Experimental analysis and computational modeling of epidermal wound closure in 3D suggests an important role for surrounding tissue in determining epithelial cell movement and fate.
Abstract
Wound healing is a complex process in which a tissue’s individual cells have to be orchestrated in an efficient and robust way. We integrated multiplex protein analysis, immunohistochemical analysis, and whole-slide imaging into a novel medium-throughput platform for quantitatively capturing proliferation, differentiation, and migration in large numbers of organotypic skin cultures comprising epidermis and dermis. Using fluorescent time-lag staining, we were able to infer source and final destination of keratinocytes in the healing epidermis. This resulted in a novel extending shield reepithelialization mechanism, which we confirmed by computational multicellular modeling and perturbation of tongue extension. This work provides a consistent experimental and theoretical model for epidermal wound closure in 3D, negating the previously proposed concepts of epidermal tongue extension and highlighting the so far underestimated role of the surrounding tissue. Based on our findings, epidermal wound closure is a process in which cell behavior is orchestrated by a higher level of tissue control that 2D monolayer assays are not able to capture.
Introduction
In human skin wound healing, reepithelialization is the most essential part, as the tissue’s primary objective is to quickly reestablish barrier function (Martin, 1997; Singer and Clark, 1999; Friedl and Gilmour, 2009). The individual cells of the skin are orchestrated to behave in such a way that skin integrity is reestablished in an evolutionarily proven, most robust way (Singer and Clark, 1999). It is highly challenging to design experiments capturing how this orchestration actually takes place. Although 2D monolayer experiments are ideal for analyzing individual cellular functions such as migration mechanistically on the single cell level, wound healing cannot be reduced merely to cell migration (Farooqui and Fenteany, 2005; Soderholm and Heald, 2005; Liang et al., 2007). Thus, for understanding wound healing, the analysis of the orchestration of the individual processes taking part in wound healing has to be performed. This can only be undertaken in 3D wound-healing models, which have to be systematically and quantitatively characterized. The goal is hereby to derive consistent computational models helping to uncover high-level tissue functions as well as to understand the roles of individual cellular processes in tissue repair. In the sense of Noble (2006), it is the question of how a repair function at the higher biological scale of the tissue is actually realized by the lower scale of the single cell level.
Choosing this systems biological approach can be expected to provide answers to several open questions of wound closure. A central question debated in the literature in skin wound healing is, for example, the mechanism of the creation and extension of the epidermal tongue. Two reepithelialization mechanisms were postulated so far. The first is the leap-frog or rolling mechanism in which migrating suprabasal cells roll over leading basal cells and dedifferentiate to form new leaders (Krawczyk, 1971; Paladini et al., 1996). The tractor-tread or sliding mechanism postulates that layered keratinocytes move forward in a block (Radice, 1980; Woodley, 1996). A variant is the model of Usui et al. (2005) in which suprabasal cells migrate out of the wound, thereby outnumbering the basal cells. It has up till now been unclear whether one of these mechanisms is correct and how such a mechanism is functionally embedded in the environment of the wound. The latter issue points to the question of the contributions of the intact surrounding tissue, which has been largely neglected so far and thus warrants a systematic analysis. Both aspects, tongue extension and the intact tissue of the wound, are linked to and realized by tightly regulated spatiotemporal processes of proliferation, migration, and differentiation, finally leading to reestablishment of the intact epidermal 3D morphology of the skin (Gurtner et al., 2008).
To build a consistent mechanistic model of wound closure, we set up a dedicated technical analysis pipeline comprising 3D organotypic wound models, standardized immunohistology, fluorescent whole-slide imaging, image analysis, multiplex protein analytics, and computational systems biological modeling. We applied our pipeline on large numbers (92) of 3D organotypic full-thickness skin wound models comprising keratinocytes and fibroblasts, which we tracked in time by a novel two-step time-lag fluorescence staining. This allowed us to dissect the epidermal 3D wound-healing process spatiotemporally in cell proliferation, migration, and differentiation and to derive the extending shield mechanism (ESM), a consistent theory of how these three processes are intertwined leading to the incremental and robust closure of human wounds.
Results
The organotypic skin wound model shows a concentric closure with rates similar to the in vivo situation
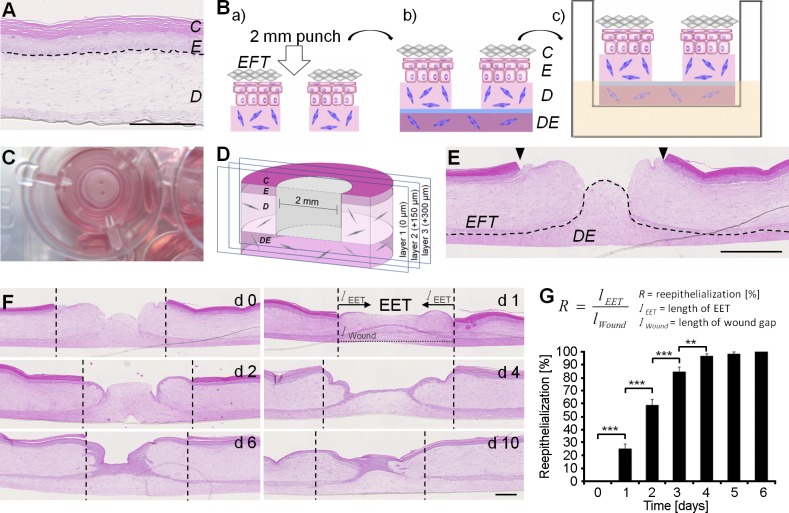
To construct a reproducible wound model, commercially obtainable epidermal full-thickness (EFT) cultures, containing epidermis and dermis, were punched and adhered to self-fabricated dermal equivalents (DEs; Fig. 1, A and B). With a handling time of 5 min per 3D culture involving wounding, attaching to the DE, and placing the wound culture into inserts, our experimental setup enables a considerable medium throughput (Fig. 1 C). For subsequent histological sectioning, we developed a defined tissue-sectioning method (see Materials and methods) to ensure consistent and comparable wound diameters for analysis (Fig. 1 D). Similarly as intact epidermis, in vivo, the unwounded epidermis of the cultures contained about five cell layers, in which basal keratinocytes showed a typical palisade structure, characterized by cubical cell shape and continual flattening with progressive differentiation, finally building up the stratum corneum. All EFT cultures seamlessly attached to the DE, providing an in vivo–like matrix for reepithelialization surrounded by clearly defined wound margins (Fig. 1 E). 1 d after wounding, keratinocytes had started migration (Fig. 1 F). At this point, the extending epidermal tongue (EET) was visible in the form of two cell layers at the margins and one cell layer at the leading edge. 4–5 d after wounding, keratinocytes had covered the whole wound area, forming a neo-epidermis, which displayed a thickness of about three cell layers near the wound margins and one cell layer at the center of the wound. From day 4–10, the neo-epidermis thickened and formed a multilayered epithelium. 1 d after wounding, 36% of the former wound area was closed (Fig. 1 G). At day 4 after wounding, reepithelialization of 100% of the wounded area was achieved, equaling complete wound closure, leading to a reepithelialization rate of 338 µm/d being in the physiological range (Laplante et al., 2001).
Figure 1.
The excisional 3D in vitro wound model using precast EFT cultures and DEs. (A) Hematoxylin and eosin–stained paraffin sections of the EFT cultures. The epidermis (E) displayed basal, spinous, granular, and cornified keratinocyte layers (C), and the dermis (D) distributed fibroblasts. Broken lines denote dermal–epidermal junction. (B) Precast EFT skin cultures were wounded twice with a 2-mm biopsy punch. (C) After wounding, cultures were placed into a 12-well ThinCert insert. (D) Cultures were halved with one punch on every tissue sample and cut until the wound was microscopically visible. Sections were marked as layer 1 (0 µm). Based on that, further defined sections in layer 2 (150 µm) and in layer 3 (300 µm) were produced. (E) Histology of the wound cultures. Hematoxylin and eosin–stained sections showed punched wounds with clearly defined wound margins (arrowheads). The underlying DE was seamlessly attached, providing the wound matrix (dotted line shows basal membrane). (F) Hematoxylin and eosin reepithelialization kinetics of the wound cultures over a period of 10 d. Broken lines denote wound margins. (G) The relative wound closure was quantified as the ratio of the migration distance (length of the EET; lEET) to the size of the wound in the respective section (lWound). The data shown are from two independent experiments; n = 8 (day 0 [d0]), 10 (day 1), 8 (day 2), 10 (day 3), 9 (day 4), 4 (day 5), and 3 (day 6); 100 slides were analyzed in total; data are means ± SEM; **, P ≤ 0.01; ***, P ≤ 0.001, Student’s t test. Bars, 500 µm.
Keratinocyte–fibroblast cross talk ensures an in vivo–like wound-healing environment
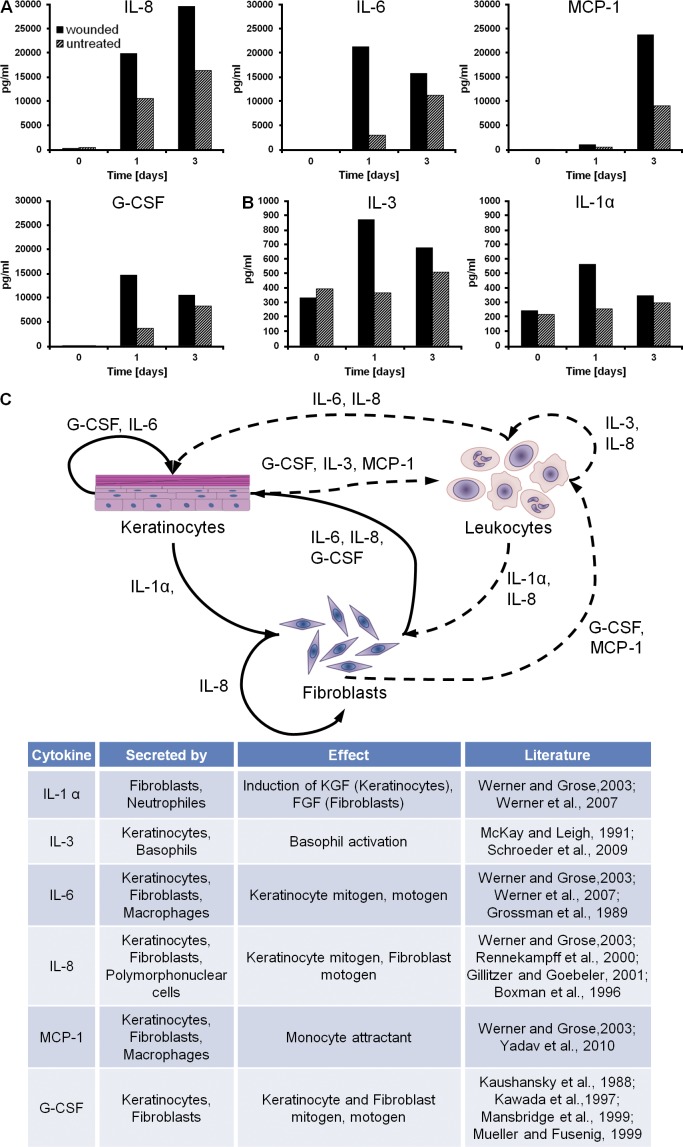
Keratinocyte–fibroblast cross talk and the underlying paracrine growth factor signaling are essential to coordinate wound healing–associated processes such as proliferation and migration. To show that our wound model shares important components of the epidermal–dermal cross talk with native skin wounds, we measured selected growth factors driving the reepithelialization mechanism. We collected supernatants of wounded and unwounded organotypic cultures and analyzed IL-1α, IL-3, IL-6, IL-8, MCP-1, and granulocyte colony-stimulating factor (G-CSF) concentrations before (day 0), as well as 1 and 3 d after wounding (Fig. 2, A and B). Most abundant were the proinflammatory cytokines IL-6, IL-8, and MCP-1, showing a mean concentration of >20,000 pg/ml. G-CSF (>12,000 pg/ml) was also present in high concentrations followed by IL-3 (>800 pg/ml) and IL-1α (>500 pg/ml). Although IL-1α, IL-3, IL-6, and G-CSF peaked 1 d after wounding, IL-8 showed an continuous increase. Elevated levels of MCP-1 concentration were only found 3 d after wounding, assuming a delayed tissue reaction. Based on the cytokine measurements, the underlying paracrine signaling network was drawn in Fig. 2 C.
