Abstract
The length and precise linkage of polyubiquitin chains is important for their biological activity. Although other ubiquitin-like proteins have the potential to form polymeric chains their identification in vivo is challenging and their functional role is unclear. Vertebrates express three small ubiquitin-like modifiers, SUMO-1, SUMO-2, and SUMO-3. Mature SUMO-2 and SUMO-3 are nearly identical and contain an internal consensus site for sumoylation that is missing in SUMO-1. Combining state-of-the-art mass spectrometry with an “in vitro to in vivo” strategy for post-translational modifications, we provide direct evidence that SUMO-1, SUMO-2, and SUMO-3 form mixed chains in cells via the internal consensus sites for sumoylation in SUMO-2 and SUMO-3. In vitro, the chain length of SUMO polymers could be influenced by changing the relative amounts of SUMO-1 and SUMO-2. The developed methodology is generic and can be adapted for the identification of other sumoylation sites in complex samples.
The ubiquitin family (1, 2) includes small ubiquitin-like modifiers (SUMOs)1 that are similar in structure to ubiquitin (3). In contrast to the well known role of ubiquitin in protein degradation by the proteasome, SUMO conjugation does not directly target proteins for destruction (4-6). In general, sumoylation regulates the function of target proteins by affecting protein-protein interactions, which can result in altered subcellular localization and activity. Sumoylation is essential for the viability of eukaryotic cells (7-12).
A significant number of target proteins have been identified for Smt3, the yeast SUMO family member, and for mammalian SUMOs (13). These proteomics studies have highlighted the broad cellular impact of SUMOs on processes including transcription, replication, RNA processing, translation, signaling, and transport.
Conjugation of SUMOs to target proteins, analogous to the ubiquitin system, involves E1, E2, and E3 enzymes (1, 2, 4-6). The E1 enzyme is a dimer that consists of SAE1 and SAE2, and in contrast to the large set of E2 enzymes involved in ubiquitination, a single E2 enzyme, Ubc9, is responsible for sumoylation. In addition, E3 enzymes, including protein inhibitor of activated signal transducer and activator of transcription family members and RanBP2, can enhance the sumoylation of target proteins but are not strictly required in vitro (14-17). Sumoylation is reversible; SUMOs can be removed from target proteins by specific SUMO proteases (4-6, 11). These proteases are also responsible for the maturation of SUMO precursors, a process that exposes the carboxyl-terminal diglycine motif that is characteristic for ubiquitin-like proteins and required for conjugation to target proteins.
Ubiquitin is able to form chains on target proteins via all seven internal lysines (18, 19). Ubiquitin chains were initially discovered by studying the role of ubiquitin in targeting protein substrates for proteolysis. These chains are Lys-48-linked polymers that mark target proteins for proteasome-mediated destruction (20). Structurally different ubiquitin chains can also play other roles in cells that are unrelated to protein degradation (18, 21). For example, Lys-63-linked chains are involved in translation, protein kinase activation, vesicle trafficking, and DNA repair. An interesting NMR study has revealed that the conformation of a Lys-63-linked ubiquitin dimer is distinct from a Lys-48-linked dimer (22). Yeast cells that express a K63R mutant of ubiquitin are compromised in DNA repair, but proteolysis is not affected in these cells (23).
In contrast to the extensive amount of data on ubiquitin chain formation (19, 24, 25), very little is known about multimerization of ubiquitin-like proteins. The single SUMO family member in Saccharomyces cerevisiae, Smt3, has been shown to form chains, but these chains are not required for viability (26). A yeast strain in which wild-type Smt3 was replaced by a lysine-deficient Smt3 mutant was viable; it had no obvious growth defects or stress sensitivities. The amount of Smt3 chains in yeast is limited due to the activity of the Smt3 protease Ulp2 (26). Interestingly Smt3 chains accumulate during meiosis (27). Here we investigated the polymerization of the three mammalian SUMOs by mass spectrometry. Trypsin digestion of ubiquitinated proteins produces diglycine-modified lysines, which are easily detected in MS and MS/MS spectra because of their predictable mass shift. In contrast, it is technically challenging to map attachment sites for human SUMO family members due to the fact that the long SUMO tryptic peptides attached to modified lysines substantially increase the mass of the peptide and also fragment during MS/MS. The resulting fragmentation patterns are very complex and not readily interpretable with currently available software for analyzing MS/MS spectra. Recently an automated pattern recognition tool (29) has been developed to overcome this limitation, but further work is needed to test its utility in vivo. Mutational strategies where trypsin cleavage sites are introduced close to the SUMO carboxyl-terminal diglycine (28, 30) simplify the mass spectrometric analysis but suffer from the use of non-physiological modifiers. Here we developed an alternative mass spectrometric strategy based on high resolution MS and the transfer of in vitro MS data to the in vivo data generated from very small sample amounts and high sample complexity. We used this strategy to identify conjugation sites for human SUMO family members and to unambiguously detect SUMO branched peptides. This approach allowed us to map the internal lysines that are used for SUMO chain formation and to demonstrate the ability of SUMOs to form chains in vivo.
EXPERIMENTAL PROCEDURES
Plasmids, Proteins, and Antibodies
SUMO-1 and SUMO-2 proteins were produced in Escherichia coli and purified as described previously (31). GST-SUMO-1, GST-SAE2-SAE1, GST-Ubc9, and control GST were produced in E. coli and purified as described previously (31, 32). The GST tag was removed from the E2 by thrombin cleavage to increase the enzymatic activity. T7-HIF-1α-His6 (aa 373-605) was produced in E. coli and purified as described previously (33).
Peptide antibody AV-SM23-0100 against SUMO-2/3 was generated in a rabbit using the peptide MEDEDTIDVFQQQTG (Eurogentec) (34). Monoclonal antibody 21C7 against SUMO-1 was obtained from Zymed Laboratories Inc., and monoclonal antibody 610958 against hypoxia-inducible factor-1α (HIF-1α) was obtained from BD Biosciences. Anti-T7 antibody coupled to HRP was obtained from Novagen (1:5000). Secondary antibodies used were anti-rabbit HRP and anti-mouse HRP (1:5000, Pierce) and Texas Red-conjugated antirabbit and fluorescein isothiocyanate-conjugated anti-mouse (1:350, Jackson ImmunoResearch Laboratories).
