chamber contained recording solution at pH 4, such that only channels with pH sensor facing trans were activated. Only channels that displayed typical KcsA behavior in these conditions were studied. The recording electrode was placed in the cis chamber, and the ground electrode was placed in the trans chamber.
Proton dose–response curves were measured by perfusing the trans chamber with at least 5 ml of recording solutions with varying pH and measuring channel activity. Bilayers were voltage-clamped using an amplifier (Axopatch 200A, 200B; Molecular Devices) or a bilayer clamp (BC-535; Warner Instruments). Single-channel currents were filtered at 2 kHz using a four-pole Bessel filter, digitized at 25 kHz using a Digidata 1440A or 1320 (Molecular Devices), and recorded in Clampex 10 or Axoscope 10 software (Molecular Devices). Channel orientation was confirmed by recording the typical conductance and open-channel noise properties of KcsA E71A. Single-channel current amplitudes and open probabilities (Po) were analyzed in Clampfit 10 software (Molecular Devices). The number of channels in the bilayer during an experiment was verified by perfusing back to pH 4.0 during the experiment. Bilayers where this was not achieved (because of bilayer rupture or gain/loss of channels) were not included in the final analysis.
Po versus pH data (not normalized) were plotted in Origin (Microcal) and fit with the model-independent Hill equation (Eq. 1):
| (1) |
where Pomax is the maximal open probability, EC50 is the proton concentration of half-activation (also represented as pH1/2), [H+] is the proton concentration (also represented as pH), and nH is the Hill coefficient, which describes the steepness of the dose–response curve.
The Po versus pH data were also fit to either a single proton-binding site MWC model (Fig. 4 B and Eq. 2):
| (2) |
where A is given by:
or a two proton–binding site MWC model (Fig. 4 C and Eq. 3):
| (3) |
where B is given by:
Figure 4.
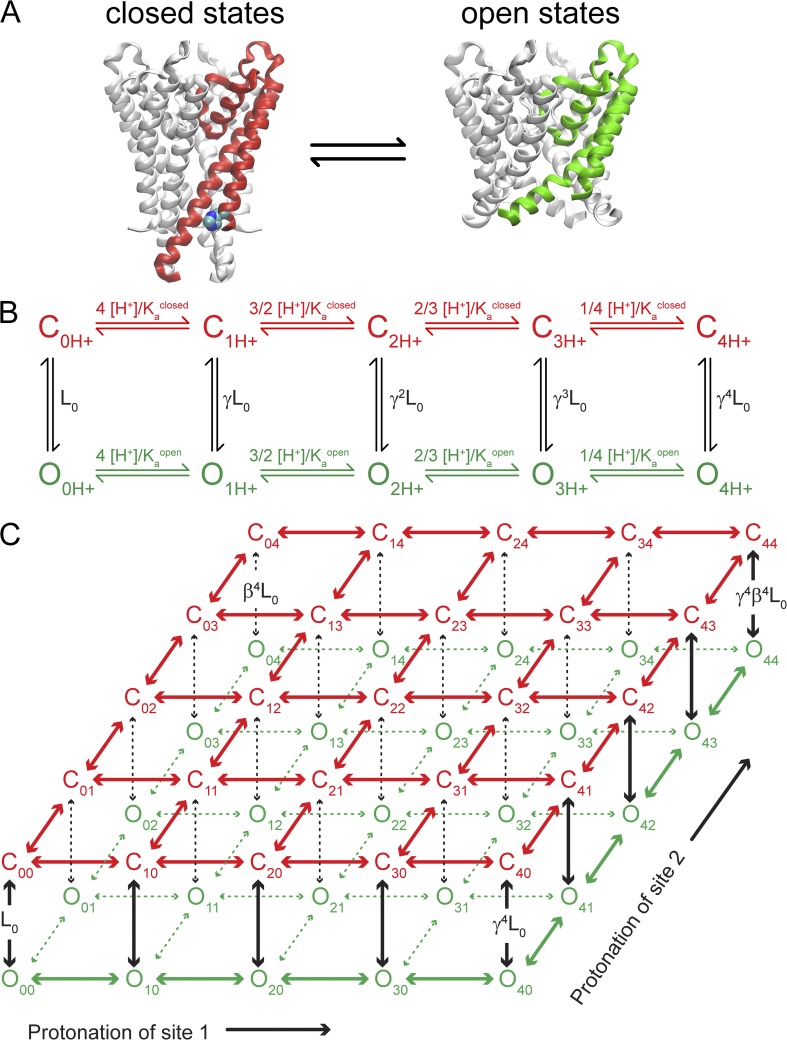
The MWC model links proton binding to the equilibrium between closed and open KcsA conformations. (A) Crystal structures of closed KcsA (Protein Data Bank accession no. 1K4C; one subunit red) and an open mutant (Protein Data Bank accession no. 3F5W; one subunit green) are shown, highlighting the position of one likely proton sensor, H25 (not modeled in 3F5W). (B) MWC model with a single independent proton-binding site per KcsA subunit. Closed states (red) are indicated as “C” and open states (green) are indicated as “O,” with subscripts indicating the number of protons bound to the channel. Lo is the gating equilibrium in the absence of protons. Kaclosed and Kaopen are the acid dissociation constants for the closed and open states. [H+] is the proton concentration. Each bound proton increases the gating equilibrium by a factor of γ = Kaclosed/Kaopen. (C) Two-site pH-sensor MWC model drawn as two tiers with protonation of site 1 along the x axis and protonation of site 2 along the y axis. Horizontal transitions depend on state-dependent pKa values as in B. Protonation of sites 1 and 2 increases the gating equilibrium (vertical transitions) by γ = Ka1closed/Ka1open or β = Ka2closed/Ka2open, respectively.