Figure 2.
Paracrine signaling of keratinocytes and fibroblasts in the excisional 3D in vitro wound-healing model. (A and B) Mean concentrations of growth factors measured in the supernatant of the 3D in vitro wound-healing model. The growth factors are divided into two groups, based on the maximum mean concentration: >10,000 pg/ml (A); <1,000 pg/ml (B). (C) The paracrine signaling of keratinocytes and fibroblasts under wound-healing conditions is shown. Arrows mark dependencies between the cell types. Dashed lines represent possible dependencies, which could not be measured in the 3D in vitro wound-healing model caused by the lack of leukocytes. Growth factor–secreting cell types and the impact of the factors on cell behavior are listed (Kaushansky et al., 1988; Grossman et al., 1989; Demetri and Griffin, 1991; McKay and Leigh, 1991; Boxman et al., 1996; Kawada et al., 1997; Mansbridge et al., 1999; Mueller and Fusenig, 1999; Rennekampff et al., 2000; Gillitzer and Goebeler, 2001; Angel and Szabowski, 2002; Werner and Grose, 2003; Schroeder et al., 2009; Yadav et al., 2010). Data are means; n = 4 from a single experiment.
Keratinocyte proliferation occurs in a concentric traveling wave after the EET
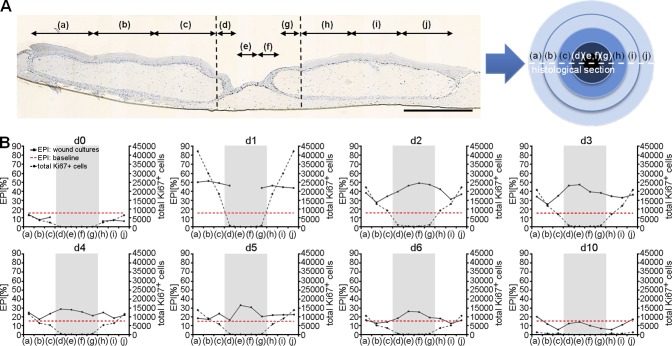
To determine the spatiotemporal profile of keratinocyte proliferation during reepithelialization, we divided histological sections of the wound cultures in 10 regions equaling five concentric rings and analyzed their epidermal proliferation index (EPI) as the fraction of proliferating basal cells over 10 d (Fig. 3 A). Comprehensive computational image analysis of the digital slides yielded an EPI baseline of 13% after averaging the EPI at day 0 and 10. Emanating from this baseline, the whole culture reacted with a proliferative burst at day 1 with an EPI of ≤50% within the wound and the surrounding tissue (Fig. 3 B). At this time point, the wound itself still showed only few in-migrated keratinocytes. With progressing time and accompanying increasing migration, the proliferative activity declined depending on the distance to the wound margin. 3 d after wounding, the EPI significantly decreased in the surrounding tissue, while remaining continuously high with 42% in the wound. The EPI then normalized until day 6 in the surrounding tissue as well as in the wound area. Based on phosphorylated histone 3 staining, the mitotic index was calculated showing a spatiotemporal profile equal to the EPI (Fig. S1, A–C). We performed systematic morphometric analysis of the unwounded tissue regions with image processing of histological sections. Right after wounding, we observed only a limited increase in the thickness of the CK10− cell layers despite nonthickening of CK10+ layers (Fig. S1 D). From day 4 on, the height of the CK10− layer was even below those of day 0. In light of the massive and sudden proliferation response of the whole tissue, we conclude that the newly produced cells must have been used to establish a massive concentric cell stream toward the wound. Collectively, the results show an immediate burst in proliferation in a concentric pattern around the punch wound, which traveled as a wave together with former and newly produced cells into the direction of the punch wound.
Figure 3.
Quantification of proliferation by Ki67 image processing of virtual slides. (A) Ki67-stained wound cultures were separated into 10 regions of identical size equaling five concentric rings. The wound area was divided into four regions (d–g), whereas unwounded areas surrounding the wound were divided into three regions (a–c; h–j) at each side of the wound. Broken lines denote the wound margin. (B) The epidermal proliferation index (EPI) and the total number of Ki67+ cells were spatially and temporally evaluated by analyzing each region independently. The wound region is highlighted in gray. The red dotted lines represent the baseline levels of the epidermal proliferation index. The data shown are from a single representative experiment out of two repeats; n = 2 (day 0 [d0]), 3 (day 1), 3 (day 2), 3 (day 3), 3 (day 4), 3 (day 5), 2 (day 6), and 3 (day 10); 113 slides were analyzed in total; data are means. Bar, 1 mm.
Collective polarization of basal keratinocytes in the intact tissue toward the wound
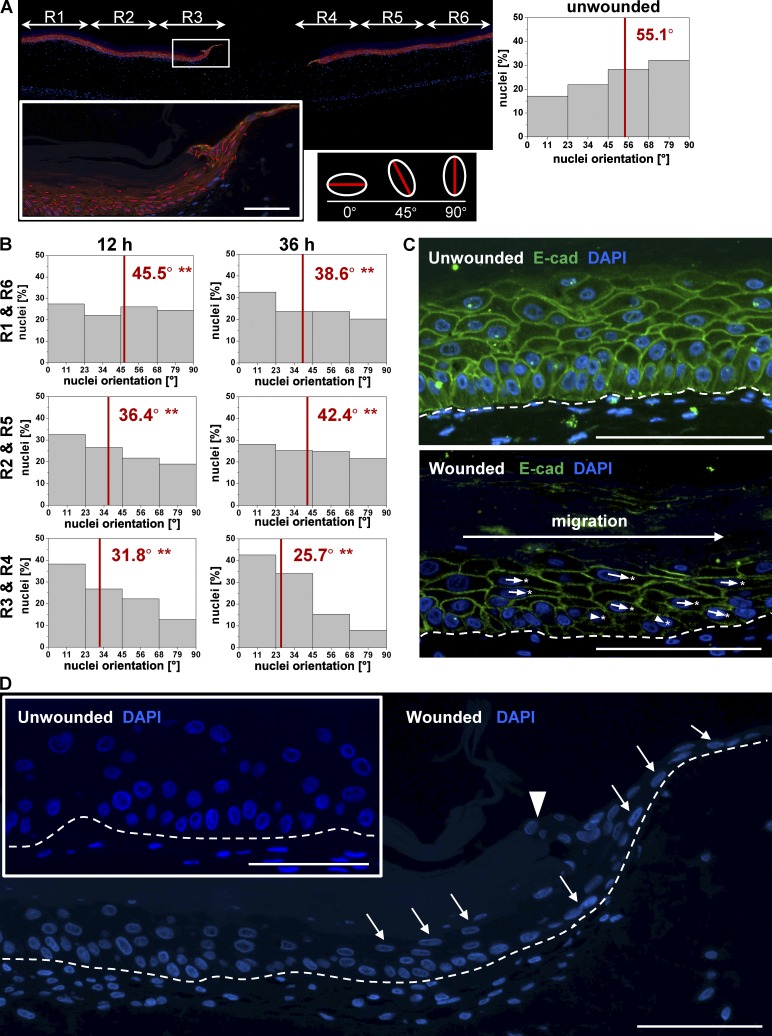
We analyzed the surrounding tissue for signs of collective polarization, indicating a potential cell stream toward the wound, indicating in turn collective migration. Therefore, we analyzed nuclear shape, positioning, and deformation as well as Golgi complex translocation during wound closure. We separated the organotypic skin cultures into six regions of equal size according to the wound distance and quantified the major axis (see Materials and methods) of every nucleus of the basal cells by computational image processing (Fig. 4 A). Unwounded cultures showed a distribution of nuclear orientations with a tendency toward 90°, in which 0° represents a horizontal orientation. After 12 h, we observed in all regions (R1–R6) of wounded cultures a distribution that significantly differed from rotation angles of nuclei in unwounded skin (Fig. 4 B). Thereby, the rotation was strongest in the inner rings R3 and R4 and quantitatively diminished with increasing distance from the punch. 36 h after wounding, this trend progressed to an even stronger rotation in which many cells took a fully horizontal orientation with their major axis parallel to the basal membrane. Only a neglectable (0.6%) fraction of cells still showed a vertical (90°) orientation. This observed shift of nuclear rotation from vertically to horizontally orientated nuclei was in line with the translocation of the Golgi complex, a cellular polarization marker, which shifted from a position underneath the nuclei in unwounded cultures to a position behind the nuclei in an opposed direction to migration in wound cultures (Fig. S2). We further qualitatively measured the position of the nucleus relative to the cell boundaries stained with E-cadherin (Fig. 4 C). Cell nuclei showed a center position as expected in unwounded cultures, whereas in wounded cultures, nuclei moved rearward, opposed to the distantly located punch wound. At the same time, nuclei of rotated keratinocytes showed an elongated, cigarlike morphology. Nuclei of keratinocytes in unwounded cultures showed a round to ellipsoid shape in basal and suprabasal regions (Fig. 4 D). We conclude that basal cells throughout the complete organotypic culture collectively change their polarization toward the wound edge in all regions of the unwounded tissue in two dimensions: in time as well as in distance to the wound margin. Together with the fact that the basal cells showed constant adhesion by E-cadherin, the collective polarization indeed points to the collective migration of the basal cell layer toward the punch wound.
Figure 4.
Polarization of collectively migrating basal cells allow shield extension. (A) Histological sections of organotypic wound cultures were separated into six different regions dependent on the distance to the wound, and the nuclear major axis of basal keratinocytes was analyzed. The inset shows the magnification of the wound margin (box) in region 3 (R3) to illustrate the change of nuclear major axis orientations (red lines). (B) Plotted nuclear orientation degrees of the major axis analysis for the respective regions 12 and 36 h after wounding. (C) E-cadherin staining of representative histological sections of unwounded tissue and wound margin. Compared with unwounded tissue cells within the wound margin show a nuclei movement according to the cell boundaries rearward in opposed direction of cell migration. The arrows indicate displacement vectors. Asterisks indicate the geometrical center of the cell. (D) Nuclei deformation of wound cultures indicated by white arrows are shown by representative DAPI stains. The arrowhead indicates the wound margin. Broken lines denote dermal–epidermal junction. The data shown are from a single representative experiment. For the experiment shown, n = 5; 2,341 cell nuclei were analyzed in total; **, P ≤ 0.01, Student’s t test. Bars, 100 µm.
