A stable monopolar array of nuclear microtubules displaces the nucleolus and kinetochores during quiescence and is required for both quiescence survival and exit.
Abstract
The microtubule cytoskeleton is a highly dynamic network. In dividing cells, its complex architecture not only influences cell shape and movement but is also crucial for chromosome segregation. Curiously, nothing is known about the behavior of this cellular machinery in quiescent cells. Here we show that, upon quiescence entry, the Saccharomyces cerevisiae microtubule cytoskeleton is drastically remodeled. Indeed, while cytoplasmic microtubules vanish, the spindle pole body (SPB) assembles a long and stable monopolar array of nuclear microtubules that spans the entire nucleus. Consequently, the nucleolus is displaced. Kinetochores remain attached to microtubule tips but lose SPB clustering and distribute along the microtubule array, leading to a large reorganization of the nucleus. When cells exit quiescence, the nuclear microtubule array slowly depolymerizes and, by pulling attached centromeres back to the SPB, allows the recovery of a typical Rabl-like configuration. Finally, mutants that do not assemble a nuclear array of microtubules are impaired for both quiescence survival and exit.
Introduction
Most cells spend the majority of their life in quiescence, a condition defined as a temporary arrest of proliferation. This cellular state is poorly understood and it has been proposed that different quiescent states may be reached depending on the cell’s history (O’Farrell, 2011). Saccharomyces cerevisiae has been a valuable model organism for studying this complex resting state (Daignan-Fornier and Sagot, 2011a; Klosinska et al., 2011; De Virgilio, 2012). When yeast cells enter quiescence after carbon source exhaustion, they assemble particular structures such as actin bodies, proteasome storage granules, chaperone-containing granules, etc. (Sagot et al., 2006; Narayanaswamy et al., 2009; Noree et al., 2010; Liu et al., 2012; Shah et al., 2013), some of which appear to be conserved in other organisms (Jensen and Larsson, 2004; Laporte et al., 2008; Noree et al., 2010; Poulter et al., 2010). Although in most cases the physiological “raison d’être” of these structures remains to be determined, it is clear that they are not simply useless amorphous aggregates (Daignan-Fornier and Sagot, 2011b).
Microtubules (MTs) are essential asymmetric polymers involved in a wide variety of cellular processes (Wade, 2009). The MT “minus” end is poorly dynamic and generally associated with a MT-organizing center (MTOC) required for MT nucleation. By contrast, the MT “plus” end is highly dynamic and alternates between periods of growth and shortening, a behavior called dynamic instability (Mitchison and Kirschner, 1984). MT dynamics is tightly controlled by a vast collection of MT-associated proteins (MAPs) and can be modulated by tubulin post-translational modifications (Hammond et al., 2008; van der Vaart et al., 2009).
In mitosis, budding yeast cells display dynamic cytoplasmic MTs (astral MTs [aMTs]) that are critical for the correct positioning of the nucleus (Markus et al., 2012). Inside the nucleus, the mitotic spindle is composed of kinetochore MTs (kMTs) that capture centromeres and pull them apart during anaphase. This process is helped by non-kinetochore MTs that drive mitotic spindle elongation through the action of MT cross-linking proteins and associated motors (Winey and Bloom, 2012). MTs are nucleated by the γ-TuC complex found on the spindle pole body (SPB), the yeast equivalent of the centrosome (Pereira and Schiebel, 1997; Erlemann et al., 2012).
In interphase, the budding yeast nucleus adopts an organization that resembles the Rabl configuration first described in salamanders and commonly found in plants and Drosophila (Rabl, 1885; Hochstrasser et al., 1986; Jin et al., 1998, 2000; Bystricky et al., 2004; Schubert and Shaw, 2011). In this configuration, the nucleolus is located opposite to the SPB and most telomeres are maintained at the nuclear periphery (Zimmer and Fabre, 2011; Taddei and Gasser, 2012). Short kMTs (∼300 nm) link each centromere to the SPB, keeping them clustered together in a “rosette” (Guacci et al., 1997; Jin et al., 1998, 2000; O’Toole et al., 1999; Bystricky et al., 2004). This clustering is lost if kMT–centromere attachment is disrupted or if kMTs are destabilized by pharmacological treatment (Jin et al., 2000). Whether MT-dependent centromere clustering in interphase is important for the fidelity of chromosome segregation in the subsequent anaphase is an open question. Besides, it was shown that in proliferating S. cerevisiae, kMT–centromere attachments are lost only very briefly, just during the time required for centromere replication in S phase (Winey and O’Toole, 2001; Kitamura et al., 2007; Tanaka et al., 2010).
Here we show that upon quiescence entry, cells assemble a long and astonishingly stable array of nuclear MTs (nMTs) that spans the entire nucleus. This nMT array emanates from the SPB, its opposite extremity being associated with several MAPs and kinetochores. Importantly, centromeres also localize on the nMT array. Consequently in quiescence, the nucleus is drastically reorganized. Upon quiescence exit, nMTs disassemble, dragging with them attached centromeres. As a result, cells go back to a Rabl-like configuration and reenter the cell cycle without losing MT–centromere attachment. Finally, mutants that do not display nMT array are impaired for both survival in quiescence and quiescence exit.