In these models, proton binding was assumed to be equivalent and independent to all KcsA subunits. Lo is the intrinsic gating equilibrium between open and closed in the absence of protons, [H+] is the proton concentration (also represented as pH), and Kaopen and Kaclosed are the acid dissociation constants for a proton sensor in the open and closed states, respectively (also represented as pKaopen and pKaclosed). Eq. 3 contains two proton sensors that are labeled with subscripts 1 and 2. In general, the absence of direct experimental constraint on the values for Lo and the very steep [H+] dependence for many of the datasets invalidated development of uniquely determined model parameters for individual datasets. We made the simplifying assumption that mutation of pH-sensor site 1 (identified as H25) did not change the pKa values for pH-sensor site 2 (identified as E118), and vice versa. In addition, we also assumed that mutation of E120 did not perturb nearby pH-sensor pKa values. Collectively, these assumptions allowed us to group several datasets together and provided significant constraint on the pKa values that could describe the data. We globally fit all mutations of H25 that retained the E118 pH sensor. Mutants H25R and H25R/E120A were fit simultaneously to a single pH-sensor model (E118) with shared values for pKaopen and pKaclosed but with two separate values for Lo. The fit was unique and well-determined but with large uncertainty in the Lo values (pKaclosed, E118 = 5.0 ± 0.1, pKaopen, E118 = 6.2 ± 0.1, LoH25R = 7 × 10−4 ± 4 × 10−4, and LoH25R/E120A = 0.08 ± 0.02). Next, we attempted to globally fit all mutations of E118 that retained the H25 pH sensor. Mutants E118A, E118Q/E120Q, and E118A/E120A were simultaneously fit to a single pH-sensor model with shared values for pKaclosed and pKaopen but with three separate Lo values. In this case, the pKa values were not uniquely determined but varied over a range of ∼0.5 pH units or less as parameter initialization was systematically varied. Therefore, the approximate pKa values were fairly well constrained by the requirement to simultaneously fit both the E118A and E118A/E120A dose–response curves that were shifted by ∼1.5 pH units. We found a global fit that overall minimized the fitting error but with large uncertainty remaining in Lo values (pKaclosed, H25 = 4.8 ± 0.6, pKaopen, H25 = 7.6 ± 0.7, LoE118A = 10−8 ± 8 × 10−8, LoE118Q/E120Q = 0.8 × 10−8 ± 5.8 × 10−8, and LoE118A/E120A = 2 × 10−4 ± 9 × 10−4). The pKa values for H25 were subsequently constrained to these best estimates, and the Lo values for all mutants were fitted using those constraints (these values are reported in Table 1). A similar procedure was followed for the unknown pH sensor in the triple mutants H25R/E118A/E120A (Fig. 1), H25R/E118Q/E120Q, and H25A/E118A/E120A (Fig. S2). Only steady-state Po data were fit to models (Figs. 1–3). No kinetic analysis of single-channel data was performed. All data fits are summarized in Table 1.
Table 1.
Summary of Hill and MWC model fits
| Mutanta | Hill fit | pH sensor site 1 H25 | pH sensor site 2 E118 | Lo (±) | |||
| pH1/2 (±) | nH (±) | pKa1closed (±) | pKa1open (±) | pKa2closed (±) | pKa2open (±) | ||
| E71Ab (control channel) | 5.3 (0.01) | 4.4 (0.1) | 4.8 (N/A)c | 7.6 | 5.0 | 6.2 | 2.5 × 10−12 (0.3 × 10−12) |
| H25Rb | 5.29 (0.02) | 1.9 (0.01) | — | — | 5.0 (0.1) | 6.2 (0.1) | 7 × 10−4 (4 × 10−4) |
| H25R/E120A | 6.3 (0.04) | 1.3 (0.1) | — | — | 5.0 (0.1) | 6.2 (0.1) | 0.08 (0.02) |
| E118A | 5.54 (0.03) | 4.5 (2.8) | 4.8 | 7.6 | — | — | 1.1 × 10−8 (0.1 × 10−8) |
| E118Q/E120Qb | 5.5 (0.004) | 4.4 (1.2) | 4.8 | 7.6 | — | — | 8.1 × 10−9 (0.5 × 10−9) |
| E118A/E120Ab | 6.6 (0.003) | 5.8 (0.1) | 4.8 | 7.6 | — | — | 1.5 × 10−4 (0.4 × 10−4) |
| H25R/E118A | — | — | — | — | — | — | 52 (11) |
| E120A | 5.3 (0.006) | 3.4 (0.1) | 4.8 | 7.6 | 5.0 | 6.2 | 2.5 × 10−12 (1.2 × 10−12) |
| H25R/E118A/E120Abd | — | — | — | — | 4.7 | 6.5 | 3.3 (0.7) |
| H25R/E118Q/E120Qbd | 6.3 (0.04) | 2.5 (0.5) | — | — | 4.7 | 6.5 | 0.021 (0.004) |
| H25A/E118A/E120Abd | 6.3 (0.01) | 2.6 (0.1) | — | — | 4.7 | 6.5 | 0.014 (0.002) |
| R121A (model fit 1) | 5.8 (0.01) | 4.0 (0.2) | 4.8 | 7.6 | 5.0 | 6.2 | 1.3 × 10−9 (0.3 × 10−9) |
| R121A (model fit 2) | — | — | 6.6 | 7.6 | 5.0 | 7.7 | 1.0 × 10−11 (0.1 × 10−11) |
| R122A (model fit 1) | 5.6 (0.01) | 6.9 (0.7) | 4.8 | 7.6 | 5.0 | 6.2 | 3.8 × 10−11 (0.6 × 10−11) |
| R122A (model fit 2) | — | — | 6 | 7.6 | 5.0 | 6.2 | 4 × 10−9 (10−9) |
| E118A/R121A (not modeled) | 6.0 (0.2) | 0.6 (0.2) | — | — | — | — | — |
| E118A/R122A (not modeled) | 6.6 (0.09) | 1.3 (0.4) | — | — | — | — | — |
| E120A/R122A | 5.7 (0.02) | 2.1 (0.2) | 6 | 6.5 | 4.5 | 5.5 | 4.2 × 10−3 (0.3 × 10−3) |
Values in parentheses are the standard error of fit.
All mutants on the background of E71A.
Data and Hill fits from Thompson et al. (2008).
Not applicable; no error given means the parameter was constrained (see Materials and methods).
Modeled as an unidentified residual pH sensor.