Basal cells collectively migrate to the tip of the EET underneath a mechanically stable suprabasal compartment
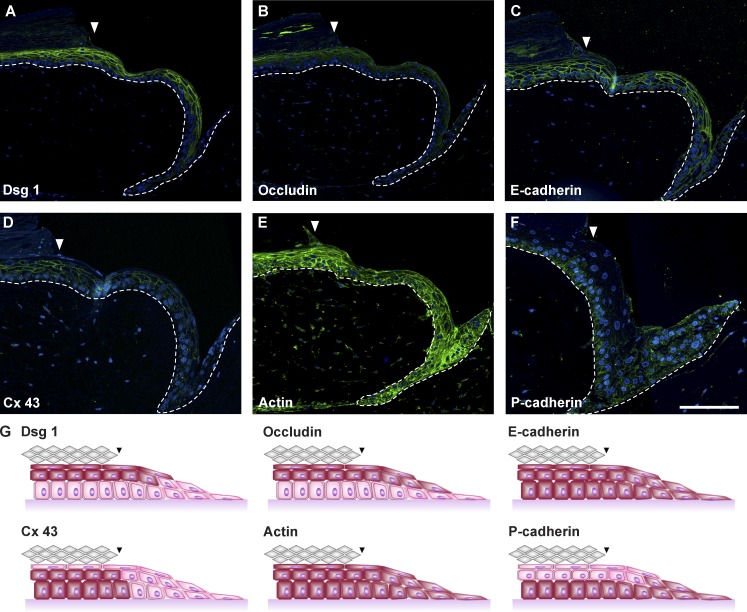
The capability of keratinocytes to collectively migrate depends on the expression of their junctions. In intact tissue and the EET, we characterized the cell junction proteins Dsg1 (desmoglein 1), occludin, and E- and P-cadherin 48 h after wounding (Fig. 5, A–C and F). Generally, we observed cell junctions normalizing quickly toward the intact tissue, whereas the leading edge showed a strong P-cadherin expression (Fig. 5 F). All suprabasal cells of the EET expressed Dsg1 and occludin, indicating intact desmosomes and tight junctions that are immediately formed after the EET becomes multilayered. E-cadherin and actin were immunolocalized in basal and suprabasal cells of the EET, showing functional adherens junctions and microfilaments in the full length of the EET (Fig. 5, C and E). The gap junction protein connexin 43 was not detected in the leading edge cells but in the rest of the EET and intact tissue (Fig. 5 D). Collectively, this indicates a mechanically stable suprabasal compartment, whereas the basal layer retains its migratory flexibility necessary for collective migration (Fig. 5 G).
Figure 5.
Spatial regulation of cell junctions and communication channels in the EET. (A–F) Histological sections of representative EETs showing the expression of desmoglein 1 (Dsg1; A), occludin (B), E-cadherin (C), connexin 43 (Cx 43; D), actin (E), and P-cadherin (F). (G) The spatial expression patterns (red colored cells) are schematically summarized. Arrowheads indicate the wound margin. Broken lines denote dermal–epidermal junction. Bar, 200 µm.
Collectively migrating cells continuously build a multilayered epithelium in which suprabasal cells never contact the ECM, thereby ruling out the leap-frog theory
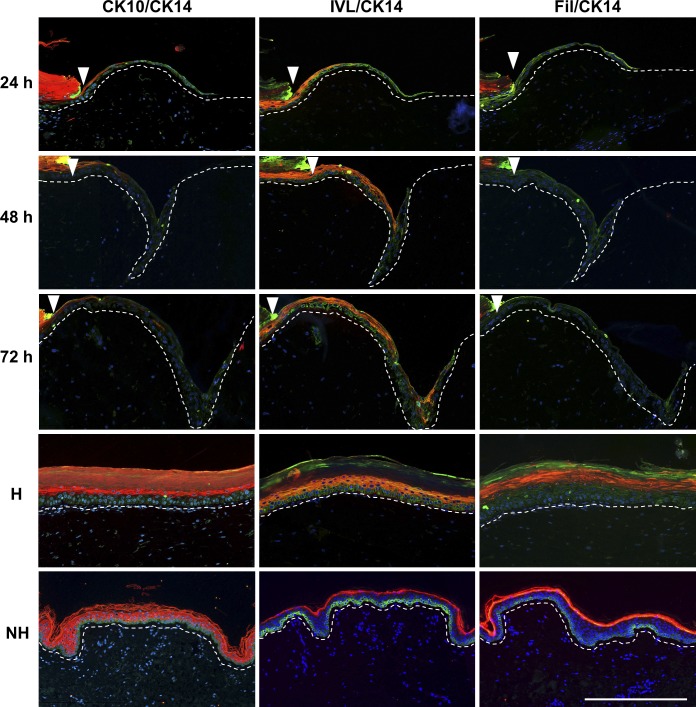
To understand how the collectively migrating cells form a multilayered epithelium, we analyzed keratinocyte differentiation (CK10, CK14, involucrin, and filaggrin) in the forming neo-epidermis 24, 48, and 72 h after wounding (Fig. 6). 24 h after wounding, suprabasal keratinocytes showed a laterally declining expression pattern of CK10 and involucrin toward the tip of the EET. With progressing time (48 and 72 h), the expression of both differentiation markers advanced laterally with extension of the tongue. Toward the intact tissue as well as in homeostatic regions of the wound models (Fig. 6, H), the differentiation patterns were equal to normal human epidermis (Fig. 6, NH). Late differentiation marker filaggrin was not expressed in the EET between 0 and 72 h but present in the intact tissue, indicating the time the EET needs to further differentiate. At all time points, the EET showed a continuously differentiated multilayered epithelium in which suprabasal keratinocytes never were in contact with the ECM or the basal membrane.
Figure 6.
Differentiated keratinocytes never contact the ECM while restoring the physiological epidermal differentiation pattern. Histological sections of representative EETs, homeostatic wound model regions (H) and normal human skin (NH) 24, 48, and 72 h after wounding. The sections show a double fluorescent staining of the differentiation markers cytokeratin 10 (CK10), involucrin (IVL), and filaggrin (Fil) stained with Alexa Fluor 594 in combination with CK14 (Alexa Fluor 488) as a marker for undifferentiated, basal keratinocytes. Arrowheads indicate the wound margin. Broken lines denote dermal–epidermal junction. Bar, 300 µm.
Cells of the wound margin become distributed on top of the EET by unstained in-migrating basal cells, indicating a lifting mechanism
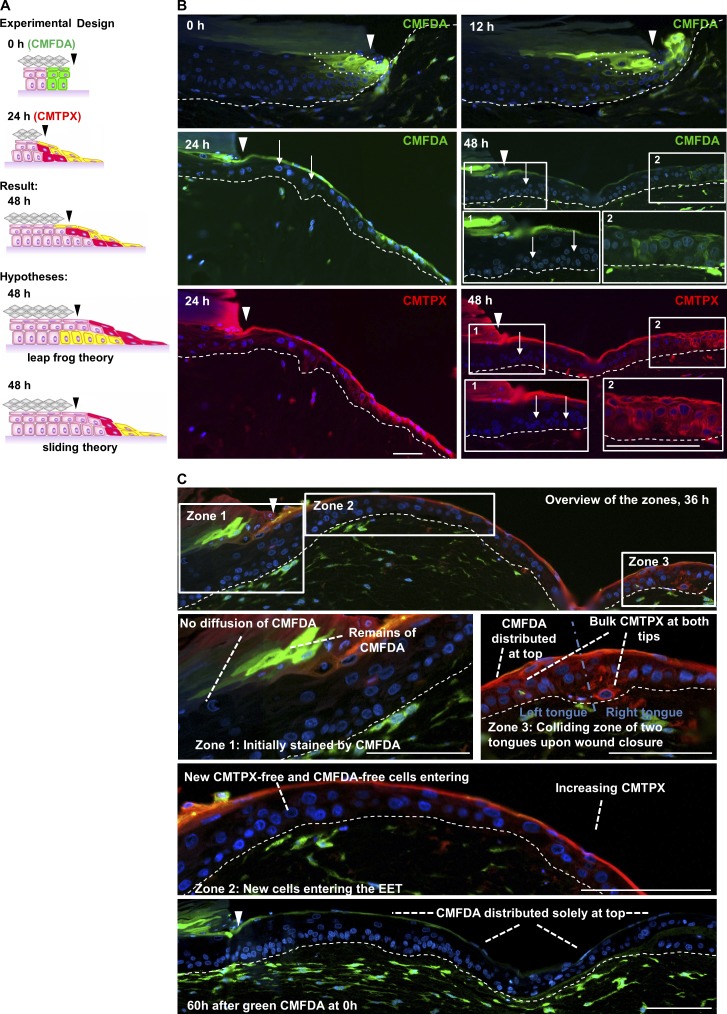
We then concentrated on the clarification of how the collectively migrating cells form a multilayered epithelium during EET extension. To this end, we designed a novel two-step staining using the cell trackers green 5-chloromethylfluorescein diacetate (CMFDA) and red CMTPX. Although green CMFDA was directly applied after wounding, red CMTPX was added 24 h after wounding into the punch (Fig. 7 A). This enabled us to track the spatial movement of a subpopulation of keratinocytes from the wound edge at day 0 (Fig. 7 B, green cells) or the further spatial movement of a completely stained EET at day 1 (Fig. 7 B, red cells) during wound closure. We showed that both cell trackers are not lost over a period of 3 d in primary human keratinocytes (Fig. S3 A). Furthermore, the cell trackers had no adverse effect on the vitality of keratinocytes in the organotypic wound models, proven by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) and TUNEL assays (Fig. S3, B and C).
Figure 7.
Capturing the spatial dynamics of the EET by applying a fluorescent time-lag double stain. (A) Experimental design of the fluorescent time-lag double stain experiment. CMFDA (green) was subsequently applied, whereas CMTPX (red) was applied 24 h after wounding, and the predicted outcomes hypothesized by the leap-frog and sliding theory were compared with the results of the experiment. (B) Sections of the EET showing the initial migration of basal CMFDA-labeled keratinocytes 0 and 12 h after wounding and the spatiotemporal dynamic behavior of CMFDA, CMTPX–labeled double-positive cells 24 and 48 h after wounding. The 0- and 24-h images shown here are presented again in Fig. 8 B to represent 1.5 and 53.3% reepithelialized wounds, respectively. The white arrows indicate CMFDA, CMTPX–negative keratinocytes, which have entered the EET from unstained regions. (C) Displays a representative EET 36 h after wounding, which can be separated into three different zones. Zone 1 shows the wound margin where labeled cells had started migration. Zone 2 displays the unstained migrating basal keratinocytes originated from tissue adjacent to the wound entering the EET. Zone 3 shows the collision of two EETs and the formation of a bulk. Arrowheads indicate the wound margin. Broken lines denote dermal–epidermal junction. The images shown are from a single representative experiment out of three repeats; 60 wound-healing models were analyzed in total. Bars, 100 µm.
At 0 h, the time-lag double stain resulted in the first two to three green vertical rows of CMFDA+ keratinocytes proximal to the wound site showing a homogeneous vertical staining in all layers of the section but a very reduced lateral spreading of the stain (Fig. 7 B). At 12 h, when keratinocytes had started to migrate, this cluster of CMFDA+ cells was stretched laterally by in-migrating CMFDA− basal cells originating from the surrounding unwounded tissue regions.
At 24 h after wounding, a long-stretched triangular EET had formed with a thin top layer of green CMFDA+ keratinocytes. This thin green CMFDA+ cell layer connected the granular cells of the intact epidermis with the leading edge keratinocytes of the EET (Fig. 7 B). In contrast, the basal cells of the EET were CMFDA−, indicating that unstained cells originating from the intact tissue had migrated into the wound region. This in turn means that while migrating into the wound, they have lifted the CMFDA+ cells and thereby formed a multilayered tongue.