Results and discussion
Microtubules form a nuclear array in quiescent S. cerevisiae
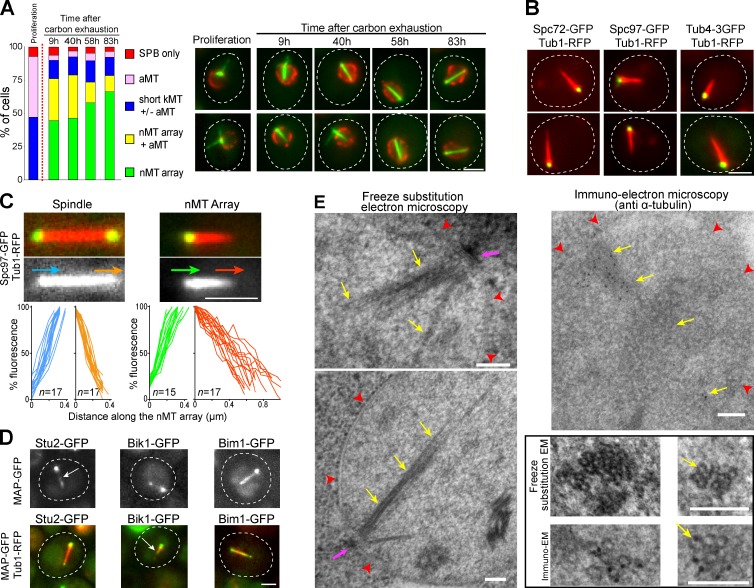
MT organization was analyzed in wild-type (WT) cells expressing GFP-Tub1. As shown in Fig. 1 A, proliferating G1 cells displayed 1–3 long cytoplasmic aMTs and short and hardly detectable nuclear kMTs. Strikingly, we observed that a few hours after carbon exhaustion, a thick array of nMTs was present in the quiescent cell’s nucleus. In parallel, as the time in quiescence increased, fewer aMTs were detected in the cytoplasm (Fig. 1 A, yellow bars). In most cells, the nMT array spanned the entire nucleus, sometimes even deforming the nuclear membrane (Fig. S1, A and B). Colocalization of Tub1-RFP with Spc72 or the γ-TuC components Tub4 and Spc97 showed that the nMT array was associated with the SPB (Fig. 1 B). Of note, the SPB did not seem to be drastically altered in quiescence (Fig. S1 C). Fluorescence line-scan slope calculation along the nMT array revealed that its extremities were unequal. Indeed, the SPB-associated end displayed a slope similar to the one measured for short mitotic spindles (Fig. 1 C). This suggested that the nMT array emanated from the SPB. By contrast, at the opposite extremity, slopes were less steep, indicating that within the nMT array MTs were heterogeneous in length (Fig. 1 C, red slopes). Next, we localized proteins known to be associated with the MT plus-end (Kumar and Wittmann, 2012). Stu2, a XMAP215 family member and Bik1, the CLIP170 homologue, localized to both extremities of the nMT array (Fig. 1 D). In some cells, few discrete dots were also detected along the nMT array (Fig. 1 D, arrows). This further supported the idea that within the array, not all the MTs were of the same length. Interestingly, Bim1, the EB1 homologue, was localized all along the nMT array (Fig. 1 D). As Bim1 specifically recognizes GTP-bound tubulin, it could be that some GTP-tubulin remnants may be present along nMTs, as shown for mammalian MTs (Dimitrov et al., 2008). Finally, nMT arrays emanating from the SPB were observed in quiescent cell nuclei by EM (Fig. 1 E and Fig. S1 D). Taken together, these experiments establish that upon quiescence entry, cells assemble an array of parallel MTs that originates from the SPB and traverses the entire nucleus.
Figure 1.
MTs are organized as a nuclear array in quiescent cells. (A) MT reorganization upon quiescence entry. Cells expressing GFP-Tub1 (green) and Nup2-RFP (red) are shown. Glucose exhaustion is indicated as a red dashed line. (B) Quiescent cells (4 d) expressing Tub1-RFP and either Spc72-GFP, Tub4-3GFP, or Spc97-GFP are shown. (C) The nMT array emerged from the SPB. Fluorescence line scan slopes along nMT arrays are shown. Extremities associated with the SPB are in green and extremities opposite to the SPB in red. For comparison, slopes for mitotic spindle extremities are shown (blue and orange). Arrows indicate line-scan directions. N = 2, n is indicated. (D) Quiescent cells (4 d) expressing Stu2-GFP, Bik1-GFP, or Bim1-GFP alone (top) or coexpressed with Tub1-RFP (bottom). Arrows point at dot along the nMT array. Bars: (A–D) 2 µm. (E) nMT array visualized in WT cells by EM. Yellow arrows, MTs; red arrowheads, nuclear membrane; pink arrow, SPB. Insets: nMT array cut transversally. Bar, 100 nm.
The nMT array is composed of stable MTs
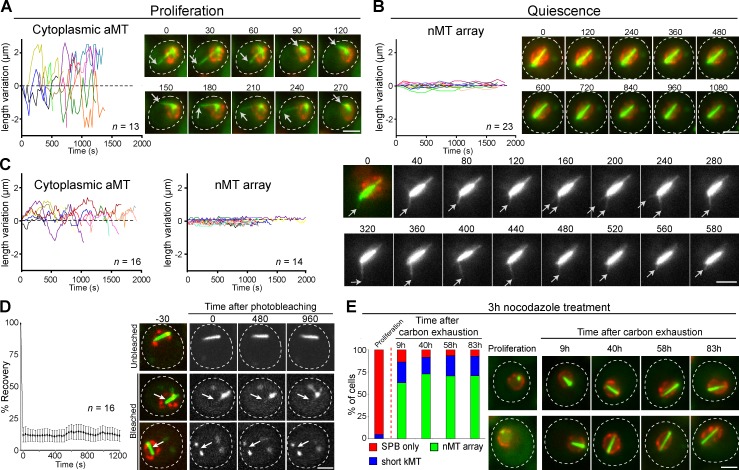
As previously shown (Carminati and Stearns, 1997), we observed highly dynamic MTs in proliferating G1 cells (Fig. 2 A and Fig. S2 A). By comparison, in 7-d-old quiescent cells the nMT array length remained remarkably constant (Fig. 2 B). This behavior was confirmed for cells in early quiescence (4 d), in which both aMTs and nMT array can be detected simultaneously (Fig. 2 C). MT stabilization was verified in quiescent cells expressing various nMT-associated MAPs (Fig. S2 B). FRAP experiments further showed that no significant GFP-Tub1 turnover could be detected within the nMT array (Fig. 2 D). Lastly, drugs that destabilize dynamic MTs had no effect on quiescent cells nMT array, whereas in proliferating G1 cells dynamic MTs were rapidly lost. Cytoplasmic aMT disappearance attested that drugs entered quiescent cells (compare Fig. 1 A with 2 E; and see Fig. S2, D and E). Importantly, drugs had no effect on nMT arrays whatever the timing of their addition after carbon exhaustion (Fig. 2 E), suggesting that MT stability was not acquired with the time spent in quiescence but was a result of quiescence entry.
Figure 2.
The nuclear array is composed of stable MTs. Variation of the length in function of time of (A) aMTs in proliferating cells, (B) nMT array in quiescent cells (7 d), and (C) the two MT structures in early quiescent cells (4 d). White arrows point at dynamic aMTs. In A–C, N = 2, n is indicated. (D) FRAP on 7-d-old quiescent cells. White arrows point to bleach areas. Error bars are SEM. In A–D, time is in seconds. (E) nMT array is not affected by nocodazole treatment. Before and various times after carbon exhaustion (red dashed line), cells were incubated for 3 h with 22.5 µg/µL nocodazole and then imaged. The corresponding control experiment is shown in Fig 1 A. In all panels, WT cells expressing GFP-Tub1 (green) and Nup2-RFP (red) are shown; bars, 2 µm.