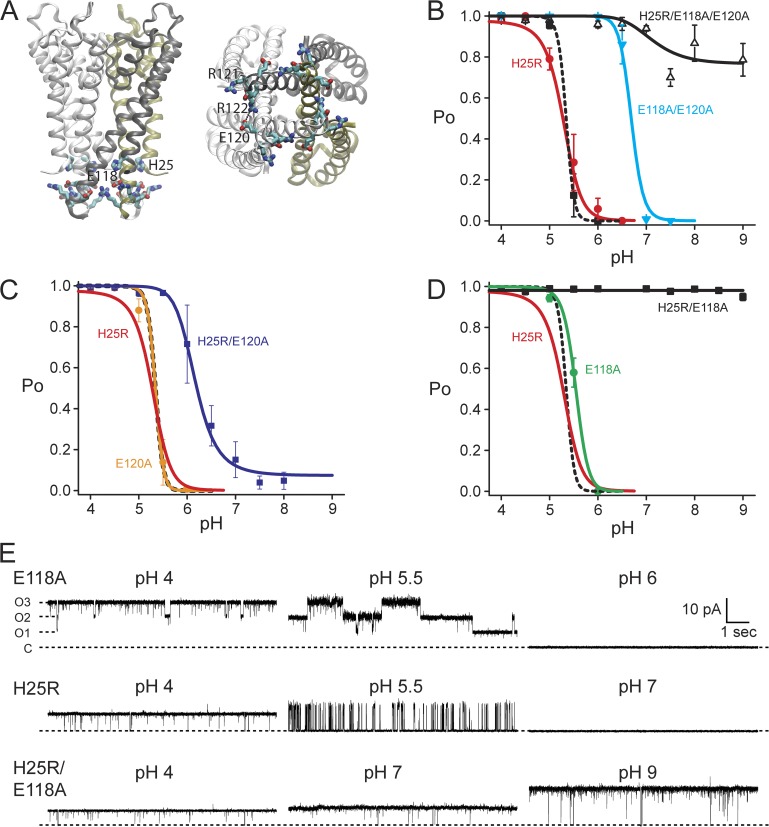
Figure 1.
Protonation of H25 and E118 may be sufficient to fully activate KcsA. (A) A high density of titratable and charged residues surrounds the cytoplasmic gate of KcsA, viewed from the side (left) and from the cytoplasm (right). H25, E118, E120, R121, and R122 are highlighted. (B) Summary of pH-dependent open probability (Po) for critical mutants reported in Thompson et al. (2008). Lines are fits with MWC models (Materials and methods; Fig. 4 and Table 1), where H25 is proton-binding site 1 and the E118 is site 2. The E71A control channel (black squares) was fit to a two pH–sensor model (black dashed line). H25R/E118A/E120A retained a minor residual pH dependence (open triangles, black line), modeled as a single proton-binding site. (C) E120 is likely not a pH sensor, and E120A was modeled similar to the control channel with two pH-sensor residues (circles, orange). H25R/E120A retained the E118 pH sensor (squares, blue line). (D) E118A was modeled with H25 pH sensor (circles, green line). H25R/E118A (squares, black line) was fitted to a pH-independent gating equilibrium (Lo = 51). Data are the mean ± SEM from three to seven bilayers, and the model for wild type is shown as a black dashed line in C and D. (E) Example single-channel recordings for E118A (top), H25R (middle), and H25R/E118A (bottom) at three pH values illustrating the pH-dependent Po. 10 s of data at 100 mV were filtered offline to 500 Hz for display.
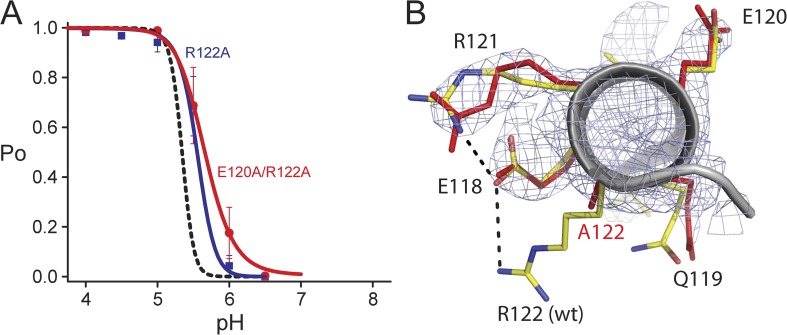
Figure 3.
An intersubunit interaction between R122 and E120 is not essential for pH-dependent gating in wild type. (A) The pH dependence of E120A/R122A (red circles) does not significantly differ from the R122A mutant (blue squares). Two pH-sensor residues are modeled for R122A (blue line) and E120A/R122A (red line), as in the control channel (black dashed line). MWC model fit parameters are given in Table 1. (B) Alignment of the intracellular end of the TM2 helix (gray ribbon) of the wild-type KcsA crystal structure (Protein Data Bank accession no. 1K4C; residues are modeled as sticks with carbon yellow, nitrogen in blue, and oxygen in red) with a model of our R122A KcsA mutant structure (red sticks). The electron density map for E118A contoured at 1.2 σ (Table 2) showed that the mutation did not significantly alter the location of E120 or the other nearby residues E118, Q119, and R121.
The change in the energy of gating caused by a single proton binding to a pH sensor is given by Eq. 4:
| (4) |
The gas constant, R, times the temperature T in Kelvin is RT = 0.59 kcal/mole at 25°C, and the coupling constant, γ, is given by,
| (5) |
For the two proton–binding site model, β is used for the second site-coupling constant.
The pH-independent equilibrium constant for the double mutant H25R/E118A was predicted from our models using the intrinsic equilibrium constants for the control channel and the single mutants H25R and E118A by,
| (6) |
Crystallography
As described previously (Zhou et al., 2001), the C-terminal domain of the KcsA E118A or the KcsA R122A mutant was removed using chymotrypsin before gel-filtration chromatography to aid in crystallization. The purified channel was then combined with a KcsA antibody Fab fragment and run over a second gel-filtration column (Zhou et al., 2001). The KcsA–Fab complex was concentrated to 8–10 mg/ml. Protein was crystallized in hanging drop vapor diffusion trays by combining 2 µl of protein solution with 2 µl of a well solution consisting of 19–22% (wt/vol) PEG 400, 25 mM Mg acetate, and 100 mM MES, pH 6.5. After crystals formed, in about 2 wk, the PEG 400 concentration in the well solution was adjusted to 40% for cryoprotection. After 24 h, the crystals were flash-frozen in liquid nitrogen.