At 48 h after wounding, also, the CMTPX+ cells that were stained 24 h after wounding now had distributed continuously on top of the EET, forming a shield completely covering the wound bed. In contrast, the cell layers below this shield were formed again by entirely unstained (CMFDA− and CMTPX−) cells. This showed for the third time (12, 24, and 48 h) how unstained basal cells migrated into the wound bed, thereby lifting leading stained cells. To exclude any artifacts caused by loss of staining, we performed a quantitative spatial analysis of CMTPX staining in EETs at three different time points (0, 12, and 24 h; Fig. S3, D–F). We thereby measured the lateral distribution of CMTPX and observed gradients with maximum staining intensity at the tip of the EET excluding a loss of the dye as a result of proliferation in the tip. The example of 36 h (Fig. 7 C) further illustrates the rearrangements of cells during wound closure when distinguishing three different zones of the EET. The viable layers of zone 1 (the wound margin) consist entirely of CMFDA− , CMTPX− cells. In contrast, CMFDA+ , CMTPX+ fibroblasts of the underlying dermal component serve as a positive staining control. In turn, highly differentiated CMFDA+ cells in zone 1 in close vicinity to CMFDA− cells negate diffusion artifacts. Zone 2 (the core EET) shows the accumulation of CMFDA+, CMTPX+ and CMFDA+, CMTPX− cells in the suprabasal and shield layer, having previously been lifted by unstained basal cells originating from the intact tissue surrounding the wound. Zone 3 (the collision zone) displays two EETs originating from the left and the right wound margin, forming a bulk of stained cells upon collision, resulting in a thickened epidermis in the collision zone. In conclusion, we showed that in epidermal wound healing, collective migration delivers cells to the tip of the EET, where they subsequently become lifted and connected to the extending shield of the neo-epidermis, thereby incrementally reconstructing a multilayered epithelium.
Computational modeling leads to the ESM
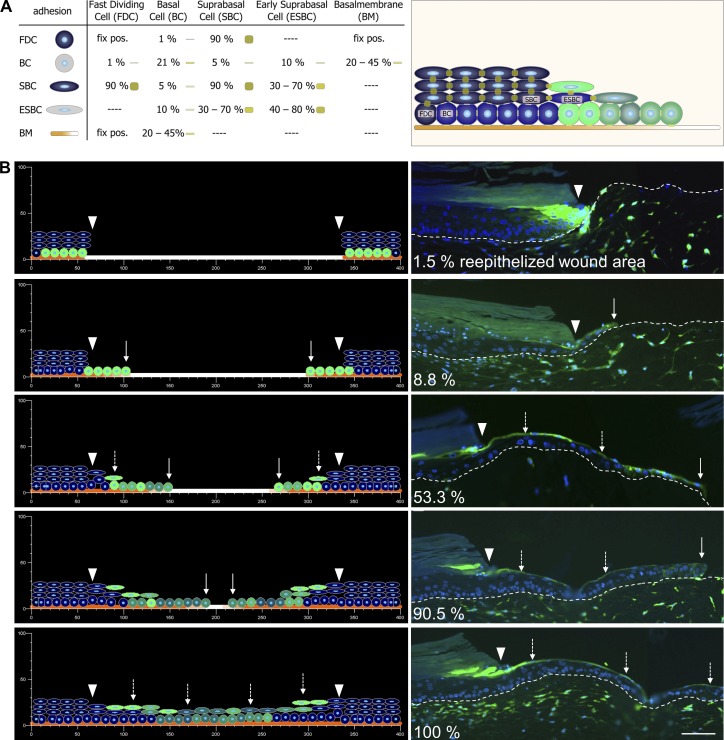
By integration of lifted CMFDA+ cells in the EET, an incrementally extending shield is created (see Fig. 10). We denote this the ESM. To test whether the ESM is capable of explaining the continuous elongation of the EET by collectively in-migrating basal cells, we developed a multicellular computational simulation of wound closure. We used our EPISIM platform (Grabe and Neuber, 2005; Sütterlin et al., 2009, 2013) to build and simulate the ESM with a computational model (see Materials and methods). This model contains four distinct cell types. (1) Fast dividing cells: fast dividing cells have a fixed position to limit basolateral cell spreading and increased proliferative activity to mimic the activated state of the keratinocytes under wound-healing conditions. (2) Basal cells: these cells were initially marked in green to simulate the cell tracker CMFDA and were placed on the basal membrane. (3) Suprabasal cells: suprabasal cells are part of the unwounded tissue region and function as an initial shield for the migratory basal cells. (4) Early suprabasal cells: if a basal cell is lifted from the basal into the suprabasal compartment, it differentiates into an early suprabasal cell. All simulated cells influence each other by exerting specific adhesion and intercellular pressure forces on neighboring cells (Fig. 8 A and Fig. S4). Generally, basal cells exert lower adhesion forces than early suprabasal cells or suprabasal cells, thereby mirroring the expression of adherens junctions (E- and P-cadherin), leading to a flexible migratory stratum basale. The expression of desmosomal cell–cell junctions (Dsg1) in turn characterizes the mechanically stable suprabasal cell layers. Impairment of active migration in the wound area is simulated with high basal adhesion (see Materials and methods). This reflects within the scope of our modeling approach the requirement of leading edge keratinocytes to adapt to the new substrate of the ECM by an integrin shift as well as the laminin-5 deposition of leading edge keratinocytes, resulting in an continuous buildup of the basal membrane.
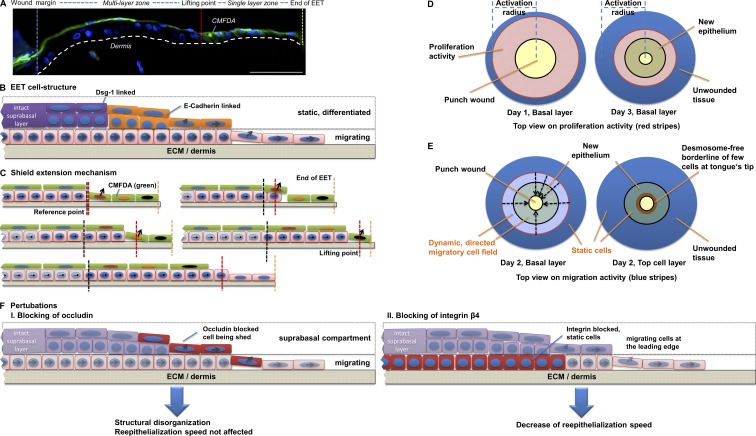
Figure 10.
The ESM is based on collectively migrating basal cells. (A) Results from the double fluorescent staining experiment show that stained cells always become distributed on top of the EET, which can be principally structured in a single layer and a multilayer zone. The broken line denotes dermal–epidermal junction. (B) Cells within the suprabasal compartments of the multilayer zone form mechanically stable tight junctions and desmosomes serving as a shield for the underlying wound bed. Because of its cell junction composition, this suprabasal layer is more static, whereas the underlying basal compartment exclusively expresses E- and P-cadherin, which can rapidly be remodeled, showing a dynamic migration behavior. (C) Because of the mechanically stable structure of the shield, extension can only be achieved by adding cells at the point at which the single layer zone connects to the multilayer zone (lifting point). At the lifting point, cells are transported upwards by following basal cells, whereas not yet lifted single-layer tip cells continue migration until they are lifted by the following cells. (D) Cells of the basal layer that supply the extending shield are in the beginning (day 1) produced in the intact unwounded tissue by a proliferative burst. With progressing wound closure (day 3), proliferation activity follows the EET to the center of the wound. (E) Viewed from top, dynamically moving cell fields can be identified characterized by directed cell migration. Although basal cells from the unwounded tissue show collective cell migration to actively supply the shield extension, static suprabasal cells shield the in-migrating basal cells by continuously building a mechanically stable barrier. (F) Depending on the contact point of perturbation, it results in a structural disorganization and shedding of occludin-blocked cells or in a decrease of reepithelialization speed if integrin β4 is blocked. Bar, 100 µm.
Figure 8.
Computational modeling of the ESM. (A) The EPISIM platform was used to build a CBM of the ESM. The model contains four distinct cell types: (1) fast dividing cells (FDC), (2) basal cells (BC), (3) suprabasal cells (SBC), and (4) early suprabasal cells (ESBC). All cells are placed on a basal membrane and exert specific adhesion forces. pos., position. (B) During simulation, keratinocytes built up the characteristic triangular structure of the EET, with CMFDA+ cells becoming distributed on top (dashed arrows), thereby reflecting the experimental results obtained by using the organotypic in vitro wound-healing model. Arrowheads indicate the wound margin, and white arrows show the leading edge of the EET. Broken lines denote dermal–epidermal junction. The 1.5 and 53.3% reepithelialized wound images are reproduced from Fig. 7 B (0 and 24 h, respectively). Bar, 100 µm.
In the simulation, we modeled cell lifting by (a) the pushing force from the proliferative compartment as well as (b) the adhesion force between the last shield cell and the newly lifted cell (Fig. 8 A). The simulation of our ESM model mirrored the cell behavior of migrating keratinocytes in our organotypic in vitro wound-healing model (Fig. 8 B, Video 1, and Video 2). In the early simulation phase, the EET is visible as a monolayer of green CMFDA+ basal keratinocytes migrating from the wound margin into the wound. This closely corresponds to the experimental situation in which the EET had covered 8% of the wound area. During simulation, cells build up the characteristic triangular structure of the EET, showing CMFDA+ cells at the tip and in the suprabasal layer of the extending tongue. Continuously, unstained basal cells migrate into the EET triggering the ESM and further driving the wound closure process. The cell distribution obtained at the end of the simulation was in accordance to the experimental results seen in our organotypic in vitro wound models. The simulated tissue kinetics proved the 2D as well as the 3D models capability to yield a fully reepithelialized and stratified neo-epidermis (Fig. S4 B).
Leader cells in the 3D situation express occludin whose blockage perturbs the ESM and results in shedding of bound cells
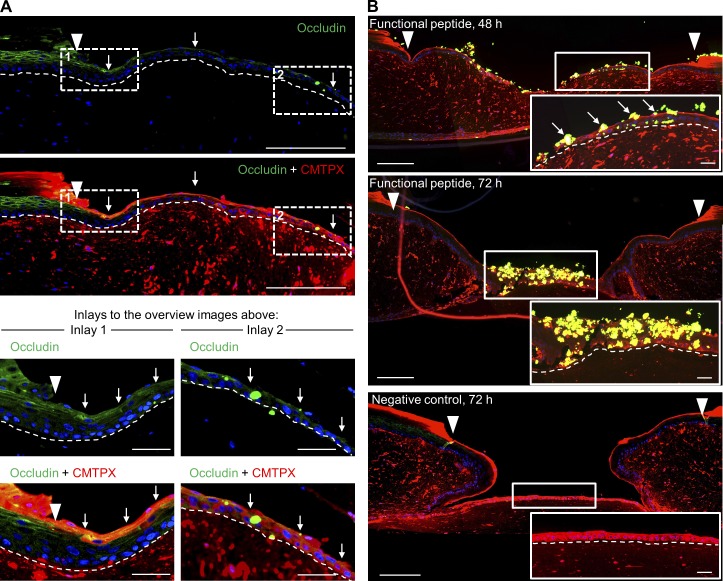
Our simulation pointed to two potential causes leading to lifting of cells: (1) the pushing force from the proliferative compartment as well as (2) cell junctions. Previously, it was shown that integrin blocking delays wound closure but does not lead to a spatially perturbed EET, indicating that probably cell junctions play a more crucial role in cell lifting (Egles et al., 2010). Our analysis of cell junctions showed that desmosomal junctions were not expressed at the tip of the EET, whereas adherens junctions were ubiquitously expressed throughout the tongue. In contrast to those junctional proteins, only occludin showed a colocalization with the cell trackers CMFDA and CMTPX (Fig. 9 A). To test whether occludin inhibition might be able to perturb the ESM, we blocked tight junction assembly during reepithelialization by using a characterized synthetic peptide (Wong and Gumbiner, 1997; Nusrat et al., 2005). This peptide is emulating the occludin loop B, whereas its scrambled version is known to still bind occludin, although with substantially less affinity. Functional and scrambled peptides tested in HaCaT cell culture confirmed their accumulation at the tight junction protein occludin (Fig. S5). 48 h after wounding, functional peptides accumulating at the leading edge and the monolayer zone of the EET demonstrated that leader cells present free occludin binding sites in the 3D situation before being included in the shield (Fig. 9 B). At 72 h, massive amounts of leader cells had been captured by the peptide and subsequently extruded. In contrast, the scrambled peptide with low occludin affinity did not completely abolish shield extension but led partly to incorporation of peptide-tagged leader cells into the extending tongue and to a small extent to the extrusion of cells. Unaffected parts of the EET as well as the EET of the negative control showed a smooth suprabasal layer with no evidence of cell shedding. Thus, we conclude that the ESM can be successfully perturbed using occludin peptides, pointing to a crucial role of occludin in wound closure.