Electrophoresis, Silver Staining, and Immunoblotting
Protein samples were size-fractionated on Novex 4–12% 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol gradient gels using 4-morpholinepropanesulfonic acid buffer (Invitrogen). Total protein was visualized by silver staining. For immunoblotting experiments, size-fractionated proteins were subsequently transferred onto Hy-bond-C extra membranes (Amersham Biosciences) using a submarine system (Invitrogen). The membranes were incubated with specific antibodies as indicated. Bound antibodies were detected via chemiluminescence with ECL Plus (Amersham Biosciences).
Cell Culture
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS and 100 units/ml penicillin and streptomycin (Invitrogen). HIF-1α was stabilized by 0.9 mm CoCl2 (Sigma) treatment for 3 h. HeLa cells stably expressing His6-SUMO-2 were described previously (34).
Purification of GST-SUMO Conjugates, His6-SUMO Conjugates, and Endogenous SUMO-2/3 Conjugates
GST-SUMO-1 conjugates were obtained by incubating 20 μg of GST-SUMO-1 or control GST with 100 μl of HeLa nuclear extract (CILBiotech) in a buffer containing 1.5 mm ATP, 5 mm creatine phosphate (Sigma), 5 mm DTT, and 2 mm MgCl2 for 2.5 h at 30 °C. GST-SUMO-1 conjugates were bound to 30 μl of glutathione beads (GE Healthcare) for 1 h at 4 °C. Beads were successively washed with conjugation buffer, PBS, PBS containing 0.1% Triton X-100, and PBS only at 4 °C. Bound proteins were eluted successively in 8 m urea, pH 7, and NuPage LDS protein sample buffer (Invitrogen).
His6-SUMO-2 conjugates were purified essentially as described previously (34). Endogenous SUMO-2/3 conjugates were purified from HeLa cells lysed in 2% SDS, 50 mm Tris-HCl, pH 7.5, and 10 mm iodoacetamide supplemented with protease inhibitor mixture 1873580 (Roche Diagnostics GmbH) (35). Lysates were sonicated and diluted 20-fold in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5 mm β-mercaptoethanol, and 0.5% Nonidet P-40 supplemented with protease inhibitor mixture. Immunoprecipitations were performed with antibody AV-SM23-0100 or preimmune serum covalently cross-linked to protein G-Sepharose beads (GE Healthcare) for 3 h at room temperature. After extensive washing, bound proteins were eluted in NuPage LDS protein sample buffer (Invitrogen).
In Vitro Sumoylation
SUMO polymer formation described in Fig. 2A was carried out in 10-μl volumes containing 120 ng of SAE1/2, 2 mm ATP, 0.6 units·ml−1 inorganic pyrophosphatase, 10 mm creatine phosphate, 3.5 units ·ml−1 creatine kinase (Sigma), 5 mm MgCl2, 50 mm Tris-HCl, pH 7.5, 800 ng of Ubc9, protease inhibitor mixture, and the amounts of Ubc9, SUMO-1, and/or SUMO-2 indicated in the figure. Experiments described in Fig. 2B were carried out in 5-μl volumes and contained the ATP regeneration mixture, 60 ng of SAE1/2, 400 ng of Ubc9, and the indicated amounts of SUMO-1 and/or SUMO-2. For mass spectrometric analysis, a similar experiment without protease inhibitors was carried out using 2 μg of SUMO-1, 2 μg of SUMO-2, 480 ng of SAE1/2, and 4 μg of Ubc9 in a total volume of 40 μl. Assays were incubated for 3 h at 37 °C before either endopeptidase Lys-C and trypsin digestion and mass spectrometric analysis or addition of SDS sample buffer for immunoblotting analysis. Aliquots representing 6% of the reaction mixtures were loaded on the gel. 5 μg of recombinant T7-HIF-1α-His6 (aa 373–605) was sumoylated in vitro and subsequently purified in 8 m urea on Talon beads for mass spectrometric analysis.
Fig. 2. SUMO-1 limits the chain length of unanchored SUMO-2 polymers.
A, in vitro SUMO-1 and SUMO-2 heteroconjugate formation. In vitro SUMO conjugation reactions were set up containing recombinant SUMO-1 labeled with 125I in the absence (lanes 1–5) and presence (lanes 6–10) of unlabeled SUMO-2 and the indicated amounts of Ubc9. Reactions were incubated for 3 h at 37 °C before addition of SDS sample buffer to halt reaction progress. Samples were fractionated by SDS-PAGE. Dried gels were subjected to phosphorimaging to detect radiolabeled species. The positions of the SUMO conjugates are indicated. B, in vitro SUMO conjugation reactions were set up containing unlabeled SUMO-1 and/or unlabeled SUMO-2. Samples contained 3 μg of SUMO-2 (lanes 1 and 4), 3 μg of SUMO-2 plus 3 μg of SUMO-1 (lanes 2 and 5), or 6 μg of SUMO-2 (lanes 3 and 6). Reactions were incubated for 3 h at 37 °C before addition of LDS sample buffer to halt reaction progress. Samples were size-fractionated by SDS-PAGE, transferred to membranes, and probed using antibody 21C7 to detect SUMO-1 (lanes 1–3) or AV-SM23-0100 to detect SUMO-2 (lanes 4–6). C-F, mixed chains of SUMO-1 and SUMO-2 were generated in vitro, digested in solution with endopeptidase Lys-C and trypsin, and analyzed by mass spectrometry. C, the LC-MS/MS analysis using LTQ-FT-ICR is represented two-dimensionally. A very similar visualization was obtained when the LTQ-Orbitrap was used (data not shown). Peptide intensities were color-coded with white representing the lowest intensities and green representing the highest intensities. The region that contains the SUMO-1/SUMO-2 branched peptide is indicated by an arrow. Isotopic distributions of the SUMO-1/SUMO-2 branched peptide (ellipse) analyzed on the LTQ-FT-ICR (D) and the LTQ-Orbitrap (E) were visualized by enlarging the corresponding region. F, MS scans with highest intensities (D and E) were directly superimposed. LTQ-FT-ICR peaks are wider and of lower intensity, indicating a better resolution and mass accuracy for the LTQ-Orbitrap at this m/z value.
Mass Spectrometry and Data Analysis
Mass spectrometric analysis was performed by nanoscale LC-MS/MS using a linear ion trap-Fourier transform-ion cyclotron resonance mass spectrometer (LTQ-FT-ICR, Thermo Fisher Scientific, Bremen, Germany) or an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark) and coupled to an Agilent 1100 nano-HPLC system (Agilent Technologies) fitted with an in-house made 75-μm reverse phase C18 column as described previously (36, 37). In-solution digestion was performed essentially as described previously (38). The 50-kDa band from a silver-stained gel containing GST-SUMO-1 conjugates (Fig. 4A) was excised, cut into 1-mm3 cubes, and subjected to in-gel digestion according to Olsen et al. (37). The resulting peptides were desalted on RP-C18 stop and go extraction tips (39). Peptides were eluted with a 140-min linear gradient of 98% solvent A (0.5% acetic acid in H2O) to 50% solvent B (80% acetonitrile and 0.5% acetic acid in H2O).