In quiescence, the nucleus is drastically reorganized
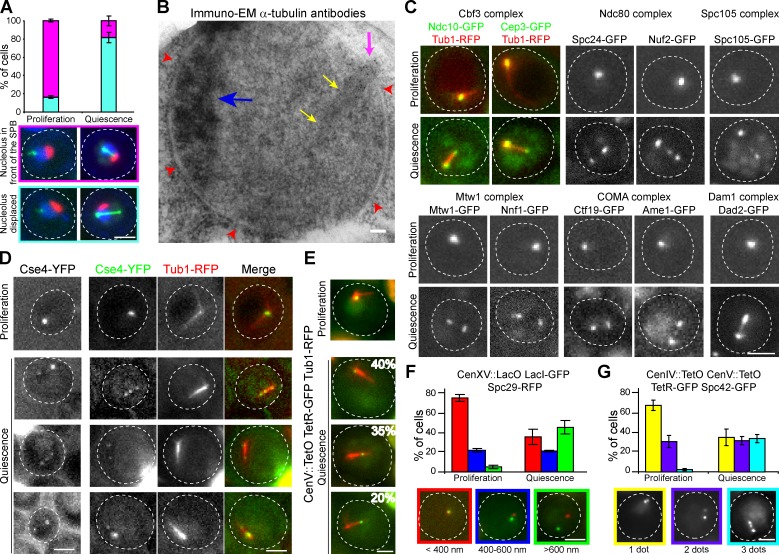
In proliferating S. cerevisiae, the nucleolus is localized opposite to the SPB (Hernandez-Verdun et al., 2010). In quiescent cells, we found that the nucleolus (Sik1-RFP) was no longer localized opposite to the SPB, but rather on the side of the nMT array (Fig. 3 A). This was confirmed by EM experiments (Fig. 3 B and Fig. S3 A). Of note, the nucleolus was very small, which was expected because in quiescence translation is drastically reduced (Fuge et al., 1994).
Figure 3.
Nucleus reorganization in quiescent cells. (A and B) The nucleolus is displaced in quiescence. (A) Relative arrangements of the nucleolus (Sik1-RFP) and the nMT array (Bim1-3GFP) in proliferating G1 and quiescent cells (4 d). (B) WT quiescent cell nucleus (7 d) visualized by immuno-EM. The electron-dense zone is the nucleolus (blue arrow), arrows are as in Fig 1 E. Bar, 100 nm. (C) Kinetochore subcomplexes relocalize in quiescent cells (4 d). Corresponding G1 proliferating cells are shown as control. (D–G) Centromeres are relocalized in quiescent cells. (D) Proliferating G1 and quiescent cells (4 d) expressing Cse4-mYFP alone (left) or in combination with Tub1-RFP (right). (E) CEN V localization in proliferating G1 and quiescent diploid cells (4 d) expressing Tub1-RFP. Percentages of cells displaying typical centromere localization are indicated. (F) Distances between CEN XV and the SBP (Spc29-RFP). (G) Centromeres can be found at different positions along the nMT array. WT cells in which TetO sequences were integrated at both CEN IV and CEN V and expressing Spc42-GFP and TetR-GFP were scored. In F and G, measurements were done in proliferating G1 cells and in 3-d-old quiescent cells. Bars: (A, and C–G) 2 µm.
We then analyzed the localization of kinetochore proteins. Interestingly, among the six kinetochore subcomplexes (Westermann et al., 2007; Gascoigne and Cheeseman, 2011), five were found localized both at the SPB and at the tip of the nMT array (Fig. 3 C and Fig. S3 B). In some cells, immobile dots of kinetochore proteins could also be detected along the nMT array (Fig. S2 C). As these dots were insensitive to MT-destabilizing drugs (see Fig. 5 D), we conjectured that they correspond to the plus end of shorter stable MTs. The sixth kinetochore subcomplex, the Dam1 complex, form rings along MTs in vitro (Miranda et al., 2005; Westermann et al., 2006; Joglekar et al., 2010; Ramey et al., 2011). In quiescent cells, it was detected all along the nMT array (Fig. 3 C and Fig. S3 B).
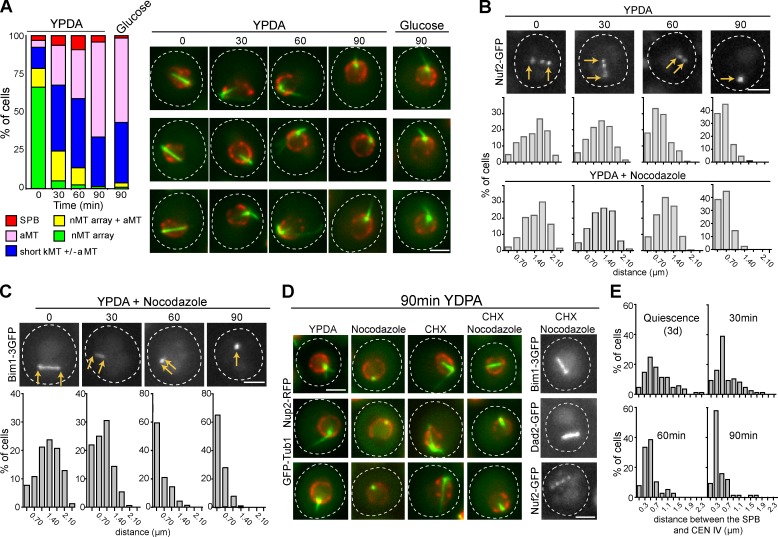
Figure 5.
nMT array slowly shrinks upon quiescence exit. (A) MT organizations before and after quiescent cells re-feeding with YPDA or with 2% of glucose. WT cells expressing GFP-Tub1 (green) and Nup2-RFP (red) are shown. (B) The nMT array slowly shrinks upon quiescence exit. The distance between the Nuf2-GFP SPB-associated dot and the nMT array Nuf2-GFP tip-associated dot was measured upon cell re-feeding with or without nocodazole (22.5 µg/µL). The distribution of the distances measured in two experiments is shown (top: n = 217 for t0, 313 for t30, 217 for t60, and 166 for t90; bottom: n = 290 for t0, 286 for t30, 225 for t60, and 76 for t90). (C) Bim1-3GFP rapidly dissociated from the nMT array upon quiescence exit in the presence of 22.5 µg/µL nocodazole. The distribution of the distances measured in two experiments is shown (n = 213 for t0, 291 for t30, 138 for t60, and 99 for t90). (D) The nMT array disassembly is slowed down in the presence of CHX. Quiescent cells (4 d) were re-fed with YPDA in the presence of the indicated drug(s) and imaged after 90 min. Bars: (A–D) 2 µm. (E) Variation of the distance between the SPB (Spc29-RFP) and CEN IV upon cell re-feeding. The distribution of the distances measured in two experiments is shown (n = 89 for t0, 88 for t30, 110 for t60, and 76 for t90). In all panels, time after re-feeding is indicated in min.