Figure 9.
Tight junction perturbation using a synthetic peptide emulating occludin loop B. (A) Histological sections of representative EETs showing occludin CMTPX colocalization (white arrows) in the suprabasal compartment over the full length of the EET. (B) A synthetic functional peptide emulating occludin loop B was used to block tight junction assembly during tongue extension. The integration of the biotin-tagged peptide into the EET was detected by applying streptavidin-FITC (green). Occludin-blocked cells failed to integrate in the ESM and were shed during tongue extension (white arrows). Insets show magnified areas of the EET to illustrate the effect of the functional peptide as well as the effect of the negative control. Arrowheads indicate the wound margin. Broken lines denote dermal–epidermal junction. Bars: (overview screens) 250 µm; (inlays and insets) 50 µm.
Discussion
To derive a consistent theory of how the cellular processes of migration, proliferation, and differentiation are intertwined in skin wound healing, we established a technology pipeline allowing to analyze 3D tissue reorganization in organotypic in vitro wound models comprising a punch wound as well as an intact wound margin over a time period of ≤10 d.
In vivo–like organotypic in vitro wound-healing model
The basis of our study is formed by commercially available, organotypic skin cultures, developed and manufactured in a good manufacturing practice–approved process. In many publications regarding different molecular and morphological readouts, they have shown a biological response equivalent to human skin (Hayden et al., 2009) comprising cytokine secretion (Mallampati et al., 2010) as well as proliferative (Black et al., 2010), inflammatory, and metabolic markers (Hu et al., 2010). To further provide evidence of the in vivo relevance of our wound model, we measured the temporal profile of key cytokines regulating cell behavior and compared it with previously published clinical skin-wounding data. In human wound fluids, the highest cytokines determined were IL-6, IL-8, MCP-1, and G-CSF (Grimstad et al., 2011). In a forensic analysis of human skin tissue also IL-6 and IL-8 emerged as the most elevated cytokines (Takamiya et al., 2008). The same cytokines and growth factors have also been reported in wound fluids (Ono et al., 1995; Angel and Szabowski, 2002). Our results are in accordance to these literature as the proinflammatory cytokines IL-6, IL-8, and MCP-1 were found most abundant, showing a mean concentration of >20,000 pg/ml. IL-1, which showed a peak 1 d after wounding, is one of the key triggers in cytokine response (Werner et al., 2007). Our findings therefore point to the double paracrine cross talk between keratinocytes and fibroblasts in wound healing (Fig. 2) as proposed by Angel and Szabowski (2002). In brief, keratinocytes secrete IL-1α, which regulates components of the AP-1 transcription factor c-Jun and JunB in fibroblasts. This in turn activates AP-1–dependent target genes in fibroblasts, leading to a secretion of keratinocyte mitogens (e.g., G-CSF and IL-6; Angel and Szabowski, 2002; Florin et al., 2004).
Cell proliferation
Regarding the dimension of proliferation in the neo-epidermis, conflicting information has been reported (Jansson et al., 1996; Laplante et al., 2001; Patel et al., 2006; Garlick, 2007). Our results show proliferative activity of >8 mm around a 2-mm punch and that the cell supply of the extending shield is nearly exclusively generated in the intact unwounded tissue and not by tongue extension. Later, during wound closure, new cell production is translocated in form of a concentric wave toward the center of the punch wound (Fig. 3 D). Hyperproliferation or a proliferative burst at the wound margins has been described previously as an initially activating, positive feedback after wounding but not as a traveling wave (Viziam et al., 1964; Garlick, 2007). Moreover, we precisely quantitatively measured this proliferative burst and showed its temporal and spatial distribution throughout the EET and the intact tissue.
Collective migration
The cell junction expression patterns seen in our in vitro wound-healing model are in accordance to in vivo wound-healing studies (Malminen et al., 2003; Becker et al., 2012; Chavez et al., 2012). These patterns indicate a concentric field of basal keratinocytes collectively migrating toward the punch wound. During this migration, cells remain physically and functionally connected via E- and P-cadherin while showing a multicellular polarity (Friedl and Gilmour, 2009; Ilina and Friedl, 2009) of the cytoskeleton toward the punch inside the intact tissue as well as throughout the full length of the EET. Polarization was confirmed by computational measurement of nuclei rotation and displacement. The capability of classical cadherins to control nucleus position and cell polarity has been described earlier (Maniotis et al., 1997; Dupin et al., 2009). We further included Golgi stainings for final proof of cell axis rotation in the intact tissue. Thus, in accordance with literature (Maniotis et al., 1997; Friedl et al., 2011), our results show basal cells collectively migrating from unwounded regions toward the punch wound. The suprabasal compartment thereby provides a mechanically robust shield with the described cell junctions (see Results).
ESM for tongue assembly
So far in literature, three mechanisms have been proposed since the last 40 yr that try to explain the recreation of a multilayered epithelium: the leap-frog, the tractor-tread, and the Usui model (Krawczyk, 1971; Radice, 1980; Paladini et al., 1996; Woodley, 1996; Usui et al., 2005). Our results show that the EET always moves as a triangular multilayered epithelium with full height toward the former wound margin and with a single cell tip at the protruding end toward the wound bed. Keratinocyte differentiation markers demonstrated at no time point that differentiated keratinocytes got into contact with the ECM, thereby ruling out the leap-frog model. We then studied cellular movement of the EET by the fluorescent time-lag double stain showing that basal cells of a stained EET with progressing time always become distributed on top of the EET (Fig. 7, B and C). Our results, which are summarized in Fig. 10, lead to the ESM thus ruling out all other reepithelialization mechanisms so far proposed in literature. Instead, directly after wounding at 12 h, green CMFDA+ cells are pushed out of the intact wound margin by following basally located CMFDA− cells. At no point did we observe CMFDA− cells at the tip of the tongue. Thus, the CMFDA+ cells formed, in the end, an incrementally extending shield. At completion of wound closure, all CMFDA+ cells had ended up in the outmost cell layer (forming the shield) or still remained at the tongue’s tip for further migration, as they were not needed for shield extension yet. It is thereby remarkable that immediately after becoming part of the shield, keratinocytes became stretched, in which one single cell shielded frequently more than five keratinocytes. By this stretching capability, a relatively small reservoir of CMFDA+ cells from the wound edge can shield large neo-epidermal surfaces.
Multicellular systems biological simulation of the ESM
Our results show that wound healing is a higher level tissue function, which only results in an intact tissue again, if the individual cellular behavior is orchestrated in a coherent way. To theoretically test the ESM for its capability in providing correct reepithelialization and to identify potential regulators, we used a multicellular computational simulation. The mechanical parameters used in the simulation reflect the adhesion characteristics of cells in the in vitro wound model, in which basal cells showed low and suprabasal cell high adhesion forces. The biological motivation for proposing lower adhesion forces in basal cells compared with suprabasal cells in our in silico model is further supported by the fact that these cell types have different mechanical stabilities caused by specific characteristics of their cell–cell junctions. In brief, basal cells have small, less organized desmosomes with a low electron density (Green and Simpson, 2007), which are further destabilized because of the low Ca2+ concentration in the basal layer (Menon, 2002; Proksch et al., 2008). The major junction of basal cells are adherens junctions binding to actin filaments with a diameter of 7 nm and therefore exhibiting less mechanical strength. In addition, basal intermediate filaments (CK5/CK14) build tonofilaments, showing a loose formation with less mechanical stability (Moll et al., 2008). Furthermore, growth factors such as EGF lead to a disruption of hemidesmosomes as a result of a PKC-α–mediated phosphorylation of β4 integrins. These integrins dissemble from the hemidesmosomes and reassemble on focal contacts, thereby leading to an increased motility of basal cells under wound healing conditions (Rabinovitz et al., 1999; Litjens et al., 2006).
Suprabasal cells instead have large desmosomes with a high organization level and a high electron density (Green and Simpson, 2007). These desmosomes have an elevated strength caused by a high Ca2+ concentration in the suprabasal layer and are linked to tightly bundled intermediate filaments (CK1/CK10) with diameters of 10 nm, therefore exhibiting maximal mechanical strength (Menon, 2002; Niessen, 2007; Moll et al., 2008; Proksch et al., 2008).
Our results show that the identified components of the wound closure process play consistently together so that, starting from proliferation, cells are transported toward the wound edge and then are assembled in such a way that, according to the ESM, a novel 3D epidermis is constructed. Thereby, wound closure is achieved with an incrementally growing stable and robust shield, under which collective migration supplies novel cells for extending the tongue being in turn stepwise displaced toward the wound center.
For realizing the simulation, passive migration through displacement by proliferation was sufficient. We explicitly do not make further assumptions regarding the actual biomechanical mode of collective keratinocyte migration in silico as this is beyond the scope of this paper.
The different functions of the apical and the basal side of the tongue
The simulation of the emerging triangular form of the EET pointed to the fact that lifting of cells in the ESM toward a multilayered epithelium can be regulated either by the lateral or the apical side of the tongue. Egles et al. (2010) provided an example of perturbing lateral movement of the EET by blocking of integrin β4, which is binding to laminin-5 (laminin-332). Although their results showed a substantial slowdown in EET extension (Egles et al., 2010), it did not lead to the structural disturbance of the tongue. To test a perturbation of the apical side of an EET, we blocked occludin as an example of a tight junction protein by using a peptide emulating occludin loop B, resulting in extruded cells during the reepithelialization process. This proves that occludin binding sites are exposed by the tip cells and that the free transmembrane occludin binding sites are further essential for integrating the in-migrating basal cells into the extending shield. Furthermore, this points to a potential key role of occludin, as its biomechanical function within the EET could be the reason for its duality being involved in cell migration as well as in sealing the epidermis during wound healing (Niessen, 2007; Du et al., 2010).
In conclusion
By revealing the ESM, this work provides a consistent theoretical model of how reepithelialization actually occurs in healing skin wounds. This shows that wound closure is not a process only happening at the leading edge of migrating cells. Instead, closure and healing appear as higher-level processes of their own right, on a higher abstraction scale than individual cellular processes.
Materials and methods
Wound model
EFT cultures.
Human EFT in vitro skin cultures were obtained from MatTek Corporation. The cultures are derived from human neonatal foreskin tissue and consist of normal human epidermal keratinocytes and normal human dermal fibroblasts (NHDFs), which have been cultured to form a multilayered, highly differentiated model of the human dermis and epidermis. The cultivation was performed according to the manufacturer’s recommendation.
Preparation of cellular DEs.
For DEs, two different NHDF populations originally isolated from ≥50-yr-old female skin were used. Obtaining skin samples was approved by the Heidelberg Ethics Commission. The cells were grown in fibroblast culture medium, consisting of DMEM supplemented with 4.5 g/liter glucose, 100 U/ml penicillin, 100 mg/ml streptomycin, 250 ng/ml amphotericin B, and 20% fetal bovine serum at 37°C in a 5% CO2 humidified incubator and harvested by trypsinization. NHDFs were used within one to three passages. Cells were incorporated into a collagen gel prepared by addition to chilled, buffered, pH 7.0, rat tail collagen I solution (4 mg/ml; Serva). 2.5 ml of the solution was transferred to each well of a 6-well plate. Polymerized collagen gels formed after incubation at 37°C for 15 min in a 5% CO2 incubator. Once manufactured, the DEs were stored, immersed in fibroblast culture medium for 10 d at 37°C in a 10% CO2 incubator. During this time, the NHDF secrete additional ECM components for remodeling the collagen matrix and DE, contracting thereby ∼90%. Medium was changed every 2 d.