Fig. 4. GST-SUMO-1 is conjugated to endogenous SUMO-2 and SUMO-3 in a nuclear extract.
A–C, GST-SUMO-1 or control GST was added to HeLa nuclear extracts in the presence of ATP and incubated at 30 °C for 2.5 h prior to purification on glutathione beads. The purified proteins were eluted in 8 m urea, and proteins that were still bound to the glutathione beads after urea elution were subsequently eluted in LDS protein sample buffer (S.B.). Purified fractions were size-separated by SDS-PAGE and analyzed by silver staining (A) or by immunoblotting using antibody 21C7 to detect SUMO-1 (B) or AV-SM23-0100 to detect SUMO-2 and SUMO-3 (C). D, the GST-SUMO-1-enriched fraction was digested in solution with endopeptidase Lys-C and trypsin. GST-SUMO-1 was conjugated to lysine 11 of endogenous SUMO-3. Shown is the MS/MS fragmentation spectrum of a tryptic peptide consisting of aa 79–97 of SUMO-1 and aa 8–21 of SUMO-3. Precursor ion mass was measured in the orbitrap mass spectrometer (m/z 937.4390 (4+); mass deviation, −0.15 ppm), and the peptide was fragmented and acquired in the LTQ mass spectrometer.
Data were acquired in the data-dependent mode: in the case of the LTQ-FT-ICR instrument, full scan spectra (m/z 300–1800, R = 50,000, and ion accumulation to a target value of 3,000,000) were acquired in the ICR cell. The three most intense ions were sequentially isolated for accurate mass measurements by selected ion monitoring scans with 10-Da mass range, R = 50,000, and a target accumulation value of 50,000 and fragmented in the linear ion trap by collisionally induced dissociation followed by MS3 analysis of the most intense product ion in the MS/MS scan.
In the case of the LTQ-Orbitrap, the precursor ion spectra were acquired in the orbitrap analyzer (m/z 300–1600, R = 60,000, and ion accumulation to a target value of 1,000,000), and the five most intense ions were fragmented and recorded in the ion trap. In a separate experiment, peptides derived from the digestion of in vitro produced SUMO polymers were fragmented in the linear ion trap, and the fragment ions were recorded in the orbitrap (R = 15,000). The lock mass option enabled accurate mass measurement in both MS and orbitrap MS/MS mode as described previously (37). Target ions already selected for MS/MS were dynamically excluded for 30 s. The detection and fragmentation of the SUMO-1/SUMO-2 parent ion, derived from the digestion of the His6-SUMO-2 and GST-SUMO-1 conjugates, were obtained with the selected ion monitoring mode with 10-Da mass range in which the dynamic exclusion option was not active and only ions with charge state equal to or larger than 4+ or unassigned were fragmented and recorded in the LTQ.
All spectra were acquired in the profile mode. The monoisotopic m/z values for the SUMO-1/SUMO-2, SUMO-1/SUMO-3, SUMO-2/SUMO-2, and SUMO-2/SUMO-3 branched precursor peptides were calculated with GPMAW software (Lighthouse Data, Hanstholm, Denmark) and used to search for the corresponding ions with Xcalibur software (Thermo Fisher Scientific). Assignment was confirmed by manually interpreting all MS/MS spectra. The corresponding “virtual” peptides were fragmented in silico with GPMAW, and the resulting m/z values were used to manually assign fragment ions to the peaks in the experimental fragmentation spectra. All reported MS/MS spectra were manually validated. Only branched peptides having an extensive coverage of y ions were considered. The peptides modified by SUMO-2, containing two prolines, were required to show pronounced cleavage amino-terminal to the proline residue. Parent ion charge and retention time were derived from a pilot LC-MS run of simple peptide mixtures from an in vitro sumoylation reaction and used together with precursor m/z values and fragmentation spectra to search for the branched peptides in a more complex mixture.
Visualization of MS Data
The LC-MS runs were visualized by using the Viewer tool of our in-house quantitative proteomics processing pipeline (40). The resulting two-dimensional LC-MS plots show the peptide m/z values (x axis) along the retention time axis. All the precursor isotope peaks were incorporated into the plots, and their signal intensities are color-coded with white representing the lowest intensities and green representing the highest intensities. The “full range” mode visualizes the peaks present in all MS spectra acquired during a 140-min chromatographic gradient. The enlarged view of specific SUMO branched peptides was obtained by selecting the corresponding m/z and time range and was used for a visual comparison of their abundances between different samples.
Microscopy and Image Analysis
HeLa cells were grown on glass coverslips and fixed for 10 min in 3.7% paraformaldehyde in 37 °C PHEM buffer (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, and 2 mm MgCl2, pH 6.9) (41). Subsequent manipulations were carried out at room temperature. Permeabilization was carried out for 20 min in PBS containing 0.5% Triton X-100. Cells were incubated with primary antibodies AV-SM23-0100 against SUMO-2/3 (1:2000) and 21C7 against SUMO-1 (1:50), washed, and incubated with secondary antibodies. DNA was stained with 0.3 μg/ml 4′,6-diamidino-2-phenylindole (Sigma). After washing, cells were mounted in Vectashield (Vector Laboratories).
Three-dimensional images and sections were recorded on a Zeiss Axiovert S100 2TV DeltaVision Restoration microscope (Applied Precision) using a Zeiss Plan-Achromat 100 × 1.40-numerical aperture objective and a CCD-1300-Y/HS camera (Roper Scientific). Images were captured and processed by constrained iterative deconvolution using SoftWorx (Applied Precision). Images presented here are maximal intensity projections of Z stacks.
RESULTS
SUMO Chain Formation in Vitro
Human SUMO-2 and SUMO-3 contain an internal consensus site for sumoylation that is absent from SUMO-1 (Fig. 1A). This allows SUMO-2 and SUMO-3 to polymerize in vitro (31). To investigate SUMO multimerization in cells by mass spectrometry, we developed a novel strategy (Fig. 1B). Our approach is based on the idea that peptide parent ion charge state, retention time, and a high quality MS/MS spectrum can be more easily, sensitively, and accurately obtained by LC-MS/MS analysis of a low complexity mixture, such as that resulting from an in vitro sumoylation assay. This information can be used subsequently to formulate a modification-specific MS method for the analysis of a complex protein mixture, such as partially purified cell lysates. The spectra from the complex mixture, usually of lower quality, can be matched to the high quality spectra derived from analysis of in vitro samples, allowing unambiguous characterization of peptides.