Diffraction data were collected at 93 K using the X25 beamline at the National Synchrotron Light Source at Brookhaven National Laboratory. Reflections were indexed, integrated, and scaled in space group I4 using HKL2000 (HKL Research). The data were phased using molecular replacement with Protein Data Bank accession number 1K4C as the starting model. Phasing, refinement, and structure quality assessment were performed with the Phenix suite of programs (Adams et al., 2010). Model building was implemented with Coot (Emsley et al., 2010). Models and structure factors for the mutant KcsA structures E118A and R122A are deposited in the Protein Data Bank under accession numbers 4LCU and 4LBE, respectively.
Online supplemental material
Fig. S1 is provided to illustrate the modest changes in KcsA gating kinetics for the individual mutations E118A, R121A, and R122A for comparison, with the results presented in Fig. 2. Fig. S2 summarizes evidence for the existence of unidentified pH sensors in two mutants with the H25 and E118 sensors removed. Fig. S3 shows the pH dependence of the H25A mutant, demonstrates the dramatic channel to channel variability in this mutant, and further strengthens the argument for the necessity to model at least two pH-sensor residues. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201311057/DC1.
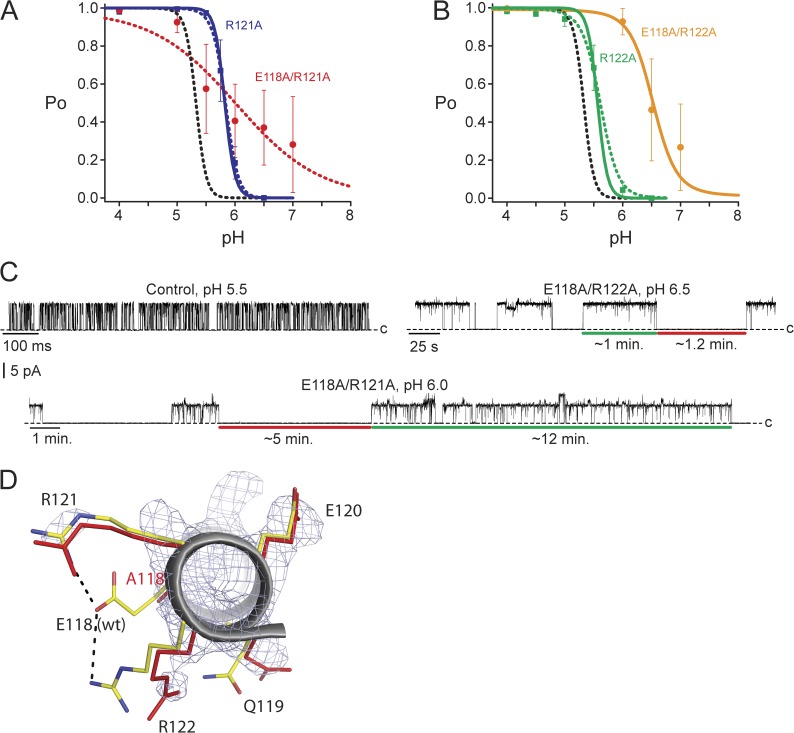
Figure 2.
Arginine residues may contribute to the intrinsic gating equilibrium as well as to the pKa values of nearby proton-binding sites. (A) R121A Po versus pH relationship (blue squares) was modeled either by changing relative to control only the intrinsic gating (solid blue line; model fit 1 in Table 1) or by also changing pKa values (dashed blue line; model fit 2 in Table 1). The E118A/R121A pH dependence (red circles) was not accurately established, and the red dotted line has no theoretical meaning. (B) R122A Po versus pH relationship (green squares) was modeled either by changing relative to control only the intrinsic gating (solid green line; model fit 1 in Table 1) or by also changing pKa values (dashed green line; model fit 2 in Table 1). The E118A/R122A pH dependence (orange circles) was not accurately established, and the orange line has no theoretical meaning. Data in A and B are the mean ± SEM from three to eight bilayers, and the model for control is shown as a black dashed line. MWC model fit parameters used in A and B are given in Table 1. (C) Example recordings of E71A control, E118A/R122A, and E118A/R121A at 100 mV and filtered offline to 50 Hz (or 20 Hz for E118A/R121A) for display. (D) The intracellular end of the TM2 helix (gray ribbon) of the E118A KcsA mutant aligned with the same region from KcsA wild type (Protein Data Bank accession no. 1K4C). The residues of interest are displayed as red sticks (E118A mutant) or with carbons in yellow, nitrogens in blue, and oxygens in red (wild type). The electron density map for the E118A KcsA mutant is shown contoured at 1.2 σ (Table 2). R121 was more disordered compared with wild type, and R122 assumed a new conformation.
RESULTS
Double mutation of histidine 25 and glutamate 118 ablated pH-dependent gating
The KcsA proton-dependent gate was identified as the cytoplasmic constriction formed by the bundle crossing of inner transmembrane helices (Perozo et al., 1998; Gross et al., 1999; Kelly and Gross, 2003). Several titratable residues are proximal to this channel gate (Fig. 1 A), indicating a possible role in pH sensing. In our previous report, we combined the mutation of two such glutamates, E118A/E120A, with the H25R mutant and established a near-pH–insensitive channel (Fig. 1 B), suggesting that the major components of pH sensing were gone (Thompson et al., 2008). H25 was implicated as a pH sensor because of a significant reduction in the steepness of the proton dose–response described by the Hill coefficient. E118/E120 were implicated as pH sensors because of a large shift relative to control in pH dependence from ∼5 to ∼6.5 pH (Fig. 1 B) (Thompson et al., 2008).
We investigated whether or not both glutamates contributed to pH-dependent gating as important proton-binding sites (Fig. 1, C and D). As in our previous report (Thompson et al., 2008), all channel mutations were made on the background of E71A, a mutation that removes selectivity filter inactivation and reveals full activation of the pH-dependent gate at low pH values (Cordero-Morales et al., 2006). In this paper, KcsA E71A is referred to as “KcsA” or “control channel.” H25R/E120A remained pH sensitive with a shifted proton dependence relative to the H25R mutant. H25R/E118A, on the other hand, exhibited no pH dependence and full activation in all pH conditions (Fig. 1, C and D). Example single-channel recordings of E118A, H25R, and H25R/E118A are shown to illustrate the combined contribution from these two residues during pH-dependent gating (Fig. 1 E). Importantly, H25R/E118A resulted in very little channel to channel variability in contrast to the previously reported H25R/E118A/E120A mutant (Thompson et al., 2008). These results suggested that E120 may not directly contribute to pH-dependent gating as a critical proton-binding site but still contributes to the gating equilibrium as seen in the double mutants E118A/E120A and H25R/E120A (Fig. 1, B and C). On the other hand, proton binding to both H25 and E118 may account for the full pH-dependent activation of KcsA, assuming that the mutations (H25R/E118A) did not alter the intrinsic gating of the channel through steric or structural changes that do not mimic protonation in wild type.