The fact that all kinetochore subcomplexes were associated with the nMT array prompted us to look at centromeres. First, we localized the CENP-A homologue Cse4, a histone-like exclusively found at centromeres (Meluh et al., 1998; Verdaasdonk and Bloom, 2011). In quiescent cells, Cse4-YFP was detected close to the SPB and at the tip of the nMT array (Fig. 3 D). Then, we localized DNA proximal to the centromere using integrated TetO arrays. In quiescent cells, DNA proximal to chromosome V centromere (hereafter refer as CEN V) was detected either at the very tip of the nMT array, along the nMT array, or close to the SPB (Fig. 3 E). The same pattern of localization was observed for CEN IV (Fig. S3 C). This was consistent with the distances measured between SPB and CEN XV (Fig. 3 F) or CEN IV (Fig. S3 D). Finally, we simultaneously localized the SPB and both CEN IV and CEN V. In one third of the population, three distinct dots could be detected (Fig. 3 G), indicating that not all centromeres are colocalized but could be found at different positions along the nMT array.
In conclusion, in quiescent cells, the overall architecture of the nucleus is drastically modified: the nucleolus is displaced and the kinetochores/centromeres, because they are still associated with the nMT plus-end, are no longer found all clustered near the SPB as in G1 cells, but rather localize along or at the tip of the nMT array (Table S1). The fact that centromeres remained attached to MT extremities while MT elongate to form the nMT array gives a mechanistic explanation for the pioneer observation of the reduced centromeres clustering at SPBs in the nucleus of early quiescent cells (Jin et al., 1998).
Several MAPs are required for nMT array formation
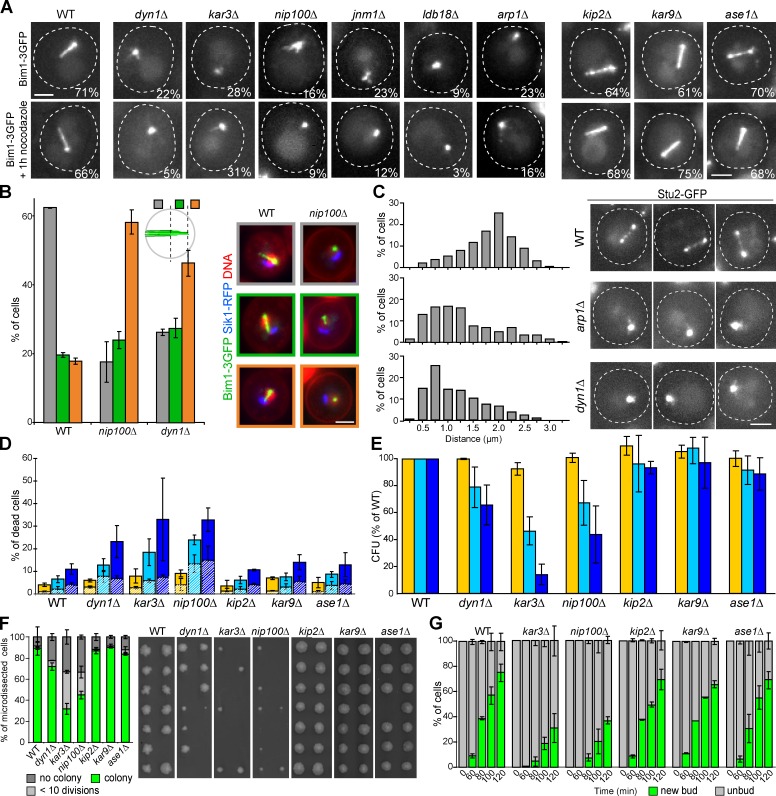
Next, we searched for MAPs that would be required for quiescent cells nMT array formation and/or stability. Deletion of various MAP-encoding genes had no effect on quiescent cell nMT array (Fig. 4 A and Fig. S3, E and F). By contrast, nMT array formation was strongly impaired in cells deleted for kar3, dynein (dyn1), or subunits of the dynactin complex (jnm1, nip100, arp1, and ldb18; Fig. 4 A and Fig. S3 F). In quiescent cells, Kar3 could not be detected, and dynein and dynactin were only detected on the cytoplasmic face of the SPB (Fig. S3 H), but we cannot rule out that a pool of these proteins may be present inside the nucleus. Importantly, in the absence of nMT array, the nucleolus was no longer displaced (Fig. 4 B) and both Stu2 and kinetochores were mainly found close to the SPB (Fig. 4 C and Fig. S3 G). These results suggested that in quiescent cells, the nMT array formation caused the observed nuclear reorganization.
Figure 4.
Kar3, dynein, and dynactin mutants are involved in nMT array formation and impaired for both quiescence survival and exit. (A) nMT formation was tested in 4-d-old cells expressing Bim1-3GFP and deleted for the indicated MAP. The nMT array stability was tested by nocodazole treatment (22.5 µg/µL). Numbers indicate the percentage of cells with nMT array. Of note, in dyn1Δ, kar3Δ, nip100Δ, jnm1Δ, ldb18Δ, and arp1Δ, most of the nMT arrays scored were shorter and thinner than in WT. (B) In the absence of nMT array, the nucleolus mainly localizes opposite to the SPB. Nucleus organization was scored as indicated: nucleolus localizing opposite to SPB (orange), in the nucleus half opposite to the SPB (green), or close to the SPB (gray). Cells expressing Bim1-3GFP (green) and Sik1-RFP (blue) stained with Hoechst (red) are shown. (C) In the absence of nMT array, Stu2-GFP localizes at SPB proximity. Distances were measured between the SPB and the Stu2-GFP dot. The distribution of the distances measured in two experiments is shown (WT, n = 241; arp1Δ, n = 274; dyn1Δ, n = 217). Bars: (A–C) 2 µm. (D) Mutants that cannot assemble nMT array have a reduced viability in quiescence. Cell viability was scored after 2 (yellow), 7 (light blue), and 14 d (dark blue) using methylene blue. Plain bars, unbudded dead cells; dashed bars, budded dead cells. (E) Colony-forming capacity measured for cells grown for 2 (yellow), 7 (light blue), or 14 d (dark blue). Percentages are given using WT cells as reference. (F) 12-d-old alive cells were micro-manipulated and tested for their ability to give rise to a colony. (G) Kinetics of new bud emergence upon quiescent exit.