Fig. 1. SUMO-2 and SUMO-3 contain internal sumoylation sites.
A, humans express three different SUMO family members, SUMO-1, SUMO-2, and SUMO-3. Lysines 11 of SUMO-2 and SUMO-3 (green) are situated within the underlined sumoylation motif ψKX(E/D) where ψ is Val, Leu, Ile, Phe, or Met. SUMO-1 lacks a consensus sumoylation motif. These polymerization sites are located within the flexible amino-terminal extensions that are absent in ubiquitin. Sites of the carboxyl-terminal trypsin cleavage sites are indicated in orange. The peptides that remain conjugated to the targets after trypsin cleavage are indicated in blue. Note that the carboxyl-terminal SUMO-2 and SUMO-3 tryptic fragments are identical. B, experimental work flow. SUMO polymers were generated in vitro and analyzed by mass spectrometry. The obtained information was subsequently used to study SUMO multimerization in purified fractions from cells.
The formation of polySUMO chains was analyzed in vitro using 125I-labeled SUMO-1, unlabeled SUMO-2, the E1 and E2 enzymes, and an ATP regeneration mixture (Fig. 2A). When large amounts of E2 enzyme were used, SUMO-1 was able to form homodimers in the absence of SUMO-2. In the presence of SUMO-2, extensive mixed chain formation was observed. To study the effect of SUMO-1 on SUMO-2 chain length, a second set of in vitro sumoylation assays was performed with 3 μg of SUMO-2 and either with or without 3 μg of SUMO-1 (Fig. 2B, lanes 1, 2, 4, and 5). We noticed that SUMO-2 polymer formation decreased when SUMO-1 was added to the reaction, indicating that SUMO-1 limits SUMO-2 polymer length in vitro. This was not due to a lack of SUMO conjugation capacity in the mixture because adding extra SUMO-2 instead of SUMO-1 resulted in increased SUMO-2 polymer formation (Fig. 2B, lane 6). Note that the SUMO-1 antibody cross-reacted to a small extent with the relatively large amounts of SUMO-2 that were used in this assay.
Next we performed in vitro sumoylation assays that contained equal amounts of SUMO-1 and SUMO-2. We digested the entire reaction mixture with endopeptidase Lys-C and trypsin and analyzed the resulting peptides by LC-MS/MS on hybrid linear ion trap-Fourier transform mass spectrometers (LTQ-FT-ICR and LTQ-Orbitrap). The complexity of the peptide mixture is depicted in Fig. 2C. The most abundant peptides were isolated in the instrument and fragmented by CID, and the fragment ions were acquired in the LTQ when LTQ-FT-ICR was used and in the orbitrap when the LTQ-Orbitrap was used. In the case of the LTQ-FT-ICR mass spectrometer, the most intense product ions in the MS/MS scans were further fragmented and analyzed in the LTQ (MS3) to confirm the identity of the peptide (data not shown). Both instruments have very high resolution for MS spectra, and the MS/MS analysis in the orbitrap also results in very high accuracy fragmentation data albeit at the cost of using more ions. The accurate m/z values of the SUMO-1/SUMO-2 and SUMO-2/SUMO-2 branched precursor peptides were calculated and used to search for the corresponding ions. To compare the resolution of the two instruments we plotted the chromatographic regions of the 4+-charged SUMO-1/SUMO-2 precursor ions (Fig. 2, D and E) and directly superimposed the MS spectra (Fig. 2F). Because under given conditions for these large peptides the LTQ-Orbitrap performed better in terms of resolution and mass accuracy, we decided to use this instrument for subsequent experiments.
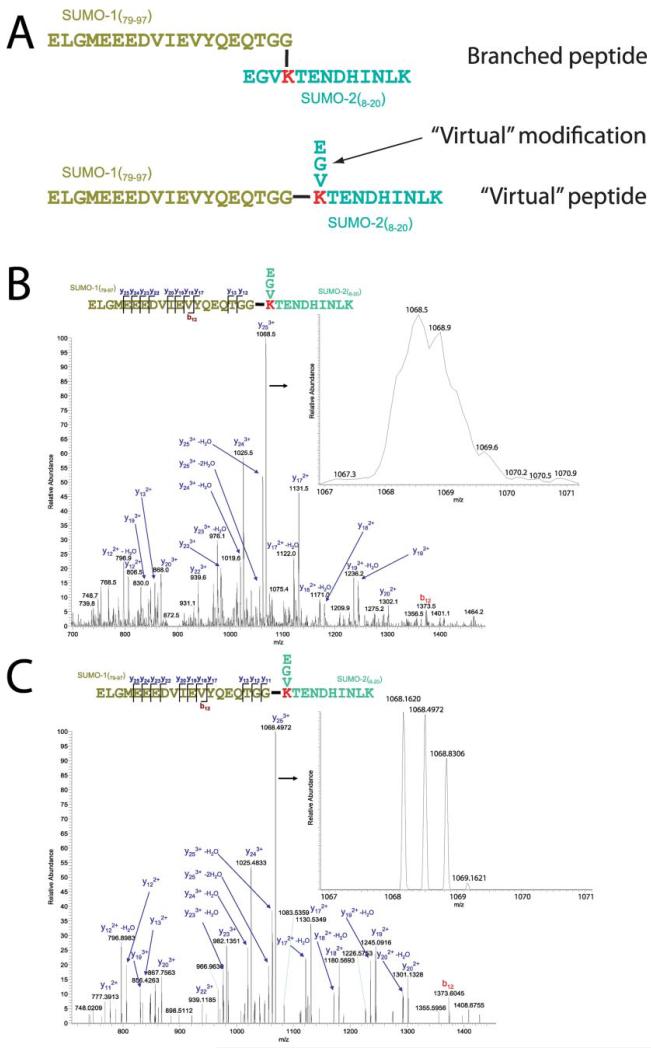
To interpret the MS/MS spectra, we needed to calculate the m/z of the fragment ions; this is not trivial for cross-linked peptides because they can have up to four different fragmentation series (for an introduction to peptide sequencing see Ref. 42). We noticed that only a few peaks could be assigned if the modifying peptides of SUMO-1 and SUMO-2 were considered as indivisible modifications. Constructing a “virtual peptide” consisting of the entire modifying peptide joined with the carboxyl terminus of the modified peptide (Fig. 3A) allowed the assignment of the majority of peaks (except the ones arising from the three amino-terminal amino acids of the modified SUMO-2 peptide) and the identification of Lys-11 in SUMO-2 as the major internal SUMO acceptor site (Fig. 3, B and C, and supplemental Fig. S3). The isopeptide bond is chemically identical to the peptide bond, therefore no mass difference between the virtual and “real” peptides is observed. It is important to note that almost all fragment ions are composed of amino acids derived from both the modifying and modified peptides; this explains why it is not possible to interpret the MS spectra by separately fragmenting the two parts of the branched peptides in silico. By using our targeted approach we also found that Lys-5 of SUMO-2 can be modified by both SUMO-1 and SUMO-2 in vitro, but we did not detect the corresponding peptides in vivo (supplemental Figs. S1 and S2) (43).