Fabrication of wound cultures.
Equilibrated EFT cultures were wounded by a circular biopsy punch (Stiefel). Depending on the respective analysis, wounding was achieved either by duplex concentric punching using a 2-mm biopsy punch (Stiefel) or by single punching using a 3-mm biopsy punch. To quantify reepithelialization and proliferation, the 2-mm punching strategy was chosen to generate high amount of data for ensuring in vivo–like wound model properties. To analyze keratinocyte migration, a 3-mm punch was opted to ensure larger wound space for tracking the newly forming EET. The cultures were connected on top to the prepared DE using 6 µl unpolymerized rat tail collagen I as “glue” and transferred into 12-well inserts (ThinCert; Greiner Bio-One). Cultures were incubated at 37°C for 5 min in 5% CO2 to allow polymerization of the collagen glue and terminal bonding of the DE with the EFT culture. The manufactured wound cultures were incubated at 37°C and 5% CO2 at the air–liquid interface in EFT-maintaining medium (MatTek Corporation). Half of the medium was replenished every 2 d. Wound cultures were harvested at the indicated time points and subsequently fixed in 4% buffered formalin for 6.5 h at RT.
Two-step staining procedure
Keratinocyte migration was observed using the fluorescent cell trackers green CMFDA and red CMTPX, which on cleavage of its acetates by cytosolic esterases produce bright fluorescence only in live cells. In total, 60 organotypic wound-healing models within three independent experiments were labeled stepwise by applying 4 µl of 25-µM CMFDA at time point 0 h or CMTPX at time point 24 h directly into the wound and incubation at 37°C for 45 min. The wound surface was washed three times with PBS, and fresh medium was added in each case. After staining, the wound cultures were incubated as long as indicated.
Histology and automated immunohistochemistry
Tissue samples were consecutively cut until the wound was microscopically visible. For immunohistochemistry, 3-µm sections were deparaffinized by reverse ethanol/xylene washes and rehydrated. The histology of the cultures was assessed by a standard hematoxylin and eosin stain.
For Immunofluorescent staining, sections were incubated overnight at 4°C with primary antibodies, washed 3× with TBS and incubated for 1 h at RT with the secondary antibody before mounting in fluorescent mounting medium (Dako). The following antibodies were used for immunofluorescence: mouse clone SPM181 anti-filaggrin (1:75), rabbit polyclonal anti–laminin 5 (1:1,000), and rabbit polyclonal anti–histone 3 phospho-S10 (1:1,500; Abcam); rabbit polyclonal anti–connexin 43 (1:300) and rabbit polyclonal anti–P-cadherin (1:200; Sigma-Aldrich); rabbit clone EP700Y anti–E-cadherin (1:100) and rabbit clone EP1612Y anti-CK14 (1:50; Epitomics); clone MIB-1 anti-Ki67 (1:50) and mouse clone HHF35 anti-actin (1:1,000; Dako); rabbit polyclonal anti-Dsg1 (1:200; Santa Cruz Biotechnology, Inc.); rabbit polyclonal anti-occludin (1:75; Invitrogen); clone VIK10 anti-CK10 (1:100; Acris); mouse clone Sy5 anti-involucrin (1:400; Novocastra). Alexa Fluor 488–conjugated goat anti–rabbit IgG and Alexa Fluor 594–conjugated donkey anti–mouse IgG (1:100; Invitrogen) were used as secondary antibodies. Fluorescent double staining was performed simultaneously. For automated immunohistochemistry using the Bond-Max system (Leica), sections were incubated for 30 min at RT with primary antibodies, washed 3× with washing solution provided by the manufacturer, and incubated for 8 min at RT with a poly-HRP anti–mouse antibody. The following primary antibodies were used for immunohistochemistry: rabbit polyclonal anti-GOLGA2 (1:500; Sigma-Aldrich) and clone MIB-1 anti-Ki67 (1:100; Dako). At least two independent consecutive sections of all cultures were stained and analyzed.
Virtual microscopy and image analysis
Sections were scanned and digitized using a virtual microscopy platform (NanoZoomer 2.0-HT; Hamamatsu Photonics). With this system, virtual images of whole microscopic tissue sections were obtained. Minor fluctuations in fluorescence intensity are sometimes apparent in the concatenated images but do not reflect differences in image acquisition parameters between sections. The imaging systems contains three 4,096 × 64–pixel time delays and integration charge-coupled device sensors (cell size 8 × 8 µm) and a 20× objective lens, NA 0.75. Using the NDP.scan software (version 2.5; Hamamatsu Photonics), slides were scanned at RT in three z layers with a spacing of 2 µm each and a resolution of 460 nm/pixel (20×). Images were analyzed manually as well as automatically by using the Visiomorph image processing software version 3.6.5.0 (Visiopharm). Microscopic capturing of whole tissue sections allows for large-scale histological evaluations with high precision across the complete wound and its outer regions.
Quantification of reepithelialization.
Tissue samples were consecutively cut until the wound was microscopically visible. This sectioning plane was defined as layer 1, and 10 consecutive sections of 3 µm each were cut. Based on that first layer, further defined sections in layer 2 (150 µm) and in layer 3 (300 µm) were also cut and stored (Fig. 1 D). As equal wound diameters are a prerequisite for sufficient and comparable reepithelialization analysis, we only considered sections having a diameter of 1,000–1,500 µm indicated by the wound margin. Reepithelialization was measured as the length of the EET (lEET) in micrometers per day. The maximal wound diameter (lWound) is indicated by the distance of the unwounded stratum corneum. Relative wound closure is obtained by dividing lEET by lWound (Fig. 1 G). As the circular punch wound is sectioned, the observed wound diameter lWound varies according to cutting depth. EET measurements were performed using the NanoZoomer virtual slide viewer software.
Quantification of epidermal thickness.
For quantification of the epidermal thickness, consecutive sections of all three section planes (see Quantification of reepithelialization) within the wound region were used. The basal and suprabasal compartment were defined on the basis of the differentiation markers CK14 and CK10. For thickness calculation, the area of the respective compartment was quantified using Visiomorph image processing software and divided by the length of the basal lamina.
Quantification of proliferation (EPI).
To analyze the general tissue proliferation in response to wounding, we quantified consecutive sections of all three section planes (see Quantification of reepithelialization) within the wound. For detecting and distinguishing negative and DAB-stained Ki67-expressing cells, appropriate algorithms were developed and implemented using the Visiomorph software. In brief, a preprocessing step using a median filter and a Bayesian classifier were applied for nuclei detection. The following postprocessing steps have been performed within the hematoxylin channel to identify nuclei: (a) Small particles beyond 3-pixel particle size have been removed. (b) Structures showing a circularity value of <2 were identified as potential nuclei. (c) Structures having an area >60 pixels were marked as potentially conglomerated nuclei and treated with an eroding step of 2 pixels to separate these objects. (iv) An additional separate-object step was applied to insufficiently separated conglomerated nuclei. (v) Separated conglomerates were revised and again tested for size and circularity, thereby removing objects showing a circularity >2.5 or a size <4 pixels. Verified nuclei were identified as proliferative if the Ki67-indicating DAB-staining was in a 1-pixel distance to hematoxylin-indicated nuclei. To calculate the EPI, the amount of Ki67-expressing basal keratinocytes was divided by the whole number of basal keratinocytes, to determine the percentage of proliferating cells as an indicator for proliferative activity. Furthermore, the total number of Ki67+ cells was determined. For this purpose, we harvested 46 tissue samples and determined Ki67 by analyzing 113 tissue sections in total. For spatial analysis of the tissue sections, virtual slides were separated into 10 different regions as follows: The wound area was separated into four regions (Fig. 3 A, d–g) of equal size, whereas the unwounded tissue surrounding the wound was separated into six regions of equal size, three regions (Fig. 3 A, a–c and h–j) of each side of the wound area (Fig. 3 A). The EPI and the total number of Ki67+ cells were calculated independently for each region (day 1–7).
Cell polarization
To assess nuclear shape, we separated the cultures into six equal regions (R1–R6) and analyzed every region independently. The epithelial and basal regions were manually marked using roipoly function in MATLAB (MathWorks, Inc.; for source code see Supplemental material), and the nuclei in the respective regions were detected in the DAPI channel. After initial cleaning and intensity adjustments, the nuclei were detected by stepwise thresholding. The detected nuclei clusters were further filtered by size and split using a watershed method with local maxima as seed points. Ringlike stained nuclei were excluded from the watershed procedure. Finally, valid nuclei were selected based on region properties such as size and solidity. To show the tilt in the nuclei with respect to the basal lamina, the angle of major axis was calculated. The orientation angle of a nucleus is measure of the angle (in degrees ranging from 90 to 0°) between the x axis (basal lamina) and the major axis of the ellipse that has the same second moments as the nucleus (Fig. 4 A, red line in nuclei scheme). The regions of eight samples were used for the calculations, and in total, 2,340 basal cells were analyzed.
Multiplex analysis of cytokine concentrations
Multiplex analysis of cytokine concentrations were conducted using the Luminex 100 platform built on xMAP technology (Luminex Corp.). The principle of this assay is similar to a capture sandwich immunoassay: By coupling an antibody directed against the target protein to internally color-coded microspheres, the target protein is bound to the individual microsphere and detected by a secondary biotinylated antibody. Streptavidin-phycoerythrin is then used to detect secondary antibodies. Based on this principle, the technology enables measuring of multiple total target proteins within one well. Supernatants of wounded and unwounded organotypic cultures before (day 0) as well as 1 and 3 d after wounding were collected, and concentrations of the following cytokines were analyzed: IL-1α, IL-3, IL-6, IL-8, MCP-1, and G-CSF.
Computational modeling using the EPISIM platform
Graphical cell behavioral models (CBMs) are built with the EPISIM graphical modeling system. The graphical modeling system allows formulating CBMs as graphical process diagrams. Such a process diagram can include deterministic as well as stochastic model elements (Sütterlin et al., 2013). Thus, pure deterministic, pure stochastic, and also hybrid models can be realized. A graphical CBM is automatically translated in highly efficient executable code by the EPISIM code generator. This code is then loaded by the EPSIM simulation environment, conducting a multiagent-based multicellular tissue simulation based on the translated executable CBM (Sütterlin et al., 2009). The computational model of the ESM, the EPISIM modeler, and the EPISIM simulator can be downloaded by using the following link: http://www.tiga.uni-hd.de/downloads.html.
Cell center–based biomechanical model.
Beside the CBM, such a tissue simulation is composed of a biomechanical model (BM). The BM is an off-lattice, center-based model, allowing cells to move freely in space. Cells are modeled as discrete objects, and all occurring forces act on a cell’s center of mass (center based; Dallon, 2007; Drasdo, 2007). In such a model, cells are commonly represented by circles and spheres (Walker et al., 2004; Galle et al., 2005; Schaller and Meyer-Hermann, 2007; Adra et al., 2010) or ellipses and ellipsoids (Palsson and Othmer, 2000; Dallon and Othmer, 2004). We developed a 2D as well as a 3D cell center–based BM with an elliptical cell shape in 2D and an ellipsoidal cell shape in 3D. As depicted in Fig. S4 A, the model aims at optimizing the distance between a cell c and its neighboring cells This optimization is performed for all cells in the multicellular tissue simulation and results in passive migration of the cells. A cell’s position is equivalent to its center of mass, r. The basal membrane is discretized into sections of equal length. The center of such a section is denoted by Fig. S4 A shows the optimal distance balancing between two consecutive simulation steps t − 1 and t, considering cell c colored in red. The distance balancing involves the calculation of intercellular pressure (see cell ) as well as cell–cell adhesion (see cells and ) and finally adhesion to the basal membrane. The calculated balance of cell distances is perturbed by cell division. A proliferating cell grows and compresses its neighboring cells. These cells in turn passively migrate away from the dividing cell because of this intercellular pressure. Further parameters of the BM are listed in Fig. 8 A.