Individual mutations of the glutamates showed that although E120A was nearly indistinguishable from the control channel (Fig. 1 C), the E118A mutation (Fig. 1 D) also resulted in only a very small shift in pH dependence, despite the dramatic effect of this mutation when combined with H25R. Considering the small effect observed when only E118 is mutated compared with the considerable effect of the H25R mutation, we propose that protonation of both E118 and H25 is necessary to gate the channel, and that H25 contributes a considerably greater amount of energy during the gating process.
Two arginine residues influence pH dependence and gating kinetics
We next investigated the role of residues near the proposed E118 and H25 pH sensors. Two arginine residues, R121 and R122, appear to interact with E118 in the closed-state KcsA crystal structure (Zhou et al., 2001; Thompson et al., 2008). We explored the effect these residues have on pH-dependent gating by mutating them individually to alanine. R121A introduced a significant shift in pH dependence (Fig. 2 A, blue squares), with no change in steepness (solid blue line) relative to control (black dashed line). The R122A mutation (Fig. 2 B, green squares) resulted in a relatively modest shift in pH dependence as well as a small decrease in slope (nH = 3.4; Table 1).
We next removed each individual arginine on the background of the E118A mutation (Fig. 2, A and B). These mutant channels displayed gating kinetics that differed remarkably from the control channel. Fig. 2 C illustrates the kinetic properties observed for these channels at intermediate pH values where the channels are near half-activation. Unlike the control channel, which opened and closed on the millisecond timescale, E118A/R122A displayed minute-long open bursts and closed durations. Even more dramatically, the E118A/R121A mutant displayed very long stretches of high activity (∼10 min) interrupted by equally long closings. Brief closures within a burst, called channel flickers, are thought to arise from residual selectivity filter gating events (Piskorowski and Aldrich, 2006) and are likely not related to movement of the pH-dependent gate. Although both mutants were clearly pH dependent, the dose–response curves were difficult to establish accurately because of the uncertain Po values measured at intermediate pH values (Fig. 2, A and B, red and orange circles). Regardless, these data have interesting implications. First, they show that residues within the pH-sensing region are able to dramatically increase the energy barriers between the open and closed states, resulting in drastically slower gating. These intermediate states during gating likely relate to the intersubunit cooperativity and may also explain why an increase in subconductance gating is also observed in these mutant channels (Fig. 2 C). Second, the individual mutations E118A and R121A both slow channel gating modestly within the time scale of seconds, whereas R122A has very little effect on gating kinetics (Fig. S1). The drastic change in the gating kinetics observed for both double mutants (E118A/R121A and E118A/R122A) indicates energetic coupling interactions between E118 and both R121 and R122 during gating transitions.
We further investigated the structural relationship between E118 and the two arginines, R121 and R122, by solving the crystal structure of the E118A mutant in the closed state (Fig. 2 D and Table 2). Supporting our conclusion that E118 interacts with both arginines, the structure of E118A shows deviations from the wild-type structure for both R121 and R122. R122 adopted a different conformation, whereas R121 became disordered in the mutant structure (Fig. 2 D, residues shown in red). The mutation also resulted in the disruption of the potential intersubunit R122–E120 interaction (Fig. 1 A). The interactions these residues make in the open state and during gating intermediate states remain unknown.
Table 2.
Crystallographic data collection and refinement statistics
| KcsA E118A | KcsA R122A | |
| Data collection | ||
| Space group | I4 | I4 |
| Cell dimensions | ||
| a, b, c (Å) | 155.95, 155.95, 75.95 | 155.94, 155.94, 75.81 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.0–2.75 (2.85–2.75)a | 50.0–2.75 (2.85–2.75) |
| Rsym | 10.8 (93.2) | 8.3 (65.5) |
| I/σI | 18.5 (2.4) | 20.8 (2.2) |
| Completeness (%) | 99.7 (99.3) | 99.9 (99.2) |
| Redundancy | 7.4 (7.1) | 7.0 (6.2) |
| Refinement | ||
| Resolution (Å) | 49.31–2.75 | 42.87–2.75 |
| No. reflections | 23,757 | 23,755 |
| Rwork/Rfree | 0.183/0.216 | 0.169/0.202 |
| No. atoms | ||
| Protein | 4,057 | 4,081 |
| Ligand/Ion | 37 | 44 |
| Water | 56 | 89 |
| B factors | ||
| Protein | 83.6 | 72.9 |
| Ligand/Ion | 79.2 | 85.0 |
| Water | 57.4 | 56.7 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.008 | 0.012 |
| Bond angles (°) | 1.481 | 1.819 |
Values in parentheses are for the highest-resolution shell.
An intersubunit salt bridge does not contribute significantly to pH sensing
We proposed previously that an intersubunit salt bridge between R122 and E120 observed in the closed-state KcsA crystal structure contributes to pH-dependent gating (Thompson et al., 2008). However, as shown above, the E120A mutation alone does not alter pH-dependent gating (Fig. 1 C). Furthermore, E120A in addition to the R122A did not further perturb pH-dependent gating (Fig. 3 A). A crystal structure of the R122A mutant showed almost no change in the structure of the bundle crossing and proposed pH-sensor residues (Fig. 3 B and Table 2). It is possible that R122 can make several molecular interactions that modestly alter the intrinsic closed–open equilibrium; however, these results further argue that E120 is not a pH-sensor residue and that proton binding to E118 and H25 alone drives gating.