Fig. 3. The SUMO-1/SUMO-2 branched peptide has a unique fragmentation pattern.
Mapping sumoylation sites by mass spectrometry is challenging due to the complex MS/MS spectra that are generated. The interpretation of MS/MS spectra resulting from the fragmentation of the modified peptide is possible if the cross-linked peptide is “reversed” (A). The virtual peptide consists of the entire modifying peptide joined with the carboxyl terminus of the modified peptide. B and C, SUMO-1 is conjugated to lysine 11 of SUMO-2 in vitro. MS/MS fragmentation spectrum of a tryptic peptide consisting of aa 79–97 of SUMO-1 and 8–20 of SUMO-2 analyzed in the LTQ (B) or analyzed in the orbitrap (C). B, precursor ion mass was measured in the FT-ICR analyzer (m/z 908.9266 (4+); mass deviation, −1.76 ppm), and the peptide was fragmented and acquired in the LTQ mass spectrometer. C, precursor ion mass was measured in the orbitrap mass spectrometer (m/z 908.9282 (4+); mass deviation, 0.03 ppm), and the peptide was fragmented in the LTQ and analyzed in the orbitrap. The insets are magnifications of the most abundant fragment ion at m/z 1068.5 (3+). Note the much higher resolution and the isotope spacing of the orbitrap (C) compared with the LTQ (B). The low resolution of the LTQ does not allow an unambiguous assignment of the charge state.
Peptide identification is unambiguous in these experiments for four reasons. (i) The enzymatic digestion of the in vitro sumoylation reaction produces a simple mixture of peptides (Fig. 2C) that decreases the possibility that two peptides share the same m/z value. (ii) Precursor and fragment ions analyzed on the orbitrap have very high mass accuracy in the low or sub-ppm range. (iii) No SUMO-1/SUMO-2 or SUMO-2/SUMO-2 branched peptides were detected when only SUMO-1 was used in the in vitro reaction. The SUMO-2/SUMO-2 ion, but not the SUMO-1/SUMO-2 ion, was detected if we used only SUMO-2 in the mixture (supplemental Fig. S4). (iv) Unique characteristics of SUMO branched peptides, such as high charge state and complex fragmentation patterns, are not shared by non-sumoylated peptides.
SUMO Chain Formation in Nuclear Extracts
To study SUMO chain formation under more physiologically relevant conditions, an assay was developed that utilizes HeLa nuclear extracts as a source of E1, E2, and E3 enzymes, SUMO target proteins, and SUMO proteases to allow dynamic sumoylation and desumoylation cycles. HeLa nuclear extracts contain significant amounts of SUMO-2/3 but hardly any detectable amounts of SUMO-1 (data not shown). Adding ATP to this reaction mixture is sufficient to stimulate the conjugation of GST-SUMO-1 to target proteins. GST-SUMO-1 conjugates were purified from the reaction mixture and analyzed by silver staining (Fig. 4A) and by immunoblotting using an antibody that detects SUMO-1 (Fig. 4B). Many different high molecular weight SUMO-1 conjugates were detected in the GST-SUMO-1-purified fraction. Interestingly immunoblotting results indicated the presence of SUMO-2 and/or SUMO-3 in the GST-SUMO-1-enriched fraction (Fig. 4C). The most prominent band detected by this antibody suggested that SUMO-2 and/or SUMO-3 are direct targets for GST-SUMO-1. Note also that the SUMO-2/3 antibody detected the relatively large amount of monomeric GST-SUMO-1 due to apparent crossreactivity, which was rather surprising because recombinant SUMO-1 and control GST were not detected by the antibody (44). A potential explanation could be that the antibody also recognizes an epitope that consists of a carboxyl-terminal fragment of GST and an amino-terminal fragment of SUMO-1.
To investigate SUMO-SUMO conjugates present under these conditions, the 50-kDa band recognized by both anti-bodies was excised from the silver-stained gel (Fig. 4A, arrow), digested with trypsin, and analyzed by mass spectrometry. The MS/MS spectrum that was obtained showed conjugation of GST-SUMO-1 to the internal sumoylation site of endogenous SUMO-2 (supplemental Fig. S5). In addition purified GST-SUMO-1 conjugates were digested in solution with endopeptidase Lys-C and trypsin and analyzed by mass spectrometry. The mass spectrometric analysis of the insolution digestion showed that Lys-11 of endogenous SUMO-3 can be modified by SUMO-1 (Fig. 4D), whereas the SUMO-1/SUMO-2 peptide was not selected for sequencing by the software.
SUMO Chains Purified from Cells
Next we addressed whether SUMO chain formation occurs in vivo in cultured mammalian cells. Due to the activity of SUMO proteases and the lack of specific inhibitors of these proteases, SUMO conjugates are best preserved in denaturing buffers. To purify SUMO conjugates, we made use of our previously published stable cell line that expresses low levels of His6-SUMO-2 (34). Nuclei were prepared from this stable cell line, and His6-SUMO-2 conjugates were purified by immobilized metal affinity chromatography. The purified fraction was size-separated on a gradient gel, transferred to a membrane, and probed with antibodies to detect SUMO-2/3 and SUMO-1. SUMO-1 was detected in the His6-SUMO-2-enriched fraction (Fig. 5A, lane 4). The SUMO-1 signal in lane 4 was not due to cross-reactivity of the antibody with SUMO-2 because monomeric SUMO-2 was not detected by the SUMO-1 antibody.
Fig. 5. SUMO polymers detected in extracts from HeLa cells.