Cells unable to assemble nMT array are impaired for both survival in quiescence and quiescence exit
To investigate the physiological function of the nMT array, we analyzed the phenotypes of mutants impaired for the assembly of this structure in quiescence. We found that a significant percentage of kar3Δ, dyn1Δ, or nip100Δ cells were dead after 7 or 14 d in culture (Fig. 4 D). Kar3, dynein, and dynactin are involved in spindle orientation during mitosis. Defects in this process cause the accumulation of budded cells with misoriented spindle or bi-nucleated cells. Consequently, activation of the spindle position checkpoint (SPOC) delays exit from mitosis (Caydasi et al., 2010; Winey and Bloom, 2012). We first hypothesized that in kar3Δ, dyn1Δ, or nip100Δ cells, defects in spindle orientation were causing SPOC activation, thus preventing quiescence establishment. However, we found that the majority of the kar3Δ, dyn1Δ, or nip100Δ dead cells were not budded cells (Fig. 4 D), and in quiescence the number of bi-nucleated cells did not suffice to account for the number of dead cells (Fig. S3 I). Moreover, kar3Δ, dyn1Δ, or nip100Δ cells displayed actin bodies (Fig. S3 J), structures specifically assembled upon quiescence entry (Sagot et al., 2006). Furthermore, activation of SPOC by other routes (deletion of KIP2, KAR9, or ASE1; Winey and Bloom, 2012) did not prevent nMT formation and had no drastic effect on cell capacity to face quiescence (Fig. 4, A, D, and E). Importantly, in the absence of nMT array, the number of dead cells increased with the time spent in quiescence (Fig. 4 D). In good agreement, kar3Δ, dyn1Δ, or nip100Δ cells that have just entered quiescence were capable of giving rise to a progeny, whereas this ability decreased with the time spent in quiescence (Fig. 4 E). This was also observed for the other dynactin mutants (Fig. S3 K). Taken together, these experiments indicate that kar3Δ, dynein, and dynactin mutant cells do enter quiescence but fail to form an nMT array. This correlates with a progressive loss of viability. Of note, dynactin is involved in a checkpoint that monitors cell wall integrity (Suzuki et al., 2004), yet, providing osmotic support by sorbitol addition did not improve dynactin mutant survivability in quiescence (Fig. S3 L).
Interestingly, in kar3Δ, dyn1Δ, or nip100Δ mutants we found a discrepancy between the number of cells alive in quiescence and those capable of giving rise to a colony upon quiescence exit (Fig. 4, compare D with E). Micro-manipulation of kar3Δ, dyn1Δ, or nip100Δ cells demonstrated that a significant percentage of cells still alive after 12 d of culture were unable to give rise to a healthy progeny (Fig. 4 F). Furthermore, among kar3Δ or nip100Δ cells that were capable of exiting quiescence, bud emergence was greatly delayed (Fig. 4 G). In conclusion, our results establish that several mutants that do not assemble an nMT array are impaired not only for survival during quiescence but also for quiescence exit.
nMT array slowly shortens upon quiescence exit
We next investigated the MT behavior upon quiescence exit. When cells were re-fed, aMT rapidly reappeared in the cytoplasm while nMT arrays slowly shortened. After 60–90 min, a typical G1 cell MT organization was recovered (Fig. 5 A). In fact, a sole addition of glucose was sufficient to trigger nMT array disassembly (Fig. 5 A). This suggested that carbon source was the signal for MT remodeling upon quiescence exit, as previously shown for other quiescence-specific structures (Laporte et al., 2011). We can envision that upon quiescence exit, the nMT array disassembly is needed to regenerate the pool of free tubulin that will be required for the mitotic spindle formation, just as it was recently proposed for each subsequent round of mitoses during proliferation (Woodruff et al., 2012). Localization of kinetochore proteins indicated the nMT array shortening was progressive and not influenced by MT-destabilizing drugs (Fig. 5 B). By contrast, Bim1-GFP was quickly lost from the nMT array (Fig. 5 C), suggesting that a likely GTP-tubulin cap was rapidly dismantled upon quiescent exit. Furthermore, efficient nMT array disassembly required de novo synthesis of proteins, as it was significantly slowed down by cycloheximide (CHX; Fig. 5 D). Interestingly, in the presence of CHX, Bim1, Dad2, and Nuf2 remained on the nMT array (Fig. 5 D), indicating that their displacement was driven by neo-synthesized proteins. Finally, upon cell re-feeding, the distance between the SBP and CEN IV decreased with a similar kinetic to the nMT array shortening (Fig. 5 E), strongly suggesting that centromeres stayed attached to the nMT array while it depolymerized.
Altogether, our findings establish that quiescent cells assemble a monopolar spindle-like structure that emanates from the SPB. This array is composed of highly stable nMTs, yet the precise molecular mechanism responsible for MT stabilization remains to be discovered. As suggested by quiescence exit experiments in the presence of CHX, one hypothesis could be that MT-destabilizing proteins are no longer present in quiescent cells. Conversely, but not exclusively, MT stabilization in the nucleus could be enhanced by the activity of one or several MAPs. The physiological function of the nMT array is still mysterious, yet it drives a drastic reshaping of the nucleus. This raises the intriguing possibility that an nMT-driven nuclear reorganization is contributing to cell survival both during quiescence and reentry into the proliferation cycle.
Materials and methods
Yeast strains and growth conditions
Integrative plasmids pTUB1-CFP-TUB1 (HIS3) or pTUB1-GFP-TUB1 (LEU2) are derivatives of PB1002 and were a gift of D. Pellman (Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA; Tirnauer et al., 1999). The RFP (tdimer 2(12); Campbell et al., 2002) sequence carried by plasmids p3695 or p4143 (pRS306 backbone, URA3) was integrated at the 3′ end of the NUP2 and TUB1 endogenous loci, respectively. Three tandem copies of GFP sequences carried by plasmid p4587 (pRS306 backbone, URA3) and p4453 (pRS305 backbone, LEU2) were integrated at the 3′ end of the BIM1 and the TUB4 endogenous loci, respectively. The plasmid p2568 (pRS304 backbone, TRP1) was used to integrate a GFP sequence at the 3′ end of the SPC42 endogenous locus and was a gift of P. Sorger (Harvard Medical School, Boston, MA; He et al., 2000). Three tandem copies of GFP or YPF sequences carried by plasmids p4754 (pRS303 backbone, HIS3) and p4753 (pRS305 backbone, LEU2), respectively, were integrated at the 5′ of Dyn1 chromosomal locus and were a gift of D. Pellman (Sheeman et al., 2003). Three tandem copies of GFP sequences carried by plasmid p4756 (pRS305 backbone, LEU2) were integrated at the 5′ of the Arp1 chromosomal locus. The functionality of Arp1-3GFP was tested by crossing Arp1-3GFP–expressing cells with a strain deleted for kar9. RFP (tdimer(2)12) sequence carried by plasmid p3696 (pRS306 backbone, URA3) was integrated at the 5′ of the Htb1 chromosomal locus. Details of the plasmid constructions are available upon request.
All the S. cerevisiae strains used in this study are isogenic to BY4741, BY4742, or BY4743 available from Euroscarf. Yeast strains carrying GFP fusions were obtained from Invitrogen. Strain KBY 8065 (Fig. 3 F) was a gift of K. Bloom (University of North Carolina at Chapel Hill, Chapel Hill, NC; Stephens et al., 2011). Strain JW2687, a gift of J-Q. Wu (The Ohio State University, Columbus, OH; Coffman et al., 2011), was transformed with p4143 (pRS306 backbone, URA3) and used in Fig. 3 D. Strains A5811 (diploid) and A15978 were gifts of A. Amon (Massachusetts Institute of Technology, Cambridge, MA; Lee et al., 2004) and were respectively transformed with p4143 (Fig. 3 E) or p2568 (Fig. 3 G). Strains A15978 and A5244, gifts of A. Amon, were transformed with p4143 (Fig. S3 C) and p2568 (Fig. S3 D).