DISCUSSION
Several previous studies have used mutagenesis targeted to modify or remove KcsA pH-dependent gating to establish the proton-binding sites and qualitatively describe the mechanism (Takeuchi et al., 2007; Thompson et al., 2008; Cuello et al., 2010c; Hirano et al., 2011). Hirano et al. (2011) proposed that the C-terminal domain contains strong pH sensors at E146 and D149. However, truncation of the bottom half of the C-terminal domain (Δ140) (Cortes et al., 2001) as well as its replacement with a cytoplasmic domain from a different ion channel (unpublished data) did not change KcsA pH dependence, suggesting that this region has little effect on KcsA gating. An NMR study (Takeuchi et al., 2007) identified the importance of H25; however, no single mutation of H25 removed pH dependence in electrophysiological assays (Fig. 1 and Thompson et al., 2008) or in radioactive flux assays (Cuello et al., 2010c). We also examined H25A (the substitution used in the NMR study) to compare the results with H25R. Unfortunately, this mutant results in a high degree of channel to channel variability, but importantly, pH dependence remains (Fig. S3). Furthermore, single point mutations at other candidate residues (H20, E118, E120, or H124) did not ablate pH dependence (Thompson et al., 2008; Cuello et al., 2010a). Therefore, functional analysis pointed to the likely contribution from at least two proton-binding sites.
A proposed model for proton binding and gating in the KcsA channel
The proton-binding and gating relationship in KcsA appears to be a highly cooperative process (nH = 4.4 for KcsA control; Table 1). As in other ligand-gated channels, this property can arise naturally from subunit cooperativity during gating. We propose an MWC model for KcsA pH sensing and the interpretation of our mutagenesis results. The fundamental assumptions underlying the MWC model are that the protein exists in two global conformations, open (O) and closed (C), to which the ligand binds with different affinities (Monod et al., 1965; Changeux, 2012). X-ray crystallography has been used previously to determine both closed and open conformations of KcsA, shown in Fig. 4 A (Zhou et al., 2001; Cuello et al., 2010b).
The simplest pH-sensor MWC model with a single proton-binding site per KcsA subunit yields a 10-state model with 5 closed and 5 open states, one each for the differently liganded states (Fig. 4 B). On the other hand, two independent proton-binding sites per KcsA subunit (as proposed above: H25 and E118) can lead to a larger MWC model, with protonation occurring along two independent directions, one for each proton-binding site (Fig. 4 C). For each differently liganded state in the single proton-sensor model, there are 5 differently liganded states for the secondary proton sensor, leading to a total of 25 closed and 25 open states.
We thus formalized the contributions from the two pH-sensor residues (H25 and E118) using a two proton–binding site MWC model that emphasizes subunit cooperativity and state-dependent pKa values that energetically drive gating. All other contributions to the protein chemical energy combine to set the intrinsic equilibrium between open and closed conformations, called Lo. In this view, descriptions of interacting amino acids and electrostatic repulsion, variously emphasized in the literature (Miloshevsky and Jordan, 2007; Thompson et al., 2008; Cuello et al., 2010c), are connected to a specific and testable chemical property, state-dependent pKa values. We used these models to describe the open probabilities (Po) versus pH (Eqs. 2 and 3; Materials and methods) of the KcsA control channel (containing the two proposed pH sensors, E118 and H25), the H25R mutant (gated by the single E118 sensor), and the E118A mutant (gated by the single H25 sensor). The individual datasets do not provide enough information to constrain the MWC model parameters partly because the Po versus pH rises steeply from 0 to 1. Thus, to constrain the pKa values, we assumed that mutation of the H25 sensor did not perturb the pKa values of the E118 sensor and vice versa. These assumptions allowed us to group our datasets into global fits that added a significant constraint on the pH-sensor pKa values (see Materials and methods). Model fits are plotted in Figs. 1–3, and parameters are summarized in Table 1.
Because mutation of H25 resulted in a significant reduction of proton sensitivity (decrease in steepness of proton dose–response) and mutation of the E118 did not, we modeled H25 as a strong proton sensor with a large pKa shift between closed (pKaclosed = 4.8) and open (pKaopen = 7.6). The single-site MWC model with these values approximated well the pH dose–response of the E118A KcsA mutant that has just the H25 proton sensor remaining (Fig. 1 D). The glutamate proton sensor was described as weak, with a more modest pKa state dependence (pKaclosed = 5.0 and pKaopen = 6.2). The single-site MWC model with these values also approximated well the pH dose–response of the H25R KcsA mutant that has just the E118 proton sensor remaining (Fig. 1 B). For the models to fit the dose–response curves of the mutant channels, we assumed that mutation of a pH-sensor site did not alter the pKa values of the remaining pH sensor, but rather changed the intrinsic gating equilibrium constant, Lo (Fig. 4 B and Table 1; see Materials and methods). This made sense because the mutant channels were expected to have increased Lo values compared with the control channel, as the pH-sensor sites were mutated in a way to mimic constitutive protonation, hence favoring channel opening. These models, proposed as a conceptual starting point for further investigation, lack sufficient constraints to define the precise energetic contributions from the two proton sensors, E118 and H25. However, we notably captured the salient feature in our data that mutation of H25 always significantly decreased the slope of the pH dose–response, suggesting that the individual energetic contribution from H25 is significantly greater than the contribution from other proposed pH-sensor sites.
In these models, the pH sensors H25 and E118 have state-dependent pKa values, such that the affinities for proton binding are higher in the open state (higher pKa values). This makes structural sense for H25, which appears buried in the closed-state crystal structure (Zhou et al., 2001) and near three arginine residues (R117, R121, and R122), likely hindering proton binding and thus shifting its closed-state pKa value to more acidic values. In contrast, a crystal structure of a mutant KcsA in a presumed open state (Fig. 4 A) showed a displacement of the TM2 helices, which would dramatically increase the solvent exposure of H25 (Cuello et al., 2010b), yielding a higher pKa value, closer to 6.5. A similar but less drastic change in pKa can be assumed for E118, as it is located very close to H25 at the bundle crossing (∼3 Å in the closed crystal structure) and is less buried.