A, SUMO-2 conjugates were purified from nuclei of HeLaHis6-SUMO-2 cells, separated by SDS-PAGE, transferred to membranes, and probed using antibody AV-SM23-0100 to detect SUMO-2/3 (lanes 1 and 2) or 21C7 to detect SUMO-1 (lanes 3 and 4). B-E, purified His6-SUMO-2 conjugates were digested in solution by endopeptidase Lys-C and trypsin and analyzed in the LTQ-Orbitrap. B, the LC-MS/MS analysis is represented two-dimensionally, and the region that contains the SUMO-1/SUMO-2 branched peptide is indicated by an arrow. Isotopic distributions of the SUMO-1/SUMO-2 branched peptide (ellipse) analyzed by the full scan method (C) or by the selected ion monitoring method (D) were visualized by enlarging the corresponding regions. E, the MS spectra of the SUMO-1/SUMO-2 branched peptide from the analysis of the in vitro and in vivo sample were directly superimposed.
The purified fraction was digested in solution with endopeptidase Lys-C and trypsin, and SUMO-SUMO conjugates in the purified fraction were studied by mass spectrometry (Fig. 5, B-E, and supplemental Fig. S7). The complexity of the peptide mixture is depicted in Fig. 5B. The intensity of the SUMO-1/SUMO-2 precursor ion was very low, almost indistinguishable from background (Fig. 5C), and as a result was not automatically selected for sequencing. However, the very low mass deviation of 0.8 ppm, agreement in retention time, and complete superimposition of the precursor isotopic distribution of this sample and of the in vitro sumoylation reaction indicated that the ion signal was derived from the SUMO-1/SUMO-2 peptide. To unequivocally confirm this finding we used a peptide-specific method based on the information obtained from the in vitro experiment (Fig. 1B and “Experimental Procedures”). Only a 10-Da range around the ion of interest was monitored in the MS mode (selected ion monitoring), dynamic exclusion of sequenced ions was disabled, and the branched peptide was repetitively fragmented and sequenced. As expected, the intensity of the precursor isotope cluster was much higher compared with the full scan detection (Fig. 5, C and D). The MS/MS spectrum confirmed the presence of the SUMO-1/SUMO-2 branched peptide in the purified fraction (supplemental Fig. S7). Note the striking similarity in MS/MS patterns in Fig. 3, B and C, and supplemental Fig. S7. Furthermore we obtained MS/MS data that confirmed the presence of SUMO-2/SUMO-2 and SUMO-2/SUMO-3 polymers (Fig. 6, A and B). Thus, we conclude that SUMOs form polymers in cultured mammalian cells.
Fig. 6. SUMO polymers detected in extracts from HeLa cells.
Purified His6-SUMO-2 conjugates were digested in solution by endopeptidase Lys-C and trypsin and analyzed in the LTQ-Orbitrap. A, SUMO-2 is conjugated to lysine 11 of another molecule of SUMO-2. Shown is the MS/MS fragmentation spectrum of a tryptic peptide consisting of aa 59–92 of SUMO-2 and aa 8–20 of another molecule of SUMO-2. Precursor ion mass was measured in the orbitrap mass spectrometer (m/z 1070.7000 (5+); mass deviation, −1.03 ppm), and the peptide was fragmented and acquired in the LTQ mass spectrometer. B, SUMO-2 is conjugated to lysine 11 of SUMO-3. Shown is the MS/MS fragmentation spectrum of a tryptic peptide consisting of aa 59–92 of SUMO-2 and aa 8–21 of SUMO-3. Precursor ion mass was measured in the orbitrap mass spectrometer (m/z 1093.5092 (5+); mass deviation, −0.45 ppm), and the peptide was fragmented and acquired in the LTQ mass spectrometer.
Using the same narrow mass range approach we were also able to obtain a high quality fragmentation spectrum of the SUMO-1/SUMO-2 branched peptide in the GST-SUMO-1 sample digested in solution (supplemental Fig. S6). Note that this peptide was not sequenced when a full range acquisition method was used.
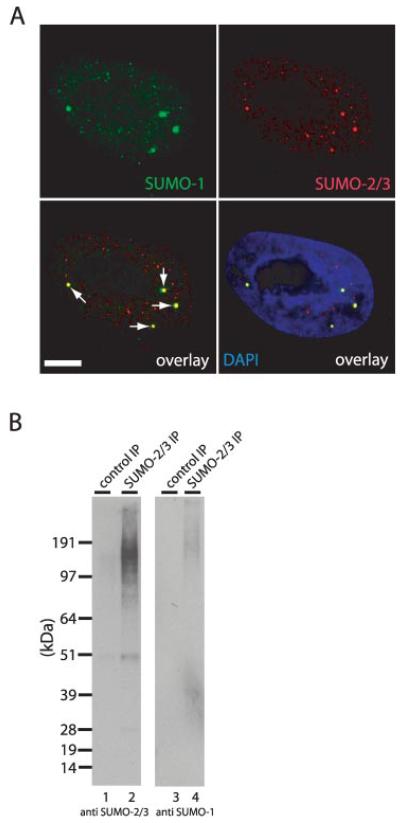
Subsequently endogenous SUMOs were studied. First, the subcellular localization of SUMO-1 and SUMO-2/3 was determined by immunostaining (Fig. 7A). SUMOs are nuclear proteins that are present throughout the nucleoplasm and also accumulate in nuclear bodies (34, 45, 46). These nuclear bodies are the most prominent sites of colocalization of SUMO-1 and SUMO-2/3 and therefore could be sites where mixed SUMO chains are present in cells.
Fig. 7. Endogenous SUMO polymers detected in extracts from HeLa cells.
A, SUMO-2/3 and SUMO-1 colocalize in nuclear bodies. HeLa cells were fixed, permeabilized, and stained with 4′,6-diamidino-2-phenylindole (DAPI) to detect DNA (blue). Immunofluorescence was performed to detect SUMO-1 (green) and SUMO-2/3 (red). Nuclear bodies are enriched for both SUMO-1 and SUMO-2 as indicated by the arrows. Scale bar, 5 μm. B, endogenous SUMO-2/3 conjugates were immunoprecipitated from total HeLa lysates, and a control immunoprecipitation (IP) was performed using preimmune serum. Proteins were separated by SDS-PAGE, transferred to membranes, and probed using antibody AV-SM23-0100 to detect SUMO-2/3 (lanes 1 and 2) or 21C7 to detect SUMO-1 (lanes 3 and 4).