For all experiments, yeast cells were grown in liquid YPDA medium at 30°C in flasks for 4 or 7 d as specified in each figure, except for Fig. 3, E–G and Fig. S3, C and D, for which strains were grown in SC-His for 3 d. In Fig. S3 L, yeast cells were grown in liquid YPDA medium with or without 1 M sorbitol (S1876; Sigma-Aldrich) at 30°C in flasks for indicated times.
For quiescence exit in the sole presence of glucose, cells were washed twice with water, then inoculated in glucose 2% at 0.6–1 OD600nm and incubated at 30°C. For quiescence exit in the presence of MT-destabilizing drugs, cells were preincubated for 1 h in the presence of the drug before quiescence exit. For scoring new bud emergence upon quiescence exit, ConA-FITC (Sigma-Aldrich) was added to a final concentration of 0.2 g/L onto a 12-d-old cell culture. Cells were then incubated for 1 h at 30°C, washed twice, and then transferred into YPDA in the absence of ConA-FITC (Sahin et al., 2008).
Cell staining
Actin phalloidin staining was done as described in Sagot et al. (2006). In brief, cells were fixed with formaldehyde (3.7% final), washed, and stained for 24 h with Alexa Fluor 568–phalloidin (Invitrogen). Cells were then washed twice, resuspended in a mounting solution containing 70% glycerol and 5 mg/L paraphenylenediamine, and imaged at room temperature using the Cy3 filter described below. DNA was stained by incubating cells for 5 min at 30°C with Hoechst (Invitrogen) at 0.5 mg/ml.
Cell viability
Cell viability in quiescence was measured using methylene blue. Colony-forming capacity was addressed after 2, 7, and 14 d at 30°C by plating 200 cells on YPDA, as measured using a particle counter (Multisizer 4; Beckman Coulter). Micro-manipulation of quiescent cells was performed as described in Laporte et al. (2011). In brief, 12-d-old cells grown in liquid YPDA were stained for 5 min with methylene blue; 10 µl of cell suspension was then deposited onto a YPDA plate. Cells that were not stained (i.e., alive cells) were micro-manipulated using a Singer MSM system (Singer Instrument Co. Ltd.).
Fluorescence microscopy
Cells were observed in a fully automated inverted microscope (model 200M; Carl Zeiss) equipped with an MS-2000 stage (Applied Scientific Instrumentation), a 300-Watt xenon light source (Lambda LS; Sutter Instrument), a 100× 1.4 NA Plan-Apochromat objective, and a five-position filter turret. For GFP imaging, we used a FITC filter (excitation, HQ487/25; emission, HQ535/40; beam splitter, Q505lp). For RFP imaging we used a Cy3 filter (Ex: HQ535/50 – Em: HQ610/75 – BS: Q565lp). For Hoechst imaging we used a DAPI filter (Ex: 360/40 – Em: 460/50 – BS: 400). For CFP imaging we use a CFP filter (Ex: HQ436/20 – Em: HQ480/40 – BS: 455dclp). For YFP imaging we use a YFP filter (excitation, HQ500/20; emission, HQ535/30; beam splitter, Q515lp). All the filters are from Chroma Technology Corp. Images were acquired using a CoolSnap HQ camera (Roper Scientific). The microscope, camera, and shutters (Uniblitz) were controlled by SlideBook software 5.0 (Intelligent Imaging Innovations). Images are, unless specified, 3D maximal projections of z-stacks performed using a 0.27-µm step. Few microliters of cell culture were deposited onto a glass slide and directly imaged at room temperature.
FRAP experiments were done on an inverted microscope (model DMI 6000; Leica) equipped with a spinning disk confocal head (CSU-X1; Yokogawa Corporation of America), a QuantEM camera (Photometrics), and a scanner FRAP system (Roper Scientific). The diode lasers used were at 408 nm and 491 nm. The objective used was an HCX PL APO CS 100× oil 1.4 NA. The z-stacks were done with an objective scanner (Piezo P721.LLQ; Physik Instrumente). This system was controlled by MetaMorph software (Molecular Devices). Few microliters of the cell culture were deposited onto a glass slide holding an agarose pad (1.5%) and the coverslip was sealed with VALAP and directly imaged at room temperature.
Data analysis
In Fig. 1 C, a line scan (i1) of 3-pixel width containing both GFP signal and background was drawn along the nMT array using ImageJ software (National Institutes of Health, Bethesda, MD). A line of 6-pixel width (at the same location) was drawn in order to calculate the intensity of the surrounding background (i2). The real intensity (ir) was calculated as follows: ib = (i2 × 6) − (i1 × 3) and ir = i1 − (ib/3). We arbitrarily set the highest fluorescence value for each line scan to 100%. On the graph, depending on the directionality of the line scan, 0 or 100% fluorescence was set to 0 µm on the x axis.
To measure MT length variation (Fig. 2 and Fig. S2), position of the SPB and the plus-end extremities of the MT structure (aMT or nMT array) were followed over time. Euclidian distances (D) between two positions were calculated as follows: D = √ ((x2 − x1)2 + (y2 − y1)2). In time-lapse series, the first measured length was set to zero and the variation was calculated according to this reference. SPB/centromere and SPB/kinetochore Euclidian distance measurements were done using the MTrackJ plugin in ImageJ, their respective positions being determined in the best focal plane.
Fluorescence recovery was corrected for background noise and continuous photobleaching using ImageJ software, and then normalized to the fluorescence measured before bleach (Vavylonis et al., 2008). The fluorescence intensity of a region of interest (5 × 4 pixels) was measured on maximal projection of z-stacks. The fluorescence intensity Ir was calculated over time as follows: I = (Iregion of interest − Ibackground). Photobleaching was measured on unbleached nMT arrays, and the intensity was corrected using Ir = Iy × (Iunbleached first point / Iunbleached point y), where y is a specific time point over time.
The relative position of the nucleolus versus the SPB was determined using a central symmetry axis along the nucleus draw from the SPB.
To determine the location of Arp1/Dyn1 toward nuclear membrane, a line scan (i1) of 5-pixel width containing both GFP/RFP signal and background was drawn along the GFP/RFP signal using ImageJ software. A line of 10-pixel width (at the same location) was drawn in order to calculate the intensity of the surrounding background (i2). The real intensity (ir) was calculated as follows: ib = (i2 × 10) − (i1 × 5) and ir = i1 − (ib/5). Individual Arp1/Dyn1/Nup2 Gaussian fits were determined and aligned using GFP fits set to 0 on the x axis. GFP/RFP average fits are displayed in Fig. S3 H.