Optimal distance calculation.
The optimal distance between two elliptical or ellipsoidal cells is the target value of the cell distance balancing process, calculated over multiple simulation steps. An ellipse’s semimajor axis is denoted by a, and the semiminor axis is denoted by b. Accordingly, the semiprincipal axes of the ellipsoid are a, b, and c. An ellipse’s or ellipsoid’s axes are aligned with the axes of the used Cartesian coordinate system.
The calculation of the optimal distance between two ellipses as well as two ellipsoids follows the same scheme: (a) calculation of the line connecting cell centers (of mass) rc and rn as well as the same line in the opposite direction (rn and rc), (b) calculation of the intersection points between these lines with the ellipses or ellipsoids, and (c) summation of the length of the line segments between a cell’s center and the according intersection point. Consequently, the optimal distance between two cells represented by ellipses located at rc and rn is
Accordingly, the optimal distance between two ellipsoidal cells is
Intercellular pressure and cell–cell adhesion.
The optimal distance is used to calculate the intercellular pressure Fpr(rc, rn) between overlapping cells:
with vnc = rc − rn. The factor δol (0 < δol ≤ 1) modulates the allowed overlap between two adjacent cells and, by that, a cell’s compressibility. We used δol = 0.95 corresponding to an allowed cell overlap of 5%.
The adhesive force Fadh(rc, rn) between adjacent cells is
| (1) |
Thereby, cell–cell adhesion is limited to cells whose distance is below δadh × dopt(rc, rn), with δadh = 1.25. Intercellular pressure Fpr(rc, rn) and cell–cell adhesion Fadh(rc, rn) are used to calculate a cell movement vector:
| (2) |
The adhesion coefficient adhkcn can be interpreted as a linear spring constant modulating the strength of the adhesive force between two cells. The adhesion coefficient can be used to model different cell–cell junctions, such as integrins, desmosomes, or tight junctions.
For each adjacent cell, a movement vector m(rc, rn) is calculated. All vectors are finally summed and lead to a cell’s final position.
Adhesion to basal membrane.
The basal membrane is discretized into segments of equal length (10 µm per segment) to model basal adhesion. A segment’s center is denoted by rbm. The adhesive force Fadh(rc, rn) and the cell movement vector m(rc, rbm) are calculated according to Eqs. 1 and 2. The used optimal distance between a cell and the basal membrane is for elliptical cells
and for ellipsoidal cells
The strength of the cell junctions to the basal membrane and the adhesion, respectively, depends on the adhesion coefficient adhkcn and the cell contact time tcon_bm. The cell contact time corresponds to the period of time a particular basal membrane segment (BMS) was covered by a cell. The higher the cell contact time, the darker the coloring of a segment (Fig. S4 A). Moreover, the lower the contact time, the less ECM is assumed to exist on this BMS. The absence of ECM, in turn, hinders active cell migration. It is a property of the chosen modeling approach that cells can migrate only passively. Because of this limitation to passive migration, the inability of active cell migration is mimicked by high adhesion on the BMSs with low ECM and accordingly low tcon_bm values. This high adhesion biologically corresponds to low adhesion and to the force feedback of cells that cannot actively migrate. We therefore define a high basal membrane adhesion factor adhkc_bm_hi. The effective adhesion coefficient reflecting the ECM level on a particular BMS is
| (3) |
Threshold Tcon_bm is the contact time assumed for reaching the maximum ECM level on a particular BMS. The range of the effective adhesion coefficient adhk′c_bm according to Eq. 3 is adhkc_bm ≤ adhk′c_bm ≤ adhkc_bm_hi ×; adhkc_bm, with adhkc_bm_hi = 2.5 and Tcon_bm = 1,050 (simulation steps corresponding to 11.5 h real time).
Peptide perturbation
For tight junction perturbation, a functional and a scrambled peptide were used showing the following amino acid sequences: Biotin-SQIYALCNQFYTPAATGLYVD-NH2 (functional peptide) and Biotin-TSPYAQIYLANFQDTALGYCV-NH2 (scrambled peptide). For 2D experiments, HaCaTs were cultured using culture slides until they reached a confluency of 70–80%. Cells were washed with HBSS−/−, and tight junctions were disrupted by using a Ca2+ switch protocol according to Nusrat et al. (2005). Peptides having a concentration of 200 µM were applied, and cells were incubated at 37°C for 5 and 24 h. Organotypic cultures were wounded, and 4 µl peptide solution having a concentration of 400 µM was applied every hour directly into the wound for a period of 6 h.
Online supplemental material
Fig. S1 shows the comparison of the EPI to the mitotic cell fraction and the morphometric analysis of the epidermal thickness during reepithelialization. Fig. S2 shows the Golgi positioning in polarized migrating cells. Fig. S3 shows the persistence of the cell trackers CMFDA and CMTPX in primary human keratinocytes and their influence on the viability of organotypic cultures. Fig. S4 shows the in silico biomechanical cell model of the ESM. Fig. S5 shows the characterization of functional and scrambled peptides emulating occludin loop B on primary keratinocytes and organotypic cultures. Video 1 shows the 2D simulation of the ESM. Video 2 shows the 3D simulation of the ESM. The MATLAB source code of the major axis analysis is provided online as a Word document. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212020/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201212020.dv.
Supplementary Material
Acknowledgments
We would like to thank Hamamatsu Photonics, Japan and Germany, for equipment and advice and Thermo Fisher Scientific for providing the histological equipment.
This work was supported by the German Federal Ministry of Education and Research Systems Biology programs FORSYS (Research Units for Systems Biology), 0315263, and MEDSYS (Medical Systems Biology), 0315401B.
Footnotes
Abbreviations used in this paper:
- BM
- biomechanical model
- BMS
- basal membrane segment
- CBM
- cell behavioral model
- CMFDA
- 5-chloromethylfluorescein diacetate
- DE
- dermal equivalent
- EET
- extending epidermal tongue
- EFT
- epidermal full thickness
- EPI
- epidermal proliferation index
- ESM
- extending shield mechanism
- G-CSF
- granulocyte colony-stimulating factor
- NHDF
- normal human dermal fibroblast
References
- Adra S., Sun T., MacNeil S., Holcombe M., Smallwood R. 2010. Development of a three dimensional multiscale computational model of the human epidermis. PLoS ONE. 5:e8511 10.1371/journal.pone.0008511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Szabowski A. 2002. Function of AP-1 target genes in mesenchymal-epithelial cross-talk in skin. Biochem. Pharmacol. 64:949–956 10.1016/S0006-2952(02)01158-9 [DOI] [PubMed] [Google Scholar]
- Becker D.L., Thrasivoulou C., Phillips A.R.J. 2012. Connexins in wound healing; perspectives in diabetic patients. Biochim. Biophys. Acta. 1818:2068–2075 10.1016/j.bbamem.2011.11.017 [DOI] [PubMed] [Google Scholar]
- Black A.T., Hayden P.J., Casillas R.P., Heck D.E., Gerecke D.R., Sinko P.J., Laskin D.L., Laskin J.D. 2010. Expression of proliferative and inflammatory markers in a full-thickness human skin equivalent following exposure to the model sulfur mustard vesicant, 2-chloroethyl ethyl sulfide. Toxicol. Appl. Pharmacol. 249:178–187 10.1016/j.taap.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxman I.L., Ruwhof C., Boerman O.C., Löwik C.W., Ponec M. 1996. Role of fibroblasts in the regulation of proinflammatory interleukin IL-1, IL-6 and IL-8 levels induced by keratinocyte-derived IL-1. Arch. Dermatol. Res. 288:391–398 10.1007/BF02507108 [DOI] [PubMed] [Google Scholar]
- Chavez M.G., Buhr C.A., Petrie W.K., Wandinger-Ness A., Kusewitt D.F., Hudson L.G. 2012. Differential downregulation of E-cadherin and desmoglein by epidermal growth factor. Dermatol. Res. Pract. 2012:309587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallon J.C. 2007. Models with lattice-free center-based cells interacting with continuum environment variables. Single-Cell-Based Models in Biology and Medicine. Anderson A.R.A., Chaplain M.A.J., Rejniak K.A., Birkhäuser, Basel, Switzerland/Boston: 197–219 [Google Scholar]
- Dallon J.C., Othmer H.G. 2004. How cellular movement determines the collective force generated by the Dictyostelium discoideum slug. J. Theor. Biol. 231:203–222 10.1016/j.jtbi.2004.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri G.D., Griffin J.D. 1991. Granulocyte colony-stimulating factor and its receptor. Blood. 78:2791–2808 [PubMed] [Google Scholar]
- Drasdo D. 2007. Center-based single-cell models: An approach to multi-cellular organization based on a conceptual analogy to colloidal particles. Single-Cell-Based Models in Biology and Medicine. Anderson A.R.A., Chaplain M.A.J., Rejniak K.A., Birkhäuser, Basel, Switzerland/Boston: 171–196 [Google Scholar]
- Du D., Xu F., Yu L., Zhang C., Lu X., Yuan H., Huang Q., Zhang F., Bao H., Jia L., et al. 2010. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Dev. Cell. 18:52–63 10.1016/j.devcel.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Dupin I., Camand E., Etienne-Manneville S. 2009. Classical cadherins control nucleus and centrosome position and cell polarity. J. Cell Biol. 185:779–786 10.1083/jcb.200812034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egles C., Huet H.A., Dogan F., Cho S., Dong S., Smith A., Knight E.B., McLachlan K.R., Garlick J.A. 2010. Integrin-blocking antibodies delay keratinocyte re-epithelialization in a human three-dimensional wound healing model. PLoS ONE. 5:e10528 10.1371/journal.pone.0010528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui R., Fenteany G. 2005. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell Sci. 118:51–63 10.1242/jcs.01577 [DOI] [PubMed] [Google Scholar]
- Florin L., Hummerich L., Dittrich B.T., Kokocinski F., Wrobel G., Gack S., Schorpp-Kistner M., Werner S., Hahn M., Lichter P., et al. 2004. Identification of novel AP-1 target genes in fibroblasts regulated during cutaneous wound healing. Oncogene. 23:7005–7017 10.1038/sj.onc.1207938 [DOI] [PubMed] [Google Scholar]
- Friedl P., Gilmour D. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Friedl P., Wolf K., Lammerding J. 2011. Nuclear mechanics during cell migration. Curr. Opin. Cell Biol. 23:55–64 10.1016/j.ceb.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle J., Loeffler M., Drasdo D. 2005. Modeling the effect of deregulated proliferation and apoptosis on the growth dynamics of epithelial cell populations in vitro. Biophys. J. 88:62–75 10.1529/biophysj.104.041459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlick J.A. 2007. Engineering skin to study human disease—tissue models for cancer biology and wound repair. Adv. Biochem. Eng. Biotechnol. 103:207–239 [DOI] [PubMed] [Google Scholar]
- Gillitzer R., Goebeler M. 2001. Chemokines in cutaneous wound healing. J. Leukoc. Biol. 69:513–521 [PubMed] [Google Scholar]