Second, HeLa cell lysates were prepared in a denaturing buffer, and these lysates were subsequently diluted with a milder buffer to allow the immunoprecipitation of endogenous SUMO-2/3 conjugates. Endogenous SUMO-1 in this immunoprecipitate could be detected by immunoblotting (Fig. 7B, lane 4) in line with our previous observations (34). To investigate whether endogenous SUMOs can form chains, we digested the SUMO-2/3 immunoprecipitate and analyzed the resulting peptide mixture by mass spectrometry. The very low peptide signals in the mass spectrometric analysis confirmed that the efficiency of the immunoprecipitation was very low (data not shown). The precursor ion of the endogenous SUMO-2/SUMO-2 conjugate and/or SUMO-3/SUMO-2 conjugate (these precursor ions are indistinguishable; Fig. 1A) was detected and sequenced (supplemental Fig. S8) showing that endogenous SUMOs polymerize. We were not able to detect the SUMO-1/SUMO-2 peptide. This may be because this peptide is below the detection limit of our instrumentation and does not in itself prove that SUMO-1 cannot modify endogenous SUMO-2 in vivo.
HIF-1α Is Conjugated to SUMO Chains
Our data clearly show that SUMO chain formation occurs in cells. However, little is known about target proteins that can be conjugated to these SUMO chains, although it has been shown that HDAC4 is attached to a SUMO-2 dimer in cells (31). Nearly all the target proteins for SUMOs reported to date are detected in mono- or disumoylated forms. This could reflect either the limited sensitivity of the detection methods used and/or the relatively low abundance of the corresponding sumoylated proteins in cells. To identify target proteins that could be conjugated to SUMO chains, we performed immunoblotting experiments using antibodies directed against previously published SUMO targets. In this screen for proteins that are conjugated to a relatively large number of SUMO molecules, we found that the transcriptional regulator HIF-1α is conjugated to up to seven SUMO molecules in cells (Fig. 8A). HIF-1α contains three sumoylation consensus sites, although only two appear to be utilized, i.e. Lys-391 and Lys-477 (47).
Fig. 8. SUMO chain formation on HIF-1α.
A, His6-SUMO conjugates were purified from nuclei of cells stably expressing His6-SUMO-2 and from nuclei of control HeLa cells, separated by SDS-PAGE, transferred to a membrane, and probed with an antibody to detect HIF-1α. B, 5 μg of recombinant HIF-1α was conjugated in vitro to a mixture of 2 μg of SUMO-1 and 2 μg of SUMO-2. HIF-1α was subsequently purified from the reaction mixture on Talon beads in 8 m urea, and a small aliquot of the purified fraction and control HIF-1α were size-fractionated by SDS-PAGE, transferred to a membrane, and probed using anti-T7 antibody to detect HIF-1α. N.S., nonspecific.
To investigate the molecular composition of the SUMO chains attached to HIF-1α by mass spectrometry, an in vitro conjugation of recombinant HIF-1α to a mixture of SUMO-1 and SUMO-2 was performed, and after sumoylation, HIF-1α was purified from the reaction mixture. Sumoylation was confirmed by immunoblotting (Fig. 8B), and the remaining amount of protein was digested with trypsin and analyzed by mass spectrometry for the presence of SUMO/SUMO chimeric peptides. SUMO-1/SUMO-2 chimeric peptides (supplemental Fig. S9) and SUMO-2/SUMO-2 chimeric peptides (supplemental Fig. S10) were found by mass spectrometry. Thus, HIF-1α is conjugated to SUMO polymers in vitro. In addition, we confirmed the conjugation of SUMO-2 to Lys-391 of HIF-1α (supplemental Figs. S11 and S12) (47).
DISCUSSION
One of the key features of ubiquitin is its ability to form chains (18). Here we show that the ubiquitin family members SUMO-1, SUMO-2, and SUMO-3 are also able to form multimers in vitro and in vivo. SUMO molecules are linked via internal sumoylation sites present in SUMO-2 and SUMO-3. SUMO-1 can also be incorporated in these chains; however, the absence of an internal consensus sumoylation site in SUMO-1 appears to limit further elongation of the chains.
MS/MS has been successfully applied to the identification of many types of post-translational modifications because of its unique advantage in providing direct evidence of the modified peptides. However, until now very few peptides modified by mammalian SUMO have been identified because the lack of an Arg or a Lys in the proximity of the carboxyl terminus leads to the uninterpretability of fragment ions spectra in a straightforward way. This has limited progress in the field toward understanding the biological roles of this class of modification. Different approaches have been proposed such as mutational strategies (28, 30) to obtain a short indivisible sequence linked to the modified lysine after digestion or a pattern recognition tool (28, 29) to interpret the complex overlapping MS/MS spectra. The mutational method has the advantage that standard sequencing software can be used successfully, but the disadvantage of working with a SUMO variant is that the conjugation efficiency and other properties could differ from the wild type in cells. The pattern recognition software has been successful in detecting peptides modified by wild-type SUMO in vitro even when using relatively low accuracy mass spectrometers. The successful identification of the mammalian SUMO modification sites mapped in this study using mass spectrometry has been achieved by first analyzing proteins sumoylated in vitro. The low abundance of SUMO conjugates in cells and the complexity of the in vivo samples represents a further challenge. In our experience not more than 20% of peptides present in a complex mixture are sequenced in an LC-MS/MS experiment. The low intensity precursor ions generally are not fragmented when a standard sequencing method is used.
By taking advantage of the fact that SUMO-2 and SUMO-3 have a known internal consensus sumoylation site, we applied a targeted approach for the specific and direct detection of the SUMO modification at this site. The same can be done with other specific proteins especially if consensus sumoylation sites are present.
It has been shown that chain formation of the single SUMO family member in S. cerevisiae is limited due to the activity of the SUMO protease Ulp2 (26). It is likely that SUMO proteases also play a role in mammalian cells to limit steady state levels of SUMO chain length (48). In addition, we have shown here that the presence of three mammalian SUMO family members in principle allows an alternative mechanism to regulate SUMO chain length.
The most obvious way for forming SUMO chains would be the sequential addition of SUMO-1, SUMO-2, or SUMO-3 to a pre-existing SUMO-2 or SUMO-3 molecule on a target protein. It will be important to determine in the future whether unanchored SUMO chains are formed in vivo. In this regard, our in vitro data demonstrate that in principle the conjugation machinery can effectively link together unanchored SUMO molecules in the absence of other protein targets even in the absence of E3 factors. Interestingly SUMO chain formation in vitro was recently found to be enhanced by noncovalent interaction between Ubc9 and SUMO (43, 49).