Data reproducibility
In all graphs, unless specified, bars indicate SD. In Figs. 1 A, 2 E, 3 (A, F, and G), 4 (B and G), 5 A, S2 (D and E), and S3 (I and J), for each time point or strain more than 200 cells were scored. In Figs. 1 A, 2 E, and 5 A the data shown are a representative kinetic experiment out of at least three experimental repeats. For Figs. 1 C, 2 (A–C), and S2 (A and B), each line represents one MT; the number of total MTs (n) is indicated. For cell viability experiments, each strain was grown in independent duplicates and each plating done in triplicate. Micro-manipulation was done twice on more than 150 cells.
Electron microscopy
Immunostaining was done as described in Sagot et al. (2006). Cells were washed for 5 min with 1 mg/ml glycine, then 5 min with fetal calf serum. Cells were then incubated for 45 min with polyclonal rabbit anti-tat1 antibodies diluted 1:250 (a gift of K. Gull, University of Oxford, Oxford, UK; Woods et al., 1989). After a rinse with Tris-buffered saline containing 0.1% bovine serum albumin, cells were incubated for 45 min at room temperature with anti–mouse IgG conjugated to 10-nm gold particles (BioCell Laboratories, Inc.). For freeze substitution, yeast were deposited on a copper grid (400 mesh) coated with Formvar. Grids were immersed in liquid propane held at −180°C by liquid nitrogen, then transferred in a 4% osmium tetroxide solution in dry acetone at −82°C for 72 h. Grids were then shifted to room temperature and washed three times with dry acetone. Cell were stained with 1% uranyl acetate then washed once with dry acetone. Samples were gradually unfiltered with araldite (Fluka). Ultra-thin sections were contrasted with lead citrate and observed using an 80-kV electron microscope (model 7650; Hitachi) at the Bordeaux Imaging Center.
Miscellaneous
Glucose concentration was measured using the d-glucose/d-fructose UV test kit (Roche). Cycloheximide (Sigma-Aldrich) was added to a final concentration of 180 µM.
Online supplemental material
Fig. S1 (A and B) shows the nuclear membrane deformation induced by nMT arrays in quiescent cells. Fig. S1 C shows fluorescence quantification of various SPB components. In Fig. S1 D another example of nMT array visualized by EM is shown. Fig. S2 A displays MT dynamics for cells in early quiescence. Fig. S2 B shows dynamics of Bim1-GFP, Dad2-GFP, Stu2-GFP, or Nuf2-GFP in quiescent cells. Fig. S2 C points at immobile Nuf2-GFP dots detected along the nMT array. Fig. S2 (D and E) displays nMT array resistance to various MT-destabilizing drugs. In Fig. S3 A, the relocalization of the nucleolus is observed by immuno-EM. In Fig. S3 B, kinetochore protein localization in quiescent cells is shown. In Fig. S3 (C and D) the distance between CEN IV and the SPB is measured. Images of MAP deletion mutants expressing Bim1-3GFP or Tub1-RFP are shown respectively in Fig. S3, E and F. In Fig. S3 G, distribution of the distances between the SBP and Spc24-GFP are displayed. Fig. S3 H shows dynein and dynactin localization in quiescent cells. Fig. S3 (I and J) indicates the percentage of dead cells, bi-nucleated cells, and cells with actin bodies in mutant without nMT array. In Fig. S3 K, the viability in quiescence of various mutants is indicated. In Fig. S3 L, the effect of sorbitol on cell survivability in quiescence is tested. Table S1 indicates the localization of various MAPs in quiescent cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201306075/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201306075.dv.
Supplementary Material
Acknowledgments
We express our profound gratitude to J.P. Javerzat for his precious comments about our work and his great help in writing this manuscript. We are grateful to L. Blanchoin, B. Daignan-Fornier, M. Gupta, A. Paoletti, D. Pellman, and P. Tran for helpful discussions about our work. We are thankful to A. Amon, K. Bloom, B. Goode, D. Pellman, and J.-Q. Wu for providing yeast strains. FRAP was done in the Bordeaux Imaging Center with the help of Christel Poujol.
This work was supported by the Université Bordeaux 2, Conseil Régional d’Aquitaine, a Young Investigator grant from the Agence Nationale pour la Recherche (JC08 310804 to I. Sagot), and an Association pour la Recherche sur le Cancer Grant (ARC, SFI20101201558 to I. Sagot).
Footnotes
Abbreviations used in this paper:
- CEN
- DNA proximal to centromere
- MAP
- MT-associated protein
- MT
- microtubule
- SPB
- spindle pole body
- WT
- wild type
References
- Bystricky K., Heun P., Gehlen L., Langowski J., Gasser S.M. 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. USA. 101:16495–16500 10.1073/pnas.0402766101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882 10.1073/pnas.082243699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J.L., Stearns T. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641 10.1083/jcb.138.3.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi A.K., Ibrahim B., Pereira G. 2010. Monitoring spindle orientation: Spindle position checkpoint in charge. Cell Div. 5:28 10.1186/1747-1028-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Wu P., Parthun M.R., Wu J.Q. 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195:563–572 10.1083/jcb.201106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B., Sagot I. 2011a. Proliferation/quiescence: the controversial “aller-retour”. Cell Div. 6:10 10.1186/1747-1028-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B., Sagot I. 2011b. Proliferation/Quiescence: When to start? Where to stop? What to stock? Cell Div. 6:20 10.1186/1747-1028-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C. 2012. The essence of yeast quiescence. FEMS Microbiol. Rev. 36:306–339 10.1111/j.1574-6976.2011.00287.x [DOI] [PubMed] [Google Scholar]
- Dimitrov A., Quesnoit M., Moutel S., Cantaloube I., Poüs C., Perez F. 2008. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science. 322:1353–1356 10.1126/science.1165401 [DOI] [PubMed] [Google Scholar]
- Erlemann S., Neuner A., Gombos L., Gibeaux R., Antony C., Schiebel E. 2012. An extended γ-tubulin ring functions as a stable platform in microtubule nucleation. J. Cell Biol. 197:59–74 10.1083/jcb.201111123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuge E.K., Braun E.L., Werner-Washburne M. 1994. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J. Bacteriol. 176:5802–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Cheeseman I.M. 2011. Kinetochore assembly: if you build it, they will come. Curr. Opin. Cell Biol. 23:102–108 10.1016/j.ceb.2010.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. 1997. Centromere position in budding yeast: evidence for anaphase A. Mol. Biol. Cell. 8:957–972 10.1091/mbc.8.6.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.W., Cai D., Verhey K.J. 2008. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20:71–76 10.1016/j.ceb.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P.K. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 101:763–775 10.1016/S0092-8674(00)80888-0 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D., Roussel P., Thiry M., Sirri V., Lafontaine D.L. 2010. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA. 1:415–431 10.1002/wrna.39 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Mathog D., Gruenbaum Y., Saumweber H., Sedat J.W. 1986. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J. Cell Biol. 102:112–123 10.1083/jcb.102.1.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.V., Larsson L.I. 2004. Actin microdomains on endothelial cells: association with CD44, ERM proteins, and signaling molecules during quiescence and wound healing. Histochem. Cell Biol. 121:361–369 10.1007/s00418-004-0648-2 [DOI] [PubMed] [Google Scholar]