- Grabe N., Neuber K. 2005. A multicellular systems biology model predicts epidermal morphology, kinetics and Ca2+ flow. Bioinformatics. 21:3541–3547 10.1093/bioinformatics/bti585 [DOI] [PubMed] [Google Scholar]
- Green K.J., Simpson C.L. 2007. Desmosomes: new perspectives on a classic. J. Invest. Dermatol. 127:2499–2515 10.1038/sj.jid.5701015 [DOI] [PubMed] [Google Scholar]
- Grimstad Ø., Sandanger Ø., Ryan L., Otterdal K., Damaas J.K., Pukstad B., Espevik T. 2011. Cellular sources and inducers of cytokines present in acute wound fluid. Wound Repair Regen. 19:337–347 10.1111/j.1524-475X.2011.00668.x [DOI] [PubMed] [Google Scholar]
- Grossman R.M., Krueger J., Yourish D., Granelli-Piperno A., Murphy D.P., May L.T., Kupper T.S., Sehgal P.B., Gottlieb A.B. 1989. Interleukin 6 is expressed in high levels in psoriatic skin and stimulates proliferation of cultured human keratinocytes. Proc. Natl. Acad. Sci. USA. 86:6367–6371 10.1073/pnas.86.16.6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner G.C., Werner S., Barrandon Y., Longaker M.T. 2008. Wound repair and regeneration. Nature. 453:314–321 10.1038/nature07039 [DOI] [PubMed] [Google Scholar]
- Hayden P.J., Petrali J.P., Stolper G., Hamilton T.A., Jackson G.R., Jr, Wertz P.W., Ito S., Smith W.J., Klausner M. 2009. Microvesicating effects of sulfur mustard on an in vitro human skin model. Toxicol. In Vitro. 23:1396–1405 10.1016/j.tiv.2009.07.021 [DOI] [PubMed] [Google Scholar]
- Hu T., Khambatta Z.S., Hayden P.J., Bolmarcich J., Binder R.L., Robinson M.K., Carr G.J., Tiesman J.P., Jarrold B.B., Osborne R., et al. 2010. Xenobiotic metabolism gene expression in the EpiDerm in vitro 3D human epidermis model compared to human skin. Toxicol. In Vitro. 24:1450–1463 10.1016/j.tiv.2010.03.013 [DOI] [PubMed] [Google Scholar]
- Ilina O., Friedl P. 2009. Mechanisms of collective cell migration at a glance. J. Cell Sci. 122:3203–3208 10.1242/jcs.036525 [DOI] [PubMed] [Google Scholar]
- Jansson K., Kratz G., Haegerstrand A. 1996. Characterization of a new in vitro model for studies of reepithelialization in human partial thickness wounds. In Vitro Cell. Dev. Biol. Anim. 32:534–540 10.1007/BF02722980 [DOI] [PubMed] [Google Scholar]
- Kaushansky K., Lin N., Adamson J.W. 1988. Interleukin 1 stimulates fibroblasts to synthesize granulocyte-macrophage and granulocyte colony-stimulating factors. Mechanism for the hematopoietic response to inflammation. J. Clin. Invest. 81:92–97 10.1172/JCI113316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada A., Hiruma M., Noguchi H., Ishibashi A., Motoyoshi K., Kawada I. 1997. Granulocyte and macrophage colony-stimulating factors stimulate proliferation of human keratinocytes. Arch. Dermatol. Res. 289:600–602 10.1007/s004030050246 [DOI] [PubMed] [Google Scholar]
- Krawczyk W.S. 1971. A pattern of epidermal cell migration during wound healing. J. Cell Biol. 49:247–263 10.1083/jcb.49.2.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante A.F., Germain L., Auger F.A., Moulin V. 2001. Mechanisms of wound reepithelialization: hints from a tissue-engineered reconstructed skin to long-standing questions. FASEB J. 15:2377–2389 10.1096/fj.01-0250com [DOI] [PubMed] [Google Scholar]
- Liang C.C., Park A.Y., Guan J.L. 2007. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2:329–333 10.1038/nprot.2007.30 [DOI] [PubMed] [Google Scholar]
- Litjens S.H.M., de Pereda J.M., Sonnenberg A. 2006. Current insights into the formation and breakdown of hemidesmosomes. Trends Cell Biol. 16:376–383 10.1016/j.tcb.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Mallampati R., Patlolla R.R., Agarwal S., Babu R.J., Hayden P., Klausner M., Singh M.S. 2010. Evaluation of EpiDerm full thickness-300 (EFT-300) as an in vitro model for skin irritation: studies on aliphatic hydrocarbons. Toxicol. In Vitro. 24:669–676 10.1016/j.tiv.2009.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malminen M., Koivukangas V., Peltonen J., Karvonen S.L., Oikarinen A., Peltonen S. 2003. Immunohistological distribution of the tight junction components ZO-1 and occludin in regenerating human epidermis. Br. J. Dermatol. 149:255–260 10.1046/j.1365-2133.2003.05438.x [DOI] [PubMed] [Google Scholar]
- Maniotis A.J., Chen C.S., Ingber D.E. 1997. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA. 94:849–854 10.1073/pnas.94.3.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansbridge J.N., Liu K., Pinney R.E., Patch R., Ratcliffe A., Naughton G.K. 1999. Growth factors secreted by fibroblasts: role in healing diabetic foot ulcers. Diabetes Obes. Metab. 1:265–279 10.1046/j.1463-1326.1999.00032.x [DOI] [PubMed] [Google Scholar]
- Martin P. 1997. Wound healing—aiming for perfect skin regeneration. Science. 276:75–81 10.1126/science.276.5309.75 [DOI] [PubMed] [Google Scholar]
- McKay I.A., Leigh I.M. 1991. Epidermal cytokines and their roles in cutaneous wound healing. Br. J. Dermatol. 124:513–518 10.1111/j.1365-2133.1991.tb04942.x [DOI] [PubMed] [Google Scholar]
- Menon G.K. 2002. New insights into skin structure: scratching the surface. Adv. Drug Deliv. Rev. 54(Suppl. 1):S3–S17 10.1016/S0169-409X(02)00121-7 [DOI] [PubMed] [Google Scholar]
- Moll R., Divo M., Langbein L. 2008. The human keratins: biology and pathology. Histochem. Cell Biol. 129:705–733 10.1007/s00418-008-0435-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M.M., Fusenig N.E. 1999. Constitutive expression of G-CSF and GM-CSF in human skin carcinoma cells with functional consequence for tumor progression. Int. J. Cancer. 83:780–789 [DOI] [PubMed] [Google Scholar]
- Niessen C.M. 2007. Tight junctions/adherens junctions: basic structure and function. J. Invest. Dermatol. 127:2525–2532 10.1038/sj.jid.5700865 [DOI] [PubMed] [Google Scholar]
- Noble D. 2006. The Music of Life: Biology Beyond Genes. First edition Oxford University Press, New York: 176 pp [Google Scholar]
- Nusrat A., Brown G.T., Tom J., Drake A., Bui T.T.T., Quan C., Mrsny R.J. 2005. Multiple protein interactions involving proposed extracellular loop domains of the tight junction protein occludin. Mol. Biol. Cell. 16:1725–1734 10.1091/mbc.E04-06-0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono I., Gunji H., Zhang J.Z., Maruyama K., Kaneko F. 1995. Studies on cytokines related to wound healing in donor site wound fluid. J. Dermatol. Sci. 10:241–245 10.1016/0923-1811(95)00454-Z [DOI] [PubMed] [Google Scholar]
- Paladini R.D., Takahashi K., Bravo N.S., Coulombe P.A. 1996. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J. Cell Biol. 132:381–397 10.1083/jcb.132.3.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson E., Othmer H.G. 2000. A model for individual and collective cell movement in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 97:10448–10453 10.1073/pnas.97.19.10448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G.K., Wilson C.H., Harding K.G., Finlay A.Y., Bowden P.E. 2006. Numerous keratinocyte subtypes involved in wound re-epithelialization. J. Invest. Dermatol. 126:497–502 10.1038/sj.jid.5700101 [DOI] [PubMed] [Google Scholar]
- Proksch E., Brandner J.M., Jensen J.-M. 2008. The skin: an indispensable barrier. Exp. Dermatol. 17:1063–1072 10.1111/j.1600-0625.2008.00786.x [DOI] [PubMed] [Google Scholar]
- Rabinovitz I., Toker A., Mercurio A.M. 1999. Protein kinase C–dependent mobilization of the α6β4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146:1147–1160 10.1083/jcb.146.5.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice G.P. 1980. The spreading of epithelial cells during wound closure in Xenopus larvae. Dev. Biol. 76:26–46 10.1016/0012-1606(80)90360-7 [DOI] [PubMed] [Google Scholar]
- Rennekampff H.O., Hansbrough J.F., Kiessig V., Doré C., Sticherling M., Schröder J.M. 2000. Bioactive interleukin-8 is expressed in wounds and enhances wound healing. J. Surg. Res. 93:41–54 10.1006/jsre.2000.5892 [DOI] [PubMed] [Google Scholar]
- Schaller G., Meyer-Hermann M. 2007. A modelling approach towards epidermal homoeostasis control. J. Theor. Biol. 247:554–573 10.1016/j.jtbi.2007.03.023 [DOI] [PubMed] [Google Scholar]
- Schroeder J.T., Chichester K.L., Bieneman A.P. 2009. Human basophils secrete IL-3: evidence of autocrine priming for phenotypic and functional responses in allergic disease. J. Immunol. 182:2432–2438 10.4049/jimmunol.0801782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A.J., Clark R.A.F. 1999. Cutaneous wound healing. N. Engl. J. Med. 341:738–746 10.1056/NEJM199909023411006 [DOI] [PubMed] [Google Scholar]
- Soderholm J., Heald R. 2005. Scratch n’ screen for inhibitors of cell migration. Chem. Biol. 12:263–265 10.1016/j.chembiol.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Sütterlin T., Huber S., Dickhaus H., Grabe N. 2009. Modeling multi-cellular behavior in epidermal tissue homeostasis via finite state machines in multi-agent systems. Bioinformatics. 25:2057–2063 10.1093/bioinformatics/btp361 [DOI] [PubMed] [Google Scholar]
- Sütterlin T., Kolb C., Dickhaus H., Jäger D., Grabe N. 2013. Bridging the scales: semantic integration of quantitative SBML in graphical multi-cellular models and simulations with EPISIM and COPASI. Bioinformatics. 29:223–229 10.1093/bioinformatics/bts659 [DOI] [PubMed] [Google Scholar]
- Takamiya M., Fujita S., Saigusa K., Aoki Y. 2008. Simultaneous detection of eight cytokines in human dermal wounds with a multiplex bead-based immunoassay for wound age estimation. Int. J. Legal Med. 122:143–148 10.1007/s00414-007-0183-5 [DOI] [PubMed] [Google Scholar]
- Usui M.L., Underwood R.A., Mansbridge J.N., Muffley L.A., Carter W.G., Olerud J.E. 2005. Morphological evidence for the role of suprabasal keratinocytes in wound reepithelialization. Wound Repair Regen. 13:468–479 10.1111/j.1067-1927.2005.00067.x [DOI] [PubMed] [Google Scholar]
- Viziam C.B., Matoltsy A.G., Mescon H. 1964. Epithelialization of small wounds. J. Invest. Dermatol. 43:499–507 [PubMed] [Google Scholar]
- Walker D.C., Southgate J., Hill G., Holcombe M., Hose D.R., Wood S.M., Mac Neil S., Smallwood R.H. 2004. The epitheliome: agent-based modelling of the social behaviour of cells. Biosystems. 76:89–100 10.1016/j.biosystems.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Werner S., Grose R. 2003. Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 83:835–870 [DOI] [PubMed] [Google Scholar]
- Werner S., Krieg T., Smola H. 2007. Keratinocyte-fibroblast interactions in wound healing. J. Invest. Dermatol. 127:998–1008 10.1038/sj.jid.5700786 [DOI] [PubMed] [Google Scholar]
- Wong V., Gumbiner B.M. 1997. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J. Cell Biol. 136:399–409 10.1083/jcb.136.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodley D.T. 1996. Reepithelialization. The Molecular and Cellular Biology of Wound Repair. Second edition Clark R.A.F., editor Plenum Press, New York: 339–354 [Google Scholar]
- Yadav A., Saini V., Arora S. 2010. MCP-1: chemoattractant with a role beyond immunity: a review. Clin. Chim. Acta. 411:1570–1579 10.1016/j.cca.2010.07.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.