Adding E3 ligases to in vitro sumoylation reactions has been reported previously to increase SUMO chain formation. A small fragment of RanBP2 was reported to be hypermodified by SUMO-1 chains in vitro (16). The SUMO-1 lysines 6, 16, 17, 37, 39, and 46 were all shown to function as acceptor sites for other SUMO-1 molecules in this case (29, 50). In addition, the small RanBP2 fragment is able to multimerize SUMO-2, a molecular event involving the internal sumoylation consensus site and two non-consensus sites (50). The yeast protein inhibitor of activated signal transducer and activator of transcription family member Siz1 was reported to enhance the multimerization of Smt3 (i.e. yeast SUMO) in an in vitro system in the presence and in the absence of septin target proteins (15). Most likely, E3 ligases will also play a role in SUMO polymerization in cells.
Ubiquitin chain formation is best known for its role in targeting conjugated proteins to the proteasome for degradation, a process that involves chain formation via Lys-48 or Lys-29 (18). Ubiquitin also forms chains via other internal lysines such as Lys-63, and for yeast ubiquitin, it has even been reported that chain formation can occur via all seven internal lysines (19). The functional relevance of SUMO chain formation remains to be determined. So far, there is little evidence for a role of sumoylation in protein degradation, although sumoylation has been linked to the degradation of DNA topoisomerase IIβ by a catalytic inhibitor (51), and SUMO modification of the promyelocytic leukemia proteinretinoic acid receptor α fusion protein is required for arsenic-induced, proteasome-dependent degradation (52).
Acknowledgments
We thank A. Groot for the HIF-1α expression construct. Juergen Cox is acknowledged for visualization of the MS data.
Footnotes
The abbreviations used are: SUMO, small ubiquitin-like modifier; E1, SUMO-activating enzyme; E2, SUMO protein carrier protein; E3, SUMO ligase; HIF, hypoxia-inducible factor; LTQ, linear quadrupole ion trap; LDS, lithium dodecyl sulfate; RanBP2, Ran-binding protein 2; SAE, SUMO-activating enzyme; Ubc9, ubiquitin-conjugating enzyme 9; aa, amino acids; HRP, horseradish peroxidase; GPMAW, General Protein/Mass Analysis for Windows.
This work was supported in part by the Netherlands Organisation for Scientific Research (NWO) (to A. C. O. V.) as part of the Innovational Research Incentives Scheme, by The European Community (RUBICON, VI Framework) (to M. M.), a grant from the Association for International Cancer Research (to R. T. H.), and by a Wellcome Trust Programme grant (to A. I. L.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental materials.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 4.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 5.Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi T, Seki M, Maeda D, Wang W, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T. Ubc9 is essential for viability of higher eukaryotic cells. Exp. Cell Res. 2002;280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 8.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones D, Crowe E, Stevens TA, Candido EP. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2001-3-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 11.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 12.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Xu P, Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim. Biophys. Acta. 2006;1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 15.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 16.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 17.Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 20.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 23.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 25.Kirkpatrick DS, Weldon SF, Tsaprailis G, Liebler DC, Gandolfi AJ. Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics. 2005;5:2104–2111. doi: 10.1002/pmic.200401089. [DOI] [PubMed] [Google Scholar]
- 26.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 27.Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomycescerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knuesel M, Cheung HT, Hamady M, Barthel KK, Liu X. A method of mapping protein sumoylation sites by mass spectrometry using a modified small ubiquitin-like modifier 1 (SUMO-1) and a computational program. Mol. Cell. Proteomics. 2005;4:1626–1636. doi: 10.1074/mcp.T500011-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Pedrioli PG, Raught B, Zhang XD, Rogers R, Aitchison J, Matunis M, Aebersold R. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat. Methods. 2006;3:533–539. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- 30.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., III Improved identification of SUMO attachment sites using C-terminal SUMO mutants and tailored protease digestion strategies. J. Proteome Res. 2006;5:761–770. doi: 10.1021/pr050451o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 32.Mohrmann L, Kal AJ, Verrijzer CP. Characterization of the extended Myb-like DNA-binding domain of trithorax group protein Zeste. J. Biol. Chem. 2002;277:47385–47392. doi: 10.1074/jbc.M202341200. [DOI] [PubMed] [Google Scholar]
- 33.Groot AJ, Verheesen P, Westerlaken EJ, Gort EH, van der Groep P, Bovenschen N, van der Wall E, van Diest PJ, Shvarts A. Identification by phage display of single-domain antibody fragments specific for the ODD domain in hypoxia-inducible factor 1α. Lab. Investig. 2006;86:345–356. doi: 10.1038/labinvest.3700395. [DOI] [PubMed] [Google Scholar]
- 34.Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 35.Jaffray EG, Hay RT. Detection of modification by ubiquitinlike proteins. Methods. 2006;38:35–38. doi: 10.1016/j.ymeth.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 36.Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Foster LJ, de Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 40.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 41.Trinkle-Mulcahy L, Andrews PD, Wickramasinghe S, Sleeman J, Prescott A, Lam YW, Lyon C, Swedlow JR, Lamond AI. Time-lapse imaging reveals dynamic relocalization of PP1γ throughout the mammalian cell cycle. Mol. Biol. Cell. 2003;14:107–117. doi: 10.1091/mbc.E02-07-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steen H, Mann M. The ABC’s (and XYZ’s) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 43.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ, Kim KW. Sumoylation increases HIF-1α stability and its transcriptional activity. Biochem. Biophys. Res. Commun. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- 48.Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J. Cell Biol. 2006;174:939–949. doi: 10.1083/jcb.200510103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper HJ, Tatham MH, Jaffray E, Heath JK, Lam TT, Marshall AG, Hay RT. Fourier transform ion cyclotron resonance mass spectrometry for the analysis of small ubiquitin-like modifier (SUMO) modification: identification of lysines in RanBP2 and SUMO targeted for modification during the E3 autoSUMOylation reaction. Anal. Chem. 2005;77:6310–6319. doi: 10.1021/ac058019d. [DOI] [PubMed] [Google Scholar]
- 51.Isik S, Sano K, Tsutsui K, Seki M, Enomoto T, Saitoh H, Tsutsui K. The SUMO pathway is required for selective degradation of DNA topoisomerase IIβ induced by a catalytic inhibitor ICRF-193(1) FEBS Lett. 2003;546:374–378. doi: 10.1016/s0014-5793(03)00637-9. [DOI] [PubMed] [Google Scholar]
- 52.Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor α degradation. J. Exp. Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]