- Jin Q., Trelles-Sticken E., Scherthan H., Loidl J. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141:21–29 10.1083/jcb.141.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q.W., Fuchs J., Loidl J. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113:1903–1912 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bloom K.S., Salmon E.D. 2010. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr. Opin. Cell Biol. 22:57–67 10.1016/j.ceb.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Kitamura Y., Tanaka T.U. 2007. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 21:3319–3330 10.1101/gad.449407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska M.M., Crutchfield C.A., Bradley P.H., Rabinowitz J.D., Broach J.R. 2011. Yeast cells can access distinct quiescent states. Genes Dev. 25:336–349 10.1101/gad.2011311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Wittmann T. 2012. +TIPs: SxIPping along microtubule ends. Trends Cell Biol. 22:418–428 10.1016/j.tcb.2012.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Salin B., Daignan-Fornier B., Sagot I. 2008. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181:737–745 10.1083/jcb.200711154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Lebaudy A., Sahin A., Pinson B., Ceschin J., Daignan-Fornier B., Sagot I. 2011. Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 192:949–957 10.1083/jcb.201009028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Kiburz B.M., Amon A. 2004. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr. Biol. 14:2168–2182 10.1016/j.cub.2004.12.033 [DOI] [PubMed] [Google Scholar]
- Liu I.C., Chiu S.W., Lee H.Y., Leu J.Y. 2012. The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol. Biol. Cell. 23:1231–1242 10.1091/mbc.E11-09-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Kalutkiewicz K.A., Lee W.L. 2012. Astral microtubule asymmetry provides directional cues for spindle positioning in budding yeast. Exp. Cell Res. 318:1400–1406 10.1016/j.yexcr.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 94:607–613 10.1016/S0092-8674(00)81602-5 [DOI] [PubMed] [Google Scholar]
- Miranda J.J., De Wulf P., Sorger P.K., Harrison S.C. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R., Levy M., Tsechansky M., Stovall G.M., O’Connell J.D., Mirrielees J., Ellington A.D., Marcotte E.M. 2009. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA. 106:10147–10152 10.1073/pnas.0812771106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C., Sato B.K., Broyer R.M., Wilhelm J.E. 2010. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190:541–551 10.1083/jcb.201003001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P.H. 2011. Quiescence: early evolutionary origins and universality do not imply uniformity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3498–3507 10.1098/rstb.2011.0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E.T., Winey M., McIntosh J.R. 1999. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 10:2017–2031 10.1091/mbc.10.6.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 1997. Centrosome-microtubule nucleation. J. Cell Sci. 110:295–300 [DOI] [PubMed] [Google Scholar]
- Poulter N.S., Staiger C.J., Rappoport J.Z., Franklin-Tong V.E. 2010. Actin-binding proteins implicated in the formation of the punctate actin foci stimulated by the self-incompatibility response in Papaver. Plant Physiol. 152:1274–1283 10.1104/pp.109.152066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. 1885. Uber zellteilung. Morphol. Jahrbuch. 10:214–330 [Google Scholar]
- Ramey V.H., Wang H.W., Nakajima Y., Wong A., Liu J., Drubin D., Barnes G., Nogales E. 2011. The Dam1 ring binds to the E-hook of tubulin and diffuses along the microtubule. Mol. Biol. Cell. 22:457–466 10.1091/mbc.E10-10-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Pinson B., Salin B., Daignan-Fornier B. 2006. Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol. Biol. Cell. 17:4645–4655 10.1091/mbc.E06-04-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin A., Daignan-Fornier B., Sagot I. 2008. Polarized growth in the absence of F-actin in Saccharomyces cerevisiae exiting quiescence. PLoS ONE. 3:e2556 10.1371/journal.pone.0002556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I., Shaw P. 2011. Organization and dynamics of plant interphase chromosomes. Trends Plant Sci. 16:273–281 10.1016/j.tplants.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Shah K.H., Zhang B., Ramachandran V., Herman P.K. 2013. Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics. 193:109–123 10.1534/genetics.112.146993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M.A., Pellman D. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13:364–372 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Stephens A.D., Haase J., Vicci L., Taylor R.M., II, Bloom K. 2011. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J. Cell Biol. 193:1167–1180 10.1083/jcb.201103138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Igarashi R., Sekiya M., Utsugi T., Morishita S., Yukawa M., Ohya Y. 2004. Dynactin is involved in a checkpoint to monitor cell wall synthesis in Saccharomyces cerevisiae. Nat. Cell Biol. 6:861–871 10.1038/ncb1162 [DOI] [PubMed] [Google Scholar]
- Taddei A., Gasser S.M. 2012. Structure and function in the budding yeast nucleus. Genetics. 192:107–129 10.1534/genetics.112.140608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Tanaka T.U. 2010. Live-cell analysis of kinetochore-microtubule interaction in budding yeast. Methods. 51:206–213 10.1016/j.ymeth.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., O’Toole E., Berrueta L., Bierer B.E., Pellman D. 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145:993–1007 10.1083/jcb.145.5.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B., Akhmanova A., Straube A. 2009. Regulation of microtubule dynamic instability. Biochem. Soc. Trans. 37:1007–1013 10.1042/BST0371007 [DOI] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Verdaasdonk J.S., Bloom K. 2011. Centromeres: unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 12:320–332 10.1038/nrm3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R.H. 2009. On and around microtubules: an overview. Mol. Biotechnol. 43:177–191 10.1007/s12033-009-9193-5 [DOI] [PubMed] [Google Scholar]
- Westermann S., Wang H.W., Avila-Sakar A., Drubin D.G., Nogales E., Barnes G. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 440:565–569 10.1038/nature04409 [DOI] [PubMed] [Google Scholar]
- Westermann S., Drubin D.G., Barnes G. 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76:563–591 10.1146/annurev.biochem.76.052705.160607 [DOI] [PubMed] [Google Scholar]
- Winey M., Bloom K. 2012. Mitotic spindle form and function. Genetics. 190:1197–1224 10.1534/genetics.111.128710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., O’Toole E.T. 2001. The spindle cycle in budding yeast. Nat. Cell Biol. 3:E23–E27 10.1038/35050663 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., Barnes G. 2012. Spindle assembly requires complete disassembly of spindle remnants from the previous cell cycle. Mol. Biol. Cell. 23:258–267 10.1091/mbc.E11-08-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500 [DOI] [PubMed] [Google Scholar]
- Zimmer C., Fabre E. 2011. Principles of chromosomal organization: lessons from yeast. J. Cell Biol. 192:723–733 10.1083/jcb.201010058 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.