The proposed model value for the wild-type intrinsic gating equilibrium suggested that KcsA is essentially fully closed in the absence of protons (Lo = 2.5 × 10−12), which is also observed in electrophysiological recordings (Cuello et al., 1998; Heginbotham et al., 1999). Despite this remarkably stable closed state in the absence of protons, the modeled H25 sensor provides enough energy when fully protonated to significantly activate the channel by itself. If H25 is the pH sensor site 1 in Fig. 4 C, then full protonation of H25 raises the gating equilibrium constant to [O40]/[C40] with a value of γ4 Lo = 0.4, corresponding to a significant open probability, Po = γ4 Lo/(1 + γ4 Lo) = 0.29. The energy contributed to gating by protonation of a single H25 residue is −3.8 kcal/mole in this model (Eqs. 4 and 5). Why then is the H25R channel completely closed rather than more open at high pH? The gating equilibrium Lo for the H25R channel is not 0.4 but is modeled as Lo = 7 × 10−4 (Table 1). This discrepancy may result from an overestimation of the “strength” of H25 and neglect of unidentified pH-sensor residues that contribute to the activation of the channel. Alternatively, this difference may reflect the fact that arginine is an imperfect mimic for protonated histidine, and therefore the value for the gating equilibrium Lo in the mutant channel can differ significantly from the value expected from the wild-type model parameters. Aside from the obvious steric differences between these residues, the charged guanidinium group of an arginine residue may not localize to the same structural position as the charged imidazole group of histidine 25. For these reasons, we did not attempt to constrain our models such that pH-sensor mutations converted the energetic contributions of proton binding in wild type into a precisely corresponding change in Lo for the mutant channels.
We modeled the secondary E118 sensor as weak, with only a 1.2–pH unit shift in pKa value between closed and open that fit the shallow proton dose–response of the H25R mutant (Fig. 1; Materials and methods). The energy contributed to gating by protonation of a single E118 residue is −1.6 kcal/mole in this model (Eqs. 4 and 5). Therefore, E118 cannot activate the wild-type channel on its own (Lo β4 = 1.5 × 10−7). However, on the background of the H25R mutant that has an elevated Lo value compared with wild type, the weak secondary sensor can fully activate the channel (Fig. 1 B, red curve). The pH-independent mutant H25R/E118A (Fig. 1 D) suggests that protonation of both H25 and E118 are sufficient to account for the full activation of the pH-dependent gate in the control channel, assuming that the H25R/E118A precisely mimics the protonated H25 and E118. Alternatively, the mutations may have inflated Lo because of steric or structural changes, resulting in an overestimation of the importance of these residues within the wild-type pH sensor. Notably, the assumption that the individual mutations H25R and E118A do not alter the remaining pH sensor but only alter Lo did not accurately predict the Lo value for the H25R/E118A double mutant (Eq. 6). In our model, LoH25R/E118A = 3.1, corresponding to a Po value of ∼0.75, was significantly different than the measured Po of ∼0.98. Additionally, mutation of E120 was assumed not to perturb the pKa values for E118 or H25 to provide necessary constraints on the model parameters (see Materials and methods). However, the model indicates that E120A increases Lo when added to the H25R channel but slightly decreases Lo when added to the H25R/E118A channel (Fig. 1 and Table 1). These discrepancies suggest that the pKa values might be somewhat altered by nearby mutations, and the total strength of each pH-sensor residue in the wild-type channel is therefore difficult to establish quantitatively using the mutagenesis approach. In addition, other contributing residues have not been conclusively ruled out.
What data supports the existence of other pH-sensor residues? Despite the dramatically pH-independent behavior of H25R/E118A, other mutations of these sites have displayed pH-dependent activity. Three triple mutations of H25/E118/E120 that displayed pH-dependent activity were reported previously. First, H25R/E118A/E120A, a mutant that induced channel to channel variability (Thompson et al., 2008), displayed a small decrease in Po at higher pH values, indicating that the addition of E120A likely lowered the value of Lo in this triple mutant relative to H25R/E118A (Fig. 1 B). More convincingly, two other triple mutants maintained robust pH-sensing capabilities. H25R/E118Q/E120Q was pH dependent with a Hill slope of ∼2.5 (Thompson et al., 2008), indicating that the mutant had a similar sensitivity to protons as the H25R single mutation. Finally, the H25A/E118A/E120A mutant was also pH dependent with a Hill slope of ∼2.5. In both these latter mutants, the channel was completely closed at high pH and was fully activated by an increase in proton concentration, indicating the presence of an unidentified pH sensor (Fig. S2 and Table 1).
If the KcsA pH sensor were distributed over a larger number of sites, the development of a realistic MWC kinetic model would be substantially more complicated as each additional site adds significantly to model complexity. For example, the case of three independent proton-binding sites increases the model to 250 states, with seven free parameters describing the steady-state Po. On a conceptual level, however, a model including three or more independent proton-binding sites directly follows from the models presented, where each pH sensor has a state-dependent pKa value corresponding to its energetic contribution to gating the channel. Other sources of complexity are also possible, including interdependence between proton-binding sites, which we ignored in this study, as there was no experimental result specifically indicating it. The physical proximity of E118 and H25 in the closed-state KcsA structure (∼3 Å), however, may suggest that proton binding at these residues (and thus the pKa values) may not be independent.
Arginine residues modulate gating kinetics and influence proton binding
In our model, mutation of residues near the pH sensor, such as the arginine residues, does not perturb the location of proton binding but must have a significant effect on either the pKa value or the gating equilibrium, Lo, to alter pH-dependent gating. The pH dependence of the arginine mutants can be easily modeled by changing only Lo (Table 1, model fit 1, and Fig. 2) or by changing both Lo and pKa values (Table 1, model fit 2, and Fig. 2).
When arginine residues were mutated in conjunction with E118A, an extremely dramatic change in gating kinetics occurred at intermediate pH values (Fig. 2 C), suggesting an important interaction between E118, R121, and R122 during gating. This interaction is also supported by the crystal structure of the E118A mutant, where the positions of the arginines seem to be altered by the absence of the glutamate (Fig. 2 D). At intermediate levels of activation, both double mutants displayed minute-long open periods interspersed with minute-long closings. This phenotype leads to an apparent channel to channel variability in pH-dependent open probabilities, although the pH dependence was clearly detectable (Fig. 2, A and B). A macroscopic measurement, sensitive only to the ensemble average activity of many channels, would be blind to these effects but would likely record a shallower dose–response curve. These results suggest that the energy of an intermediate state is greatly increased relative to the energy of both the closed and open states in the two double mutants. In other words, the cooperative transition of subunits, fundamental to the MWC model, is altered or disrupted in these mutants relative to wild type.
Conclusions
In summary, we have shown that the pH dependence of KcsA arises in part from a histidine (H25) and a glutamate (E118) located next to the closed bundle-crossing gate. A second glutamate (E120) does not appear to contribute to pH-dependent gating, and an apparent interaction between E120 and a nearby arginine (R122) does not significantly contribute to pH sensing. We described the KcsA pH sensor using a two proton–binding site MWC model. The model offered a conceptual framework to rationalize the effects of mutagenesis and to propose thermodynamic contributions of proton binding to channel gating. Histidine 25 is likely the central pH-sensor residue contributing the most energy to gating, whereas E118 and possibly other unidentified residues probably enhance activation with smaller energetic contributions. R121 and R122 likely perturb the state-dependent pKa values for nearby pH-sensor residues and also participate in setting the energy barrier crossed during the cooperative transition between closed and open.
Supplementary Material
Acknowledgments
We thank the staff of the X-25 beamline for their help.
The National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy (USDOE) under contract DE-AC02-98CH10886. Beamline X25 is funded by the Offices of Biological and Environmental Research and of Basic Energy Sciences of the USDOE and from the National Center for Research Resources of the National Institutes of Health (NIH). This work was supported by a National Research Service Award postdoctoral fellowship from the NIH (F32GM087865 to D.J. Posson) and an NIH grant (GM088352 to C.M. Nimigean).
Sharona E. Gordon served as editor.
Footnotes
Abbreviations used in this paper:
- MWC
- Monod–Wyman–Changeux
- NMR
- nuclear magnetic resonance
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux J.P. 2012. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys. 41:103–133 10.1146/annurev-biophys-050511-102222 [DOI] [PubMed] [Google Scholar]
- Colquhoun D. 1998. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br. J. Pharmacol. 125:923–947 10.1038/sj.bjp.0702164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Cuello L.G., Zhao Y., Jogini V., Cortes D.M., Roux B., Perozo E. 2006. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13:311–318 10.1038/nsmb1069 [DOI] [PubMed] [Google Scholar]
- Cortes D.M., Cuello L.G., Perozo E. 2001. Molecular architecture of full-length KcsA: Role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 117:165–180 10.1085/jgp.117.2.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L.G., Romero J.G., Cortes D.M., Perozo E. 1998. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry. 37:3229–3236 10.1021/bi972997x [DOI] [PubMed] [Google Scholar]
- Cuello L.G., Cortes D.M., Jogini V., Sompornpisut A., Perozo E. 2010a. A molecular mechanism for proton-dependent gating in KcsA. FEBS Lett. 584:1126–1132 10.1016/j.febslet.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L.G., Jogini V., Cortes D.M., Perozo E. 2010b. Structural mechanism of C-type inactivation in K+ channels. Nature. 466:203–208 10.1038/nature09153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L.G., Jogini V., Cortes D.M., Sompornpisut A., Purdy M.D., Wiener M.C., Perozo E. 2010c. Design and characterization of a constitutively open KcsA. FEBS Lett. 584:1133–1138 10.1016/j.febslet.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A., Columbus L., Hideg K., Altenbach C., Hubbell W.L. 1999. Structure of the KcsA potassium channel from Streptomyces lividans: a site-directed spin labeling study of the second transmembrane segment. Biochemistry. 38:10324–10335 10.1021/bi990856k [DOI] [PubMed] [Google Scholar]
- Heginbotham L., LeMasurier M., Kolmakova-Partensky L., Miller C. 1999. Single Streptomyces lividans K+ channels: Functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114:551–560 10.1085/jgp.114.4.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. 2001. Ion Channels of Excitable Membranes. Third edition Sinauer Associates, Inc., Sunderland, MA: 814 pp [Google Scholar]
- Hirano M., Onishi Y., Yanagida T., Ide T. 2011. Role of the KcsA channel cytoplasmic domain in pH-dependent gating. Biophys. J. 101:2157–2162 10.1016/j.bpj.2011.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., MacKinnon R. 2002a. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522 10.1038/417515a [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Cadene M., Chait B.T., MacKinnon R. 2002b. The open pore conformation of potassium channels. Nature. 417:523–526 10.1038/417523a [DOI] [PubMed] [Google Scholar]
- Kelly B.L., Gross A. 2003. Potassium channel gating observed with site-directed mass tagging. Nat. Struct. Biol. 10:280–284 10.1038/nsb908 [DOI] [PubMed] [Google Scholar]
- Li Y., Berke I., Chen L., Jiang Y. 2007. Gating and inward rectifying properties of the MthK K+ channel with and without the gating ring. J. Gen. Physiol. 129:109–120 10.1085/jgp.200609655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.S., Sompornpisut P., Perozo E. 2001. Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Biol. 8:883–887 10.1038/nsb1001-883 [DOI] [PubMed] [Google Scholar]
- Miloshevsky G.V., Jordan P.C. 2007. Open-state conformation of the KcsA K+ channel: Monte Carlo normal mode following simulations. Structure. 15:1654–1662 10.1016/j.str.2007.09.022 [DOI] [PubMed] [Google Scholar]
- Monod J., Wyman J., Changeux J.P. 1965. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12:88–118 10.1016/S0022-2836(65)80285-6 [DOI] [PubMed] [Google Scholar]
- Perozo E., Cortes D.M., Cuello L.G. 1998. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol. 5:459–469 10.1038/nsb0698-459 [DOI] [PubMed] [Google Scholar]
- Piskorowski R.A., Aldrich R.W. 2006. Relationship between pore occupancy and gating in BK potassium channels. J. Gen. Physiol. 127:557–576 10.1085/jgp.200509482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf H., Schmidt O., Kümmerlen R., Hinnah S., Müller D., Betzler M., Steinkamp T., Wagner R. 1995. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 14:5170–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Takahashi H., Kawano S., Shimada I. 2007. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J. Biol. Chem. 282:15179–15186 10.1074/jbc.M608264200 [DOI] [PubMed] [Google Scholar]
- Thompson A.N., Posson D.J., Parsa P.V., Nimigean C.M. 2008. Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. USA. 105:6900–6905 10.1073/pnas.0800873105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadek B., Nimigean C.M. 2006. Calcium-dependent gating of MthK, a prokaryotic potassium channel. J. Gen. Physiol. 127:673–685 10.1085/jgp.200609534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Morais-Cabral J.H., Kaufman A., MacKinnon R. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48 10.1038/35102